Introduction
Prostate cancer (PCa) is among the most common
malignant tumors among Western men, ranking second only to lung
cancer regarding cancer-related mortality (1). The incidence of PCa among Asian men is
significantly lower; however, the incidence of PCa in China has
increased significantly over the last few years (2). The etiology of PCa has not been fully
elucidated. It is a multifactorial disease, with genetic and
environmental factors contributing to its incidence. In addition,
several risk factors, such as ethnicity, family history and age,
have been associated with an increased PCa risk (3,4). The
exponential increase in the risk of PCa associated with aging may
reflect the accumulation of DNA damage resulting from a series of
processes, such as oxidative stress, inflammation and environmental
carcinogens, or a decrease in the DNA damage-repair response
capacity. DNA repair is essential to the ability of response to
environmental carcinogen-induced damage. DNA repair mechanisms are
important pathways in the removal of oxidative DNA compounds or DNA
adducts from damaged genomic sites (5). The key DNA repair pathways that are
often associated with cancer risk are listed as follows: The base
excision repair (BER) removes simple base modifications, such as
single-strand breaks, oxidative DNA damage and alkylation and
non-bulky adducts (6). The
nucleotide excision repair (NER) removes larger lesions, which
often result from environmental damage, such as UV radiation and
external carcinogens (7). The
mismatch repair is considered to involve MLH1, MSH2, PMS2 and MSH6
in damage recognition, followed by excision, polymerization and
ligation. The double-strand-break repair consists of the homologous
recombination and the non-homologous end joining pathways. Among
these pathways, NER and BER are the most significant (8).
In this study, we investigated the polymorphisms of
the excision repair cross-complementing rodent repair deficiency,
complementation group 2/Xeroderma pigmentosum
complementation group D (ERCC2/XPD) and the human homolog of the
8-oxoguanine DNA glycosylase 1 (hOGG1) genes. ERCC2/XPD, a gene
involved in NER and basal transcription, may influence individual
DNA repair capacity, particularly of bulky adducts (9). A previous epidemiological study
(10) evaluated the potential role
of the ERCC2 Arg156Arg polymorphism (rs238406) in cancer; however,
cross-study results have been conflicting.
hOGG1 is a protein involved in the BER pathway,
which is responsible for repairing one of the most mutagenic
lesions among base modifications, 8-hydroxyguanine (8-oxoG).
Previous epidemiological studies (11–13)
investigated the association between the Ser326Cys polymorphism
(rs1052133) in the hOGG1 gene and the risk for different types of
cancer. Significant increases were observed in the risk of
esophageal (11), lung (12) and colon cancer (13) in association with the hOGG1 326
polymorphism.
A previous study by Song et al (14) investigated the trend of PCa
incidence in the urban area of Tianjin, China, between 1981 and
2004, to provide a scientific rationale for the prevention and
control of PCa. The authors concluded that, despite the currently
low incidence of PCa in Tianjin, PCa is increasing rapidly.
Consequently, we investigated the association of PCa occurrence and
progression with the variants of the ERCC2/XPD and hOGG1 genes in a
southeastern Chinese population. The codon 156 (Arg156Arg)
polymorphism in XPD and the codon 326 (Ser326Cys) polymorphism in
hOGG1 were investigated.
Materials and methods
Study population
A total of 100 patients with pathologically
confirmed PCa and 100 age-matched control individuals were enrolled
in this study (Table I). The mean
age of the patient and control groups was 69.99±8.59 and
66.91±11.89 years, respectively. The 100 patients and the 100
cancer-free control subjects were enrolled at the First Affiliated
Hospital, School of Medicine, Zhejiang University and the Ningbo
First Hospital, between May, 2011 and October, 2012. The PCa
patient age range was 46–90 years. All the cases were diagnosed
within 1 year of enrollment. All the controls were non-cancer
patients admitted to the other wards (day and cadre wards) in the
same region, or were enrolled among individuals undergoing regular
physical exams in the hospital. Randomly selected controls were
age- and gender-matched to the cases (∼1:1). Written informed
consent was obtained from all patients and the study was approved
by the hospital boards and Ethics Committees.
 | Table I.Patient and tumor characteristics. |
Table I.
