Folate is a type of water-soluble B vitamin and an
essential nutrient required for human metabolism. Folate plays a
key role in the formation of S-adenosylmethionine, which is the
universal methyl donor for DNA methylation, as well as in the
formation of purine and thymidine for DNA synthesis (4). Folate deficiency increases the risk of
tumorigenesis through one of the following mechanisms: by leading
to aberrant DNA methylation, which may in turn lead to an altered
expression of critical tumor suppressor genes and proto-oncogenes;
or by causing imbalances in the pools of nucleotide precursors,
leading to DNA strand breaks and mutations and disruption of DNA
integrity and repair (4,5).
The methylenetetrahydrofolate reductase (MTHFR) is
an enzyme that is crucial in the metabolism of folate (6). MTHFR catalyzes the irreversible
conversion of 5,10-methylenetetrahydrofolate to
5-methyltetrahydrofolate, the primary methyl donor for the
remethylation of homocysteine to methionine. The gene encoding
MTHFR is located on chromosome 1p36.3 (7). Two common single-nucleotide
polymorphisms of MTHFR are MTHFR C677T (Ala222Val, rs1801133) and
A1298C (Glu429Ala, rs1801131). The C677T variant enhances enzyme
thermolability and is associated with decreased activity of the
MTHFR enzyme (8). The A1298C
variant (Glu429Ala, rs1801131) is a missense mutation leading to
reduced MTHFR enzyme activity (9,10). The
homozygous genotypes of MTHFR C677T and A1298C are associated with
higher homocysteine levels, which may lead to DNA hypomethylation
and increased cancer prevalence. However, the decreasing enzyme
activity results in higher 5,10-methylenetetrahydrofolate and
thymidine levels and, thus, increased DNA synthesis and repair.
Therefore, MTHFR polymorphisms are regarded as a protective factor
against tumor development (8,11).
A meta-analysis of all published studies was
conducted to determine the effect of MTHFR mutants on CRC risk. In
a subgroup analysis, the study subjects were classified by
ethnicity and tumor location to provide comprehensive evidence on
the association of MTHFR C677T and A1298C with CRC.
A literature search was conducted on PubMed, Medline
and China National Knowledge Infrastructure (January,
1991-September, 2012) databases, using the following keywords and
subject terms: 'MTHFR', 'polymorphism' and 'colon cancer' or
'rectal cancer'. All the studies in our meta-analysis were required
to meet the following inclusion criteria: i) case-control studies;
ii) raw data to calculate odds ratios (ORs) with 95% confidence
intervals (95% CIs); iii) in case of the same results published in
multiple studies, the most recent publication or the largest sample
was considered. A given study was excluded from this meta-analysis
when: i) the genotype or allele frequencies were not reported, ii)
the study design was not case-control, iii) the association between
MTHFR polymorphisms and colorectal adenoma was investigated.
Data were carefully and independently collected
according to the genotypes MTHFR C677T or A1298C. Two authors
extracted the following information from the eligible studies:
first author's name, publication year, country, ethnicity of
participants and number of cases and controls. In our study,
ethnicities were classified as European and American, Asian,
African and mixed.
The association between MTHFR C677T and A1298C gene
polymorphisms and CRC risk was assessed by using the codominant
(677CT vs. CC; 677TT vs. CC; 1298AC vs. AA; 1298CC vs. AA), the
dominant (677CT+TT vs. CC; 1298AC+CC vs. AA) and the recessive
(677TT vs. CC+CT; 1298CC vs. AA+AC) models. The same procedures
were applied for the MTHFR A1298C genotype. Subgroup analyses were
performed by tumor location and ethnicity of the control
groups.
The strength of association of the MTHFR gene
polymorphisms with CRC was measured by the ORs (1) together with the 95% CIs. The
significance of the pooled ORs was determined by the Z-test and
P<0.05 was considered to indicate a statistically significant
difference. The Chi-square test was first used to assess whether
the distribution of genotypes among controls conformed to the
Hardy-Weinberg equilibrium (HWE), with P<0.05 considered a
departure from HWE. The Q-test was used to assess heterogeneity
among the studies. When P<0.05, the heterogeneity was considered
to indicate a statistically significant difference. The
I2 index was used to quantify the percentage of the
total variation among studies when heterogeneity was calculated.
The I2 value ranged from 0 to 100%, with 25, 50 and 75%
expressing low, moderate and high heterogeneity, respectively. When
I2<50%, a fixed-effects model (the Mantel-Haenszel
method) was applied to estimate the pooled results. Otherwise, the
random-effects model (the DerSimonian-Laird method) was used.
Publication bias was visually investigated in a
funnel plot of log (OR) against its standard error (SE). An
asymmetric plot suggested possible publication bias. The degree of
asymmetry was assessed via the Egger's test (P<0.05 was
considered publication bias). A sensitivity analysis was performed
by omitting each study in turn to assess the stability of the
results.
All the analyses were performed with Stata software
version 11.0 (StataCorp, College Station, TX, USA). All the
P-values were two-sided.
A total of 302 studies were identified during the
literature search and 232 were excluded due to departures from the
inclusion criteria. Eventually, 70 studies were included (1,2,6,10,12–77).