Patient and tumor characteristics.
| Characteristics | Cases (n=100) | Controls (n=100) |
|---|
| Age (mean ±
SD)a | 69.99±8.59 | 66.91±11.89 |
| PSA (ng/ml) | | |
| <10 | 25 | |
| 10–20 | 35 | |
| >20 | 40 | |
| Gleason score | | |
| ≤6 | 24 | |
| 7 | 46 | |
| ≥8 | 30 | |
| pT stage | | |
| ≤T2a | 54 | |
| T2b | 33 | |
| ≥T2c | 13 | |
Genotype
DNA was isolated from blood samples collected from
PCa patients and control subjects using the QIAamp DNA Blood Mini
kit (Qiagen Inc., Hilden, Germany) following the manufacturer's
protocol, quantified using the GeneQuant™ pro spectrophotometer
(Amersham Biosciences, Piscataway, NJ, USA) and stored at −20°C.
The XPD Arg156Arg (rs238406) polymorphism was detected using a
polymerase chain reaction-restriction fragment length polymorphism
(PCR-RFLP)-based method (5). Each
PCR was performed in a total volume of 20 μl, consisting of 0.3 μl
of a 10-μM solution of each primer, 1.5 mM MgCl2, 0.8 mM
dNTP, 0.5 units RedTaq DNA polymerase (Sigma, St. Louis, MO, USA),
1 μl genomic DNA (80 ng/μl) and 15.6 μl H2O, using a
PTC-200 Thermal Cycler (MJ Research Inc., Watertown, MA, USA). The
thermal cycling conditions were as follows: an initial denaturation
step at 95°C for 5 min, 30 cycles at 95°C for 30 sec, annealing at
60°C for 30 sec and extension at 72°C for 1 min. The final
extension was performed at 72°C for 7 min. The PCR primers were
synthesized by Takara Biotechnology Co., Ltd., (Dalian, China) and
were as follows: forward, 5′-AGGGTTTGAAGAGTGGTTGG-3′ and reverse
5′-TCAGGTCATCCAGGTTGTAG-3′. The primers were digested with
TfiI enzyme (New England Biolabs, Beverly, MA, USA). An
aliquot of 10 μl PCR product was digested with 3 units TfiI
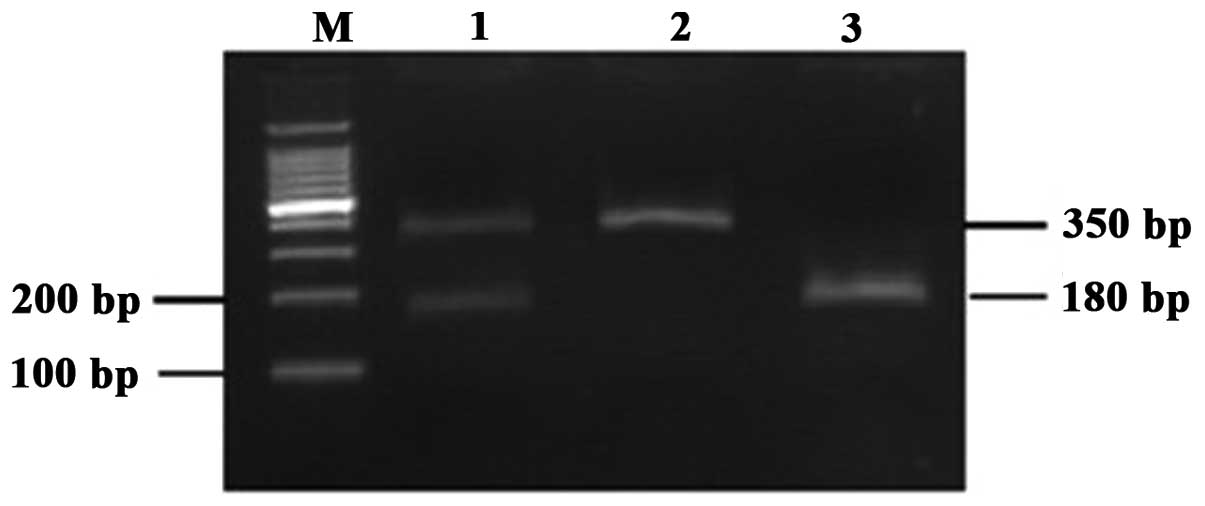
enzyme. The three possible genotypes were defined by three distinct
banding patterns: CC (350 bp fragments), AC (350, 170 and 180 bp
fragments) and AA (170 and 180 bp fragments) (Fig. 1). For hOGG1 Ser326Cys (rs1052133),
the PCR products were amplified with the primers
5′-ACTGTCACTAGTCTCACCAG-3′ (sense) and 5′-GGAAGGTGCTTGGGGAAT-3′
(antisense), then digested with Fnu4HI (New England
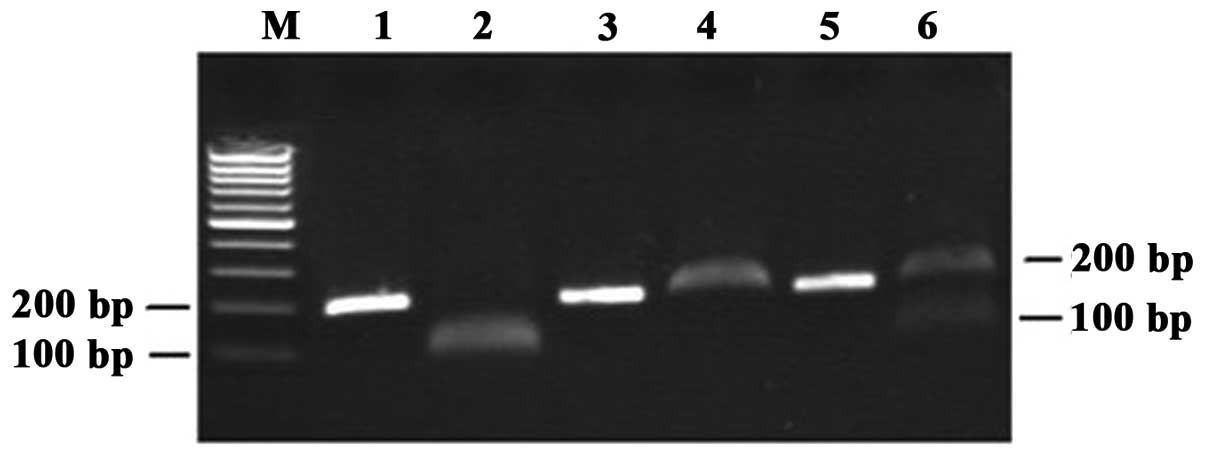
Biolabs). The PCR product was 200 bp in length. It was digested by
the Fnu4HI restriction enzyme into two 100-bp fragments for
the 326Cys allele but could not be digested for the 326Ser allele.
Fragments were separated on a 2% agarose gel and stained with
ethidium bromide. The Cys/Cys homozygote was cleaved by
Fnu4HI and yielded a 100-bp band. The Ser/Ser homozygote
could not be cleaved by Fnu4HI and remained a single 200-bp
band. The Ser/Cys heterozygote contained both the 200- and 100-bp
bands. The image of a representative gel is presented in Fig. 2. The results of RFLP-PCR were
validated by DNA sequencing.
Statistical analysis
The demographic data of the study groups were
compared by the Chi-square and Student's t-tests. Each polymorphism
in the control group was tested for deviation from the
Hardy-Weinberg equilibrium (HWE) by comparing the observed and
expected genotype frequencies using the Chi-square test. Analyses
were performed using SPSS software version 16.0 (SPSS, Inc.,
Chicago, IL, USA).