One study, conducted by Lee et al(41), consisted of three individual
case-control studies and was handled as three populations;
furthermore, the studies by Keku et al(12) and Lima et al(28), included two populations each. This
meta-analysis included a larger population compared to previous
meta-analyses. Of the 74 case-control studies included in the 70
publications, 74 studies investigated MTHFR C677T (29,783 cases and
41,772 controls) and 39 investigated MTHFR A1298C (13,285 cases and
20,164 controls). In total, 33 populations were from Europe and
America; 31 were from Asia; 4 were from Africa and the remaining
were mixed populations. In addition, a subanalysis was conducted by
tumor location. Twenty-one of the 74 studies provided detailed data
on colon and 14 on rectal cancer. The characteristics of the
studies are listed in Table I.
The association of MTHFR C677T and A1298C
polymorphisms with CRC was further stratified by ethnicity. As
shown in Table II, no significant
association was observed between MTHFR C677T and the risk of CRC
under any genetic models, in any of the populations. However,
Asians carrying the MTHFR 1298CC genotype exhibited a reduced risk
of CRC. The OR of CC vs. AA was 0.69 (95% CI: 0.54–0.89) and the OR
of CC vs. AA+AC was 0.69 (95% CI: 0.54–0.88). In the mixed
populations, the pooled analysis demonstrated that carriers of
MTHFR 677TT and 1298CC were more common among CRC patients than
among controls when compared to individuals with wild-type
genotypes (677TT vs. CC: OR=0.86; 95% CI: 0.76–0.96; TT vs. CC+CT:
OR=0.85; 95% CI: 0.76–0.95) (1298CC vs. AA: OR=0.82; 95% CI:
0.69–0.98; and CC vs. AA+AC: OR=0.83; 95% CI: 0.70–0.99).
Furthermore, the participants were stratified by
tumor location. A significantly decreased risk of CRC was observed