Results
Characteristics of PCa patients and
control subjects
The mean age, serum prostate-specific antigen (PSA),
Gleason score and pT stage of individual PCa patients are shown in
Table I. The two-tailed Student's
t-test was used to compare the distribution of age between patients
and control subjects. There was no significant difference in mean
age between cases and controls (Table
I). The Chi-squareHWE for genotype distributions was
>0.05 among controls. The genotype frequencies among controls
did not deviate from the expected distributions based on the HWE
(PXPD= 0.65 vs. PhOGG1=0.096)
Association of XPD Arg156Arg and hOGG1
Ser326Cys polymorphisms with PCa
There was no significant association between the
genotypes or alleles of the two polymorphisms and the occurrence of
PCa (Table II). A previous study by
Zheng et al (15)
demonstrated that each single-nucleotide polymorphism (SNP) is only
moderately associated with PCa; however, when SNPs are combined,
the association may be more significant. The combined effect of XPD
Arg156Arg and hOGG1 Ser326Cys was also investigated and no
statistically significant association was observed.
 | Table II.Ser326Cys polymorphism of the hOGG1
gene and Arg156Arg polymorphism of the ERCC2/XPD gene in PCa
patients. |
Table II.
Ser326Cys polymorphism of the hOGG1
gene and Arg156Arg polymorphism of the ERCC2/XPD gene in PCa
patients.
| Polymorphism genotype
or allele | Patients (n=100) | Controls (n=100) | OR | (95% CI) |
|---|
| Ser326Cys | | | | |
| CC | 22 | 29 | 1.00 | |
| CG | 52 | 57 | 1.20 | (0.59–2.49) |
| GG | 26 | 14 | 2.45 | (0.96–6.31) |
| GG+CG | 78 | 71 | 1.45 | (0.73–2.90) |
| C | 104 | 85 | 1.47 | (0.97–2.22) |
| G | 96 | 115 | 1.00 | |
| Arg156Arg | | | | |
| CC | 26 | 38 | 1.00 | |
| AC | 53 | 49 | 1.58 | (0.80–3.13) |
| AA | 21 | 13 | 2.36 | (0.93–6.08) |
| AA+AC | 74 | 62 | 1.74 | (0.92–3.34) |
| C | 95 | 75 | 1.51 | (0.99–2.29) |
| A | 105 | 125 | 1.00 | |
To evaluate the role of these polymorphisms with
regard to susceptibility to PCa, PCa patients were stratified
according to risk levels. The distribution of genotypes and
frequency of alleles among the groups of patients with different
risk levels is shown in Table
III.
 | Table III.Association of Ser326Cys polymorphism
of the hOGG1 gene and Arg156Arg polymorphism of the ERCC2/XPD gene
with PCa aggressiveness. |
Table III.
Association of Ser326Cys polymorphism
of the hOGG1 gene and Arg156Arg polymorphism of the ERCC2/XPD gene
with PCa aggressiveness.
| Polymorphism
genotype or allele | HR | OR | 95% CI | P-value | MR | OR | 95% CI | P-value | LR | OR | 95% CI | P-value |
|---|
| Arg156Arg | | | | | | | | | | | | |
| AA | 13 | 3.80 | 1.19–12.18 | 0.017 | 7 | 1.86 | 0.50–6.60 | 0.366 | 1 | 0.58 | 0.01–6.02 | 1.000 |
| AC | 32 | 2.48 | 1.02–6.35 | 0.033 | 13 | 0.92 | 0.34–2.54 | 1.000 | 8 | 1.24 | 0.33–5.22 | 0.773 |
| CC | 10 | 1.00 | | | 11 | 1.00 | | | 5 | 1.00 | | |
| AA+AC | 45 | 2.76 | 1.18–6.84 | 0.011 | 20 | 1.11 | 0.45–2.87 | 0.835 | 9 | 1.10 | 0.30–4.51 | 1.000 |
| A | 58 | 1.86 | 1.13–3.06 | 0.012 | 27 | 1.29 | 0.69–2.38 | 0.456 | 10 | 0.93 | 0.36–2.25 | 1.000 |
| C | 52 | 1.00 | | | 35 | 1.00 | | | 18 | 1.00 | | |
| Ser326Cys | | | | | | | | | | | | |
| GG | 17 | 2.93 | 1.00–8.74 | 0.033 | 6 | 1.78 | 0.41–7.48 | 0.510 | 3 | 2.07 | 0.24–17.26 | 0.406 |
| CG | 26 | 1.10 | 0.47–2.76 | 1 | 18 | 1.31 | 0.45–4.13 | 0.637 | 8 | 1.36 | 0.30–8.51 | 1.000 |
| CC | 12 | 1.00 | | | 7 | 1.00 | | | 3 | 1.00 | | |
| GG+CG | 43 | 1.46 | 0.64–3.49 | 0.447 | 24 | 1.40 | 0.51–4.27 | 0.646 | 11 | 1.46 | 0.35–8.69 | 0.754 |
| G | 60 | 1.62 | 1.00–2.67 | 0.044 | 30 | 1.27 | 0.69–2.34 | 0.465 | 14 | 1.35 | 0.56–3.24 | 0.543 |
| C | 50 | 1.00 | | | 32 | 1.00 | | | 14 | 1.00 | | |
According to the European Association of Urology
(EAU) guidelines for prostate cancer (16), patients were divided into three
groups: i) low-risk (localised PCa: cT1-T2a or Gleason score 2–6 or
PSA<10); ii) intermediate-risk (localised PCa: cT2b-T2c or
Gleason score 7 or PSA 10–20); and iii) high-risk (localised PCa:
≥cT3a or Gleason score 8–10 or PSA>20).
For the XPD156 C→A polymorphism, the frequency of
the XPD156 AA genotype in high-risk PCa patients was significantly
higher in comparison to controls (OR=3.80; 95% CI: 1.19–12.18;
P=0.017), conferring an ∼3-fold increase in the risk for cancer.
Individuals with the heterozygous AC genotype were associated with
increased risk of PCa (OR=2.48; 95% CI: 1.02–6.35; P= 0.033). In
addition, significant differences were observed (OR=2.76; 95% CI:
1.18–6.84; P=0.011) when variant-containing genotypes were combined
(AC+AA) and compared to the homozygous wild-type (CC). The
frequency of the XPD156 A allele was 0.527 among the high-risk
cases and 0.375 among the controls, a difference described as
statistically significant (Chi-square P=0.012).
For Ser326 C→G, the high-risk individuals with the
homozygous GG genotype exhibited a significantly increased risk of
PCa (OR=2.93; 95% CI: 1–8.74; P=0.033), which reduced the rate of
DNA repair compared to the CC or GC genotypes. However, no
significant difference was observed (OR=1.46; 95% CI: 0.64–3.49;
P=0.447) when variant-containing genotypes were combined (GC+GG).
The frequency of the Ser326 G allele was 0.545 among the high-risk
cases and 0.425 among the controls, a difference described as
statistically significant (Chi-square P= 0.044).
For the low- and intermediate-risk groups, no
differences were observed in the distribution of the genotypes of
either polymorphism between the two groups (Table III).
Discussion
In this population-based study, the association
between the XPD and hOGG1 gene polymorphisms and PCa risk was
investigated in 100 cases and 100 controls. No association was
observed between any of these SNPs and the overall risk of PCa. As
regards the clinical characteristics of PCa, the roles of the
Arg156Arg polymorphism of the ERCC2/XPD gene and the Ser326Cys
polymorphism of the hOGG1 gene in PCa aggressiveness were assessed.
Significant differences were identified between the two DNA repair
genes and the high-risk level group (Table III).
As regards ERCC2/XPD SNPs, several studies
investigating the possible association between ERCC2 variants in
DNA repair genes and cancer risk have been published (17,18).