under the recessive model of MTHFR 677TT (OR=0.83; 95% CI:
0.72–0.96) and 1298CC (OR=0.81; 95% CI: 0.69–0.95) in the colon
cancer group. In addition, the stratified analysis revealed that
MTHFR C677T was associated with reduced risk of rectal cancer. The
OR under the recessive model was 0.86 (95% CI: 0.77–0.97). The main
results are presented in Table
II.
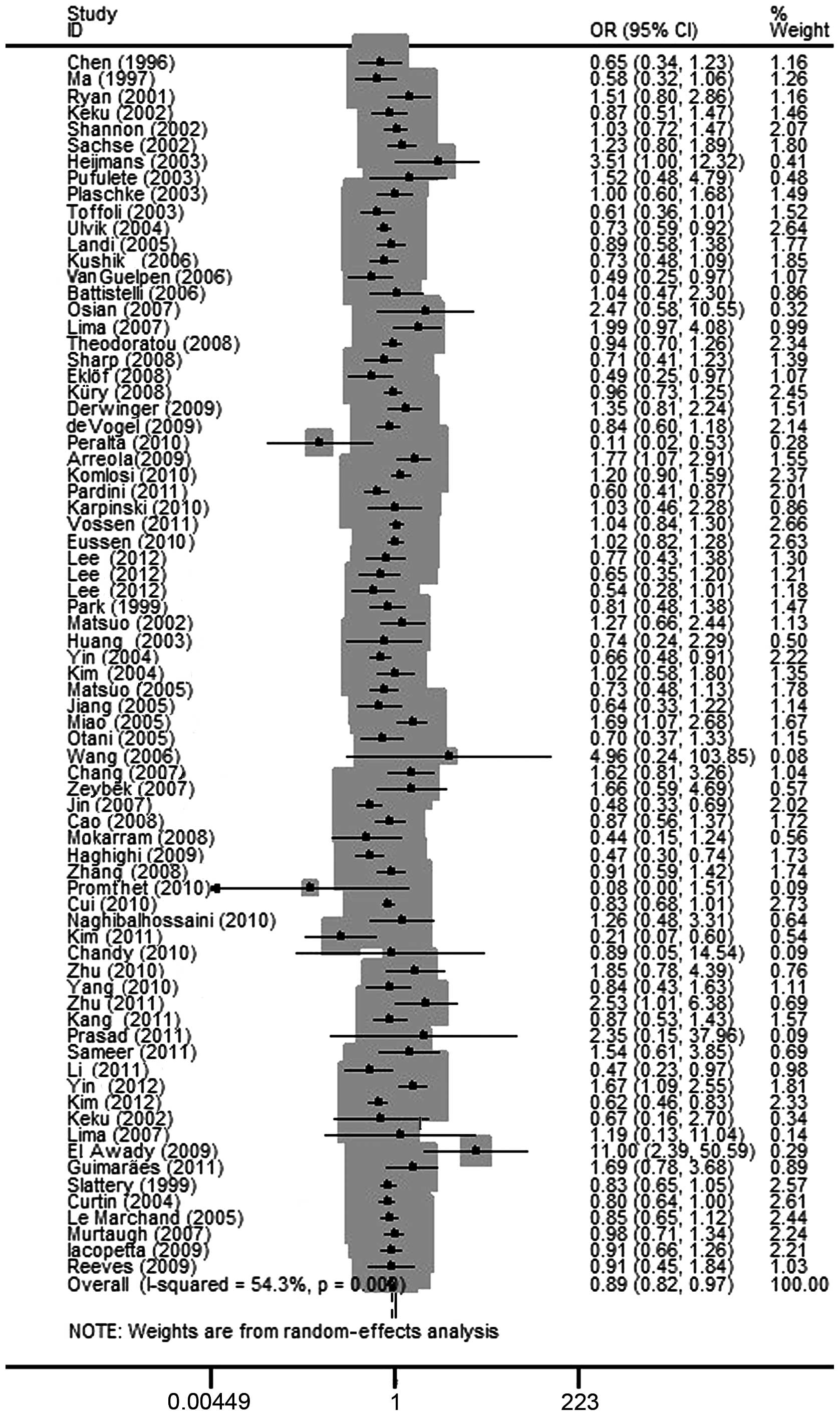
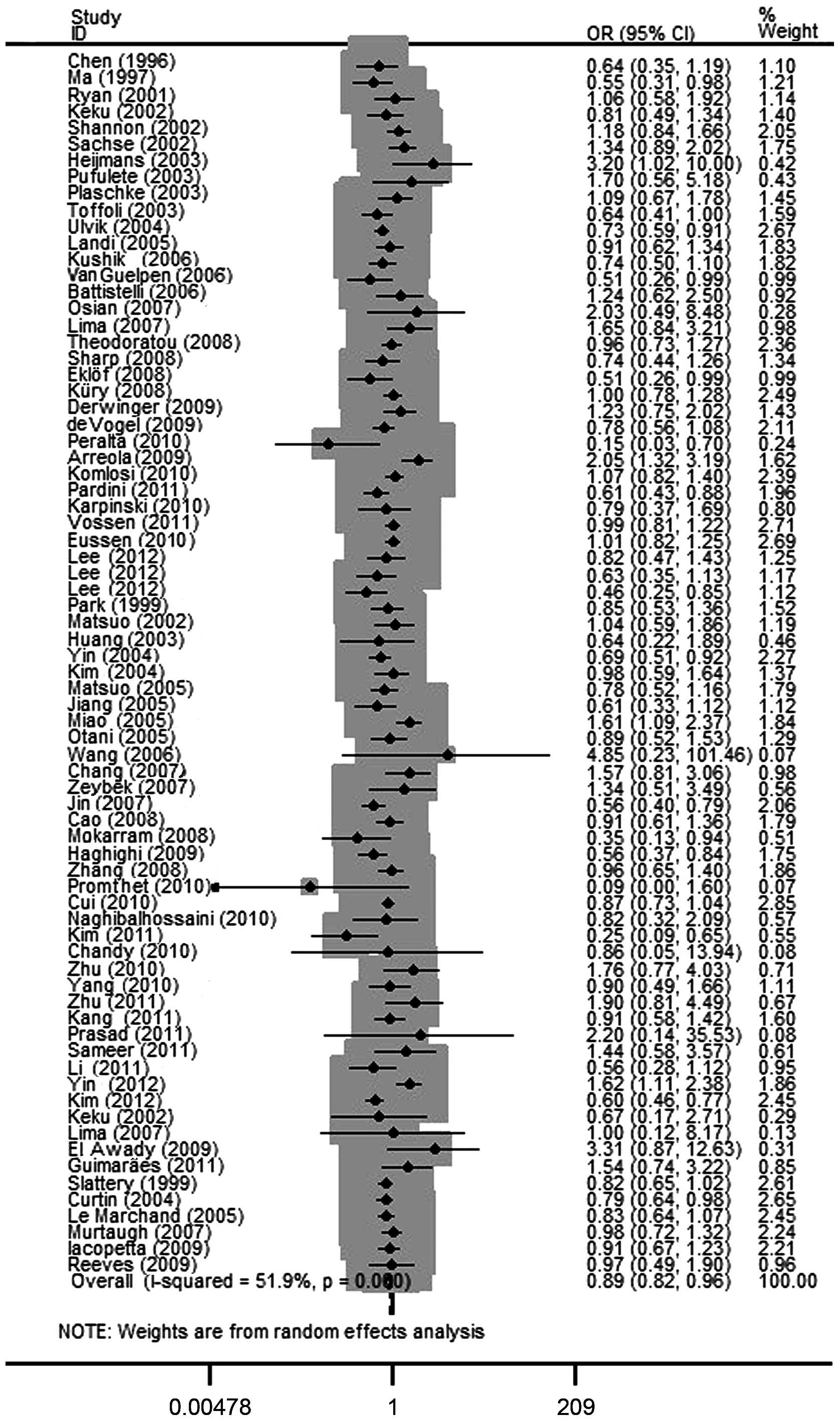
The elimination of each individual study imparted no
qualitative difference on the pooled OR values, indicating that the
final results of the meta-analysis were relatively stable. The
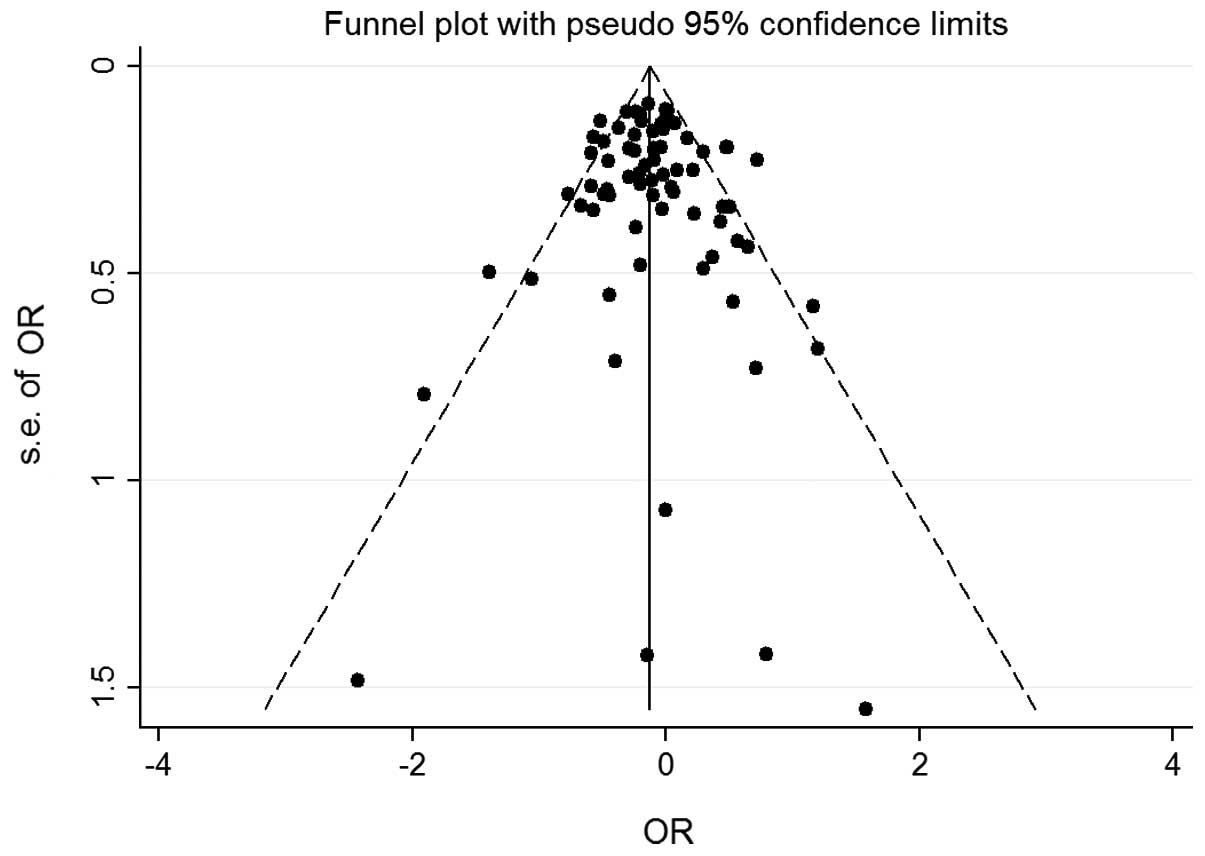
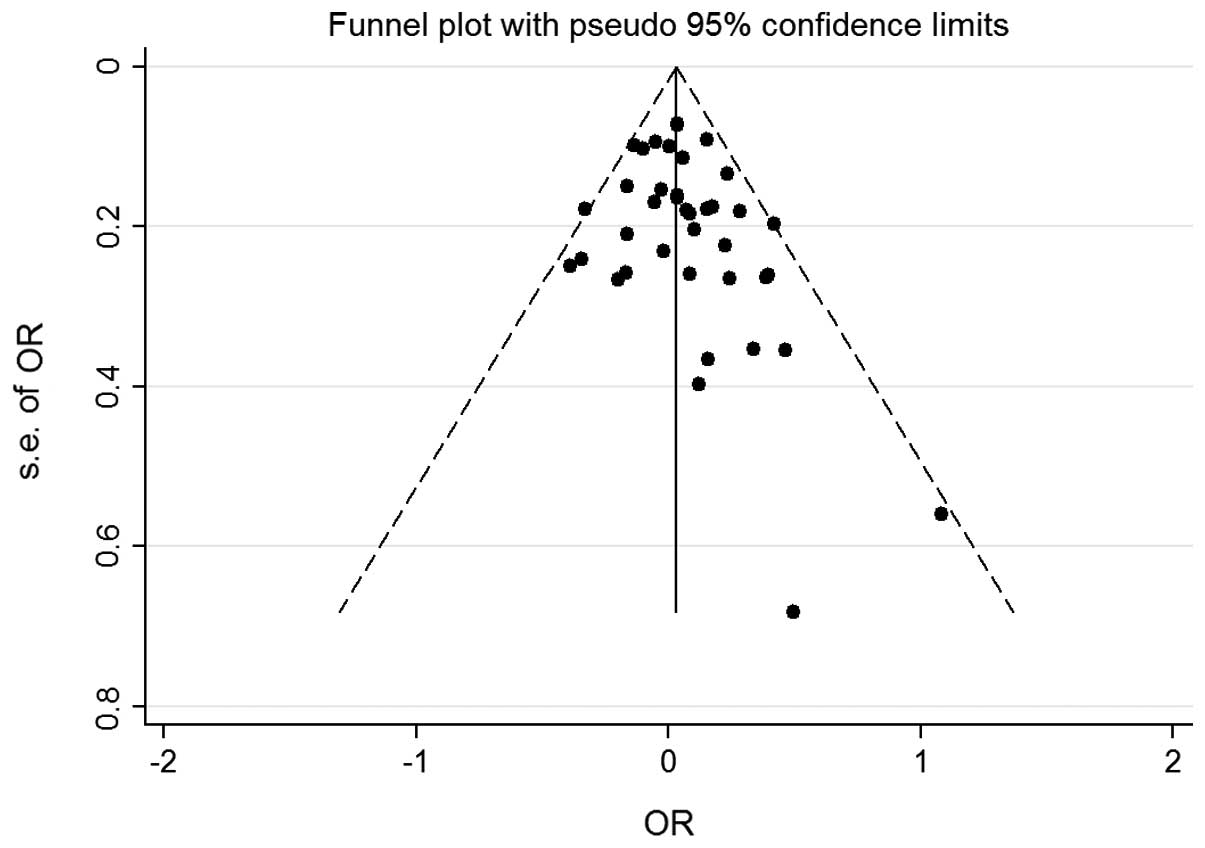
publication bias of the studies was determined by a funnel plot and
the Egger's test. The shapes of the funnel plot for each comparison
indicated no obvious asymmetry (Figs.