The most extensively investigated ERCC2 polymorphisms were
Asp312Asn and Lys751Gln. ERCC2 Arg156Arg is a silent C/A
polymorphism and thus it may exert an effect only at the
transcriptional level, if ERCC2 Arg156Arg is the biologically
effective polymorphism. The reason for a higher expression level of
ERCC2 being associated with an increased cancer risk remains to be
elucidated. According to a previous study by Vogel et al
(19), several genes in that
region, including the apoptosis-controlling gene RAI, ERCC1 and
ERCC2/XPD, correlate strongly with each other in their expression
levels. Therefore, an accompanying higher expression of another
gene may confer increased cancer risk. Previous studies
demonstrated that XPD codon 156 polymorphisms were associated with
lung (20), bladder (21) and breast cancer (22) in Chinese populations. To the best of
our knowledge, the present study was the first to demonstrate that
the silent polymorphisms of XPD156 affect the risk of PCa. Our main
finding was that the A allele of XPD Arg156Arg is associated with
predisposition to high-risk PCa (AA: OR=3.80; 95% CI: 1.19–12.18;
P=0.017; AC: OR=2.48; 95% CI: 1.02–6.35; P=0.033; A+AC: OR=2.76;
95% CI: 1.18–6.84; P= 0.011; and A:OR=1.86; 95% CI: 1.13–3.06;
P=0.012).
Previous epidemiological studies associated the
Ser326Cys polymorphism in the hOGG1 gene with the risk of various
types of cancer (23,24). Significant risk increases were
reported in subjects with the hOGG1 326 polymorphism for these
cancers. In a study by Srivastava et al (25), the homozygous variant genotypes of
hOGG1 Ser326Cys polymorphisms exhibited a statistically significant
increased risk for gallbladder carcinogenesis (OR=2.5; 95% CI:
1.1–5.4; P=026). The risk for variant-containing genotypes (CG+GG)
of hOGG1 Ser326Cys was also significant (OR=1.8; 95% CI: 1.2–2.6;
P=002) when compared to the homozygous wild-type CC genotype. A
previous study by Arizono et al (26) indicated that the frequency of the
hOGG1 codon 326 GG genotype was significantly higher in bladder
cancer cases compared to the controls. The homozygous GG genotype
exhibited a significant association with lung cancer compared to
the C allele carrier status in previous studies (27,28).
In the present study, individuals who were homozygous for the GG
genotype were at a markedly increased risk for developing PCa
(OR=2.93; 95% CI: 1–8.74; P=0.033).
Although Ser326Cys polymorphisms have been
extensively investigated in PCa, the conclusions are contradictory.
Xu et al (29) observed that
men with the CC genotype (Ser326) had an increased risk of PCa,
particularly the sporadic form. Zhang et al (30) demostrated that, following adjustment
for confounders, subjects who were heterozygous or homozygous for
the variant allele of the hOGG1 Ser326Cys polymorphism appeared to
experience a lower risk of PCa compared to those who were
homozygous for the wild-type allele (OR=0.72 and 95% CI:
0.46–1.10). Nam et al (31)
investigated a panel of 13 polymorphisms in 13 different genes in
patients with the GG and the CC genotype of the hOGG1-326 gene
(OR=0.68; 95% CI: 0.5–1.0; P=0.05). By contrast, Chen et al
(32) demonstrated that a
significantly increased PCa risk was observed in subjects with at
least one hOGG1326Cys allele (ORadj=2.1; 95% CI:
1.2–3.8). These significant risk changes were observed for subjects
with the heterozygous hOGG1 326Ser/Cys genotype
(ORadj=1.8; 95% CI: 1.01–3.3) and with the homozygous
hOGG1 326Cys/Cys genotype (ORadj=7.8; 95% CI: 1.7–36.2).
To the best of our knowledge, our study is the first to provide
evidence that men bearing the GG genotype (Cys326) are susceptible
to the development of high-risk PCa. Our results may encourage
studies on larger populations. Xu et al (29) also indicated that Cys326 confers
increased cancer risk in Asian populations, a finding that was
similar to ours.
Our study has certain limitations. First, increasing
the number of individuals may increase the statistical power of the
study. Second, the follow-up study may provide more precise
results, as we were not able to ensure a lifelong cancer-free
status in the controls.
In conclusion, the findings of our study suggest
that the ERCC2/XPD Arg156Arg and hOGG1 Ser326Cys polymorphisms
contribute to high-risk PCa susceptibility in a Chinese
population.
References
|
1
|
Jemal A, Siegel R, Ward E, Hao Y, Xu J,
Murray T and Thun MJ: Cancer statistics, 2008. CA Cancer J Clin.
58:71–96. 2008. View Article : Google Scholar
|
|
2
|
McCracken M, Olsen M, Chen MS Jr, Jemal A,
Thun M, Cokkinides V, Deapen D and Ward E: Cancer incidence,
mortality, and associated risk factors among Asian Americans of
Chinese, Filipino, Vietnamese, Korean, and Japanese ethnicities. CA
Cancer J Clin. 57:190–205. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Pienta KJ and Esper PS: Risk factors for
prostate cancer. Ann Intern Med. 118:793–803. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chan JM, Holick CN, Leitzmann MF, et al:
Diet after diagnosis and the risk of prostate cancer progression,
recurrence, and death (United States). Cancer Causes Control.
17:199–208. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Vogel U, Olsen A, Wallin H, Overvad K,
Tjonneland A and Nexo BA: Effect of polymorphisms in XPD, RAI,
ASE-1 and ERCC1 on the risk of basal cell carcinoma among
Caucasians after age 50. Cancer Detect Prev. 29:209–214. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Thompson LH and West MG: XRCC1 keeps DNA
from getting stranded. Mutat Res. 459:1–18. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sancar A and Tang MS: Nucleotide excision
repair. Photochem Photobiol. 57:905–921. 1993. View Article : Google Scholar
|
|
8
|
Ishikawa T, Zhang SS, Qin X, et al: DNA
repair and cancer: lessons from mutant mouse models. Cancer Sci.
95:112–117. 1993. View Article : Google Scholar
|
|
9
|
Terry MB, Gammon MD, Zhang FF, Eng SM,
Sagiv SK, Paykin AB, Wang Q, Hayes S, Teitelbaum SL, Neugut AI and
Santella RM: Polymorphism in the DNA repair gene XPD, polycyclic
aromatic hydrocarbon-DNA adducts, cigarette smoking, and breast
cancer risk. Cancer Epidemiol Biomarkers Prev. 13:2053–2058.
2004.PubMed/NCBI
|
|
10
|
Goode EL, Ulrich CM and Potter JD:
Polymorphisms in DNA repair gene and associations with cancer risk.
Cancer Epidemiol Biomarkers Prev. 11:1513–1530. 2002.PubMed/NCBI
|
|
11
|
Xing D, Tan W and Lin D: Genetic
polymorphisms and susceptibility to esophageal cancer among Chinese
population (Review). Oncol Rep. 10:1615–1623. 2003.PubMed/NCBI
|
|
12
|
Park J, Chen L, Tockman MS, Elahi A and
Lazarus P: The human 8-oxoguanine DNA N-glycosylase 1 (hOGG1) DNA
repair enzyme and its association with lung cancer risk.
Pharmacogenetics. 14:103–109. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kim JI, Park YJ, Kim KH, Kim JI, Song BJ,
Lee MS, Kim CN and Chang SH: hOGG1 Ser326Cys polymorphism modifies
the significance of the environmental risk factor for colon cancer.