3 and 4) and the Egger's test
was then used to provide statistical evidence for the funnel plot
symmetry. No significant publication bias was detected in the
studies. The results are presented in Table III.
The first study to evaluate the association between
the MTHFR C677T polymorphism and CRC was conducted by Chen et
al(10). The findings of that
study suggested that the MTHFR C677T mutation affected enzyme
activity and was involved in abnormal methylation as well as DNA
synthesis, leading to colorectal tumorigenesis. Similar results
were subsequently reported by Ma et al(16), Slattery et al(70) and Le Marchand et al(77). In addition, Le Marchand et
al(77) observed that the MTHFR
1298C allele was weakly protective against CRC. However, Guimarães
et al(69) reported that the
carriers of the combined variants MTHFR 1298AC+CC and 677CT+TT
exhibited an increased risk of CRC, whether in isolation or in
combination. According to Shannon et al(14) and Prasad et al(63), the MTHFR polymorphism C677T was a
risk factor for CRC development. Furthermore, several other
published studies failed to support an effect of MTHFR gene
polymorphisms on CRC risk (43,49,60),
due to statistically non-significant results.
The conflicting conclusions among the studies
mentioned above may be attributed to several causes. First, the
sample sizes of the populations included in several studies were
relatively small (19,27,69),
which may result in false-positive or false-negative outcomes.
Second, the eligibility criteria for inclusion of control subjects
differed among the studies. Certain studies were hospital-based
(35,45,50),
whereas others were population-based (12,13,47,63).
Therefore, some controls were non-cancer cases, whereas others were
healthy individuals. The inclusion of individuals from different
ethnic backgrounds should also be considered.
As regards the conflicting results, we performed a
meta-analysis to elucidate the association of MTHFR C677T and
A1298C polymorphisms with CRC risk and to provide a comprehensive
assessment. In this meta-analysis, the pooled results indicated
that the homozygous variant of MTHFR C677T polymorphism exerted a
protective effect against CRC development (OR=0.89; 95% CI:
0.82–0.97). However, when analysis was performed by ethnicity, this
effect was not observed in all the subgroups, except for the mixed
population (OR=0.86; 95%CI: 0.76–0.96). Additionally, a significant
association was observed between MTHFR 1298CC and CRC in the mixed
population (OR=0.82; 95% CI: 0.69–0.98). When this analysis was
restricted by limiting studies to Asian populations, the MTHFR
1298CC genotype exhibited a decreased risk of CRC, with an OR of
0.69 (95% CI: 0.54–0.89). In the subanalysis by tumor location it
was demonstrated that individuals with the MTHFR 677TT genotype
exhibited a decreased risk of colon and rectal cancer, with an OR
of 0.83 (95% CI: 0.72–0.96) and 0.86 (95% CI: 0.77–0.97),
respectively. It was also confirmed that the MTHFR A1298C
polymorphism is involved in colon cancer development (CC vs. AA+AC:
OR=0.81; 95% CI: 0.69–0.96). Our findings in this meta-analysis
were consistent with the results reported by the majority of the
published studies.
According to Begg's funnel plots and Egger's test,
there was no significant publication bias in the present
meta-analysis. However, there was obvious heterogeneity, which was
a potential problem when interpreting the results of the
meta-analysis. Several sources of heterogeneity should be
considered. First, different selection criteria of cases and
controls, as well as gender and age distribution, may affect
between-study heterogeneity. Second, there was some diversity among
the studies regarding design, sample size and family history.
Ethnic variations were also crucial as it was demonstrated that the
heterogeneity was decreased when analyzed by ethnicity in this
meta-analysis. The different ethnicities were distinguished
according to geography; however, potential confounding factors,
such as genetic background, lifestyle and dietary habits, could not
be excluded.
The present study has certain limitations. First,
the criteria for inclusion of controls differed among the studies.
The controls in certain studies were selected from healthy
individuals, whereas the controls in other studies were selected
from non-cancer cases. Second, our results were based on unadjusted
OR values, which may lead to relatively low power of the estimation
of the real association. The gender and age distribution of the
participants, the dietary pattern, the folate status, alcohol
consumption and other risk factors may affect between-study
heterogeneity. An analysis should be conducted to obtain adjusted
ORs for other covariates, such as age, gender and folate status,
provided more individual study data are available. Third, the
subgroup analyses had insufficient statistical power to detect the
association. The gene-gene and gene-environment interactions may
affect the association between MTHFR polymorphisms and CRC.
According to the study by Keku et al(12), the combination of MTHFR 677CC and
1298AA genotypes exhibited an increased risk of CRC. In addition,
Ma et al(16) and Kim et
al(67) demonstrated that a
high folate intake was associated with reduced risk of CRC and high
alcohol consumption was associated with an increased risk of CRC.
The gene-gene and gene-environment interactions could not be
examined due to unavailability of individual data.
Despite the limitations described above, our
meta-analysis also has certain advantages. The substantial number
of cases and controls were pooled from different studies, which
provided more reliable evidence on the association between MTHFR
polymorphisms and the risk of CRC. In addition, the study subjects
were classified into colon and rectal cancer groups, in order to
exclude certain confounding factors. The pooled data clearly
demonstrated that MTHFR C677T significantly affected carcinogenesis
in the colon and rectum, whereas A1298C appeared to be mainly
associated with rectal tumorigenesis. This finding may be
attributed to different carcinogenic mechanisms underlying colon
and rectal cancer.
In conclusion, the results of this meta-analysis
indicated a significant association of the MTHFR C677T and A1298C
polymorphisms with the risk of CRC. Particularly, the MTHFR 677T
and 1298C alleles were associated with a low risk of CRC.
This study was supported by grants no. 81102194 and
no. 81272293 from the National Natural Science Foundation of China,
grant no. LS2010168 from the Liaoning Provincial Department of
Education and grant no. 00726 from the China Medical Board. The
authors are grateful to all the participants in this study.
|
1
|
Pardini B, Kumar R, Naccarati A, Prasad
RB, Forsti A, Polakova V, Vodickova L, Novotny J, Hemminki K and
Vodicka P: MTHFR and MTRR genotype and haplotype analysis and
colorectal cancer susceptibility in a case-control study from the
Czech Republic. Mutat Res. 721:74–80. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cao HX, Gao CM, Takezaki T, Wu JZ, Ding
JH, Liu YT, Li SP, Su P, Cao J, Hamajima N and Tajima K: Genetic
polymorphisms of methylenetetrahydrofolate reductase and
susceptibility to colorectal cancer. Asian Pac J Cancer Prev.
9:203–208. 2008.PubMed/NCBI
|
|
3
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar
|
|
4
|
Blount BC, Mack MM, Wehr CM, MacGregor JT,
Hiatt RA, Wang G, Wickramasinghe SN, Everson RB and Ames BN: Folate
deficiency causes uracil misincorporation into human DNA and
chromosome breakage: implications for cancer and neuronal damage.
Proc Natl Acad Sci USA. 94:3290–3295. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Choi SW and Mason JB: Folate and
carcinogenesis: an integrated scheme. J Nutr. 130:129–132.
2000.PubMed/NCBI
|
|
6
|
Cui LH, Shin MH, Kweon SS, Kim HN, Song
HR, Piao JM, Choi JS, Shim HJ, Hwang JE, Kim HR, et al:
Methylenetetrahydrofolate reductase C677T polymorphism in patients
with gastric and colorectal cancer in a Korean population. BMC
Cancer. 10:2362010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Goyette P, Sumner JS, Milos R, Duncan AM,
Rosenblatt DS, Matthews RG and Rozen R: Human
methylenetetrahydrofolate reductase: isolation of cDNA, mapping and
mutation identification. Nat Genet. 7:195–200. 1994. View Article : Google Scholar
|
|
8
|
Pereira AC, Schettert IT, Morandini Filho
AA, Guerra-Shinohara EM and Krieger JE: Methylenetetrahydrofolate
reductase (MTHFR) c677t gene variant modulates the homocysteine
folate correlation in a mild folate-deficient population. Clin Chim
Acta. 340:99–105. 2004. View Article : Google Scholar
|
|
9
|
Ogino S and Wilson RB: Genotype and
haplotype distributions of MTHFR677C>T and 1298A>C single
nucleotide polymorphisms: a meta-analysis. J Hum Genet. 48:1–7.
2003.PubMed/NCBI
|
|
10
|
Chen J, Giovannucci E, Kelsey K, et al: A
methylenetetrahydrofolate reductase polymorphism and the risk of
colorectal cancer. Cancer Res. 56:4862–4864. 1996.PubMed/NCBI
|
|
11
|
van der Put NM, Gabreels F, Stevens EM,
Smeitink JA, Trijbels FJ, Eskes TK, van den Heuvel LP and Blom HJ:
A second common mutation in the methylenetetrahydrofolate reductase
gene: an additional risk factor for neural-tube defects? Am J Hum
Genet. 62:1044–1051. 1998.PubMed/NCBI
|
|
12
|
Keku T, Millikan R, Worley K, Winkel S,
Eaton A, Biscocho L, Martin C and Sandler R:
5,10-Methylenetetrahydrofolate reductase codon 677 and 1298
polymorphisms and colon cancer in African Americans and whites.
Cancer Epidemiol Biomarkers Prev. 11:1611–1621. 2002.PubMed/NCBI
|
|
13
|
Heijmans BT, Boer JM, Suchiman HE,
Cornelisse CJ, Westendorp RG, Kromhout D, Feskens EJ and Slagboom
PE: A common variant of the methylenetetrahydrofolate reductase
gene (1p36) is associated with an increased risk of cancer. Cancer
Res. 63:1249–1253. 2003.PubMed/NCBI
|
|
14
|
Shannon B, Gnanasampanthan S, Beilby J and
Iacopetta B: A polymorphism in the methylenetetrahydrofolate
reductase gene predisposes to colorectal cancers with
microsatellite instability. Gut. 50:520–524. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yin G, Kono S, Toyomura K, Hagiwara T,
Nagano J, Mizoue T, Mibu R, Tanaka M, Kakeji Y, Maehara Y, et al:
Methylenetetrahydrofolate reductase C677T and A1298C polymorphisms
and colorectal cancer: the Fukuoka Colorectal Cancer Study. Cancer
Sci. 95:908–913. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ma J, Stampfer MJ, Giovannucci E, et al:
Methylenetetrahydrofolate reductase polymorphism, dietary
interactions, and risk of colorectal cancer. Cancer Res.
57:1098–1102. 1997.PubMed/NCBI
|
|
17
|
Ryan BM, Molloy AM, McManus R, Arfin Q,
Kelleher D, Scott JM and Weir DG: The methylenetetrahydrofolate
reductase (MTHFR) gene in colorectal cancer: role in tumor
development and significance of allelic loss in tumor progression.
Int J Gastrointest Cancer. 30:105–111. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sachse C, Smith G, Wilkie MJ, Barrett JH,
Waxman R, Sullivan F, Forman D, Bishop DT and Wolf CR; Colorectal
Cancer Study Group. A pharmacogenetic study to investigate the role
of dietary carcinogens in the etiology of colorectal cancer.
Carcinogenesis. 23:1839–1849. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Pufulete M, Al-Ghnaniem R, Leather AJ,
Appleby P, Gout S, Terry C, Emery PW and Sanders TA: Folate status,
genomic DNA hypomethylation, and risk of colorectal adenoma and
cancer: a case control study. Gastroenterology. 124:1240–1248.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Plaschke J, Schwanebeck U, Pistorius S,
Saeger HD and Schackert HK: Methylenetetrahydrofolate reductase
polymorphisms and risk of sporadic and hereditary colorectal cancer
with or without microsatellite instability. Cancer Lett.
191:179–185. 2003. View Article : Google Scholar
|
|
21
|
Toffoli G, Gafà R, Russo A, Lanza G,
Dolcetti R, Sartor F, Libra M, Viel A and Boiocchi M:
Methylenetetrahydrofolate reductase 677 C → T polymorphism and risk
of proximal colon cancer in north Italy. Clin Cancer Res.
9:743–748. 2003.
|
|
22
|
Ulvik A, Vollset SE, Hansen S, Gislefoss
R, Jellum E and Ueland PM: Colorectal cancer and the
methylenetetrahydrofolate reductase 677C → T and methionine
synthase 2756A → G polymorphisms: a study of 2,168 case-control
pairs from the JANUS cohort. Cancer Epidemiol Biomarkers Prev.
13:2175–2180. 2004.
|
|
23
|
Landi S, Gemignani F, Moreno V,
Gioia-Patricola L, Chabrier A, Guino E, Navarro M, de Oca J,
Capellà G and Canzian F; Bellvitge Colorectal Cancer Study Group. A
comprehensive analysis of phase I and phase II metabolism gene
polymorphisms and risk of colorectal cancer. Pharmacogenet
Genomics. 15:535–546. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Koushik A, Kraft P, Fuchs CS, Hankinson
SE, Willett WC, Giovannucci EL and Hunter DJ: Nonsynonymous
polymorphisms in genes in the one-carbon metabolism pathway and
associations with colorectal cancer. Cancer Epidemiol Biomarkers
Prev. 15:2408–2017. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Van Guelpen B, Hultdin J, Johansson I,
Hallmans G, Stenling R, Riboli E, Winkvist A and Palmqvist R: Low
folate levels may protect against colorectal cancer. Gut.
55:1461–1466. 2006.PubMed/NCBI
|
|
26
|
Battistelli S, Vittoria A, Stefanoni M,
Bing C and Roviello F: Total plasma homocysteine and
methylenetetrahydrofolate reductase C677T polymorphism in patients
with colorectal carcinoma. World J Gastroenterol. 12:6128–6132.
2006.PubMed/NCBI
|
|
27
|
Osian G, Procopciuc L and Vlad L: MTHFR
polymorphisms as prognostic factors in sporadic colorectal cancer.
J Gastrointestin Liver Dis. 16:251–256. 2007.PubMed/NCBI
|
|
28
|
Lima CS, Nascimento H, Bonadia LC, Teori
MT, Coy CS, Goes JR, Costa FF and Bertuzzo CS: Polymorphisms in
methylenetetrahydrofolate reductase gene (MTHFR) and the age of
onset of sporadic colorectal adenocarcinoma. Int J Colorectal Dis.
22:757–763. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Theodoratou E, Farrington SM, Tenesa A,
McNeill G, Cetnarskyj R, Barnetson RA, Porteous ME, Dunlop MG and
Campbell H: Dietary vitamin B6 intake and the risk of colorectal
cancer. Cancer Epidemiol Biomarkers Prev. 17:171–182. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sharp L, Little J, Brockton NT, Cotton SC,
Masson LF, Haites NE and Cassidy J: Polymorphisms in the
methylenetetrahydrofolate reductase (MTHFR) gene, intakes of folate
and related B vitamins and colorectal cancer: a case-control study
in a population with relatively low folate intake. Br J Nutr.
99:379–389. 2008. View Article : Google Scholar
|
|
31
|
Eklöf V, Van Guelpen B, Hultdin J,
Johansson I, Hallmans G and Palmqvist R: The reduced folate carrier
(RFC1) 80G > A and folate hydrolase 1 (FOLH1) 1561C > T
polymorphisms and the risk of colorectal cancer: a nested
case-referent study. Scand J Clin Lab Invest. 68:393–401. 2008.
|
|
32
|
Küry S, Buecher B, Robiou-du-Pont S, Scoul
C, Colman H, Le Neel T, Le Houérou C, Faroux R, Ollivry J, Lafraise
B, Chupin LD, Sébille V and Bézieau S: Low-penetrance alleles
predisposing to sporadic colorectal cancers: a French
case-controlled genetic association study. BMC Cancer.
8:3262008.PubMed/NCBI
|
|
33
|
Derwinger K, Wettergren Y, Odin E,
Carlsson G and Gustavsson B: A study of the MTHFR gene polymorphism
C677T in colorectal cancer. Clin Colorectal Cancer. 8:43–48. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
de Vogel S, Wouters KA, Gottschalk RW, van
Schooten FJ, de Goeij AF, de Bruïne AP, Goldbohm RA, van den Brandt
PA, Weijenberg MP and van Engeland M: Genetic variants of methyl
metabolizing enzymes and epigenetic regulators: associations with
promoter CpG island hypermethylation in colorectal cancer. Cancer
Epidemiol Biomarkers Prev. 18:3086–3096. 2009.PubMed/NCBI
|
|
35
|
Fernández-Peralta AM, Daimiel L, Nejda N,
Iglesias D, Medina Arana V and González-Aguilera JJ: Association of
polymorphisms MTHFR C677T and A1298C with risk of colorectal
cancer, genetic and epigenetic characteristic of tumors, and
response to chemotherapy. Int J Colorectal Dis. 25:141–151.
2010.PubMed/NCBI
|
|
36
|
Gallegos-Arreola MP, Garcia-Ortiz JE,
Figuera LE, Puebla-Perez AM, Morgan-Villela G and Zuniga-Gonzalez
GM: Association of the 677C → T polymorphism in the MTHFR gene with
colorectal cancer in Mexican patients. Cancer Genomics Proteomics.
6:183–188. 2009.
|
|
37
|
Komlosi V, Hitre E, Pap E, Adleff V, Reti
A, Szekely E, Biro A, Rudnai P, Schoket B, Muller J, et al: SHMT1
1420 and MTHFR 677 variants are associated with rectal but not
colon cancer. BMC Cancer. 10:5252010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Karpinski P, Myszka A, Ramsey D, Misiak B,
Gil J, Laczmanska I, Grzebieniak Z, Sebzda T, Smigiel R, Stembalska
A and Sasiadek MM: Polymorphisms in methyl-group metabolism genes
and risk of sporadic colorectal cancer with relation to the CpG
island methylator phenotype. Cancer Epidemiol. 34:338–344. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Vossen CY, Hoffmeister M, Chang-Claude JC,
Rosendaal FR and Brenner H: Clotting factor gene polymorphisms and
colorectal cancer risk. J Clin Oncol. 29:1722–1727. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Eussen SJ, Vollset SE, Igland J, Meyer K,
Fredriksen A, et al: Plasma folate, related genetic variants, and
colorectal cancer risk in EPIC. Cancer Epidemiol Biomarkers Prev.
19:1328–1340. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Lee JE, Wei EK, Fuchs CS, Hunter DJ, Lee
IM, Selhub J, et al: Plasma folate, methylenetetrahydrofolate
reductase (MTHFR), and colorectal cancer risk in three large nested
case-control studies. Cancer Causes Control. 23:537–545. 2012.
View Article : Google Scholar
|
|
42
|
Park KS, Mok JW and Kim JC: The 677C >
T mutation in 5,10-methylenetetrahydrofolate reductase and
colorectal cancer risk. Genet Test. 3:233–236. 1999.
|
|
43
|
Matsuo K, Hamajima N, Hirai T, Kato T,
Inoue M, Takezaki T and Tajima K: Methionine synthase reductase
gene A66G polymorphism is associated with risk of colorectal
cancer. Asian Pac J Cancer Prev. 3:353–359. 2002.PubMed/NCBI
|
|
44
|
Huang P, Zhou ZY, Ma HT, Liu JY, Zhou YH,
Cao J, Ge HY, Yu PW and Takezaki T: MTHFR polymorphisms and
colorectal cancer susceptibility in Chongqing people. Acta
Academiae Medicinae Militaris Tertiae. 25:1710–1713. 2003.(In
Chinese).
|
|
45
|
Kim DH, Ahn YO, Lee BH, Tsuji E, Kiyohara
C and Kono S: Methylenetetrahydrofolate reductase polymorphism,
alcohol intake, and risks of colon and rectal cancers in Korea.
Cancer Lett. 216:199–205. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Matsuo K, Ito H, Wakai K, Hirose K, Saito
T, Suzuki T, Kato T, Hirai T, Kanemitsu Y, Hamajima H and Tajima K:
One-carbon