World J Gastroenterol. 9:956–960. 2003.PubMed/NCBI
|
|
14
|
Song FJ, Zhang BL, He M, et al: Trend
analysis of the incidence of prostate cancer in Tianjin between
1981 and 2004. Zhong hua Yi Xue Za Zhi. 90:2811–2814. 2010.(In
Chinese).
|
|
15
|
Zheng SL, Sun J, Wiklund F, et al:
Cumulative association of five genetic variants with prostate
cancer. N Engl J Med. 358:910–919. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Heidenreich A, Aus G, Bolla M, et al:
Guidelines on Prostate Cancer. European Association of Urology
(EAU); Arnhem, The Netherlands: 2010
|
|
17
|
Yuan H, Niu YM, Wang RX, Li HZ and Chen N:
Association between XPD Lys751Gln polymorphism and risk of head and
neck cancer: a meta-analysis. Genet Mol Res. 22:3356–3364. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ding DP, Ma WL, He XF and Zhang Y: XPD
Lys751Gln polymorphism and esophageal cancer susceptibility: a
meta-analysis of case-control studies. Mol Biol Rep. 39:2533–2540.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Vogel U, Nexo BA, Tjonneland A, et al:
ERCC1, XPD and RAI mRNA levels in lymphocytes are not associated
with lung cancer risk in a prospective study of Danes. Mutat Res.
593:88–96. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wu W, Zhang W, Qiao R, et al: Association
of XPD polymorphisms with severe toxicity in non-small cell lung
cancer patients in a Chinese population. Clin Cancer Res.
15:3889–3895. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shao J, Gu M, Xu Z, et al: Polymorphisms
of the DNA gene XPD and risk of bladder cancer in a Southeastern
Chinese population. Cancer Genet Cytogenet. 177:30–36. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yin J, Liang D, Vogel U, et al: The
polymorphism of DNA repair gene ERCC2/XPD Arg156Arg and
susceptibility to breast cancer in a Chinese population. Biochem
Genet. 47:582–590. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Guan P, Huang D, Yin Z and Zhou B:
Association of the hOGG1 Ser326Cys polymorphism with increased lung
cancer susceptibility in Asians: a meta-analysis of 18 studies
including 7592 cases and 8129 controls. Asian Pac J Cancer Prev.
12:1067–1072. 2011.PubMed/NCBI
|
|
24
|
Ni M, Qiu J, He W and Wang X: The
functional Ser326Cys polymorphism in hOGG1 is associated with
gastric cancer risk: evidence from 1180 cases and 2444 controls.
Eur J Gastroenterol Hepatol. 24:683–687. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Srivastava K, Srivastava A and Mittal B:
Polymorphisms in ERCC2, MSH2, and OGG1 DNA repair genes and
gallbladder cancer risk in a population of Northern India. Cancer.
116:3160–3169. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Arizono K, Osada Y and Kuroda Y: DNA
repair gene hOGG1 codon 326 and XRCC1 codon 399 polymorphisms and
bladder cancer risk in a Japanese population. Jpn J Clin Oncol.
38:186–191. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Okasaka T, Matsuo K, Suzuki T, et al:
hOGG1 Ser326Cys polymorphism and risk of lung cancer by
histological type. J Hum Genet. 54:739–745. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li H, Hao X and Zhang W: The hOGG1
Ser326Cys polymorphism and lung cancer risk: a meta-analysis.
Cancer Epidemiol Biomarkers Prev. 17:1739–1745. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Xu J, Zheng SL, Turner A, et al:
Associations between hOGG1 sequence variants and prostate cancer
susceptibility. Cancer Res. 62:2253–2257. 2002.PubMed/NCBI
|
|
30
|
Zhang J, Dhakal IB, Greene G, Lang NP and
Kadlubar FF: Polymorphisms in hOGG1 and XRCC1 and risk of prostate
cancer: effects modified by plasma antioxidants. Urology.
75:779–785. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Nam RK, Zhang WW, Jewett MA, et al: The
use of genetic markers to determine risk for prostate cancer at
prostate biopsy. Clin Cancer Res. 11:8391–8397. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chen L, Elahi A, Pow-Sang J, Lazarus P and
Park J: Association between polymorphism of human oxoguanine
glycosylase 1 and risk of prostate cancer. J Urol. 170(6 Pt 1):
2471–2474. 2003. View Article : Google Scholar : PubMed/NCBI
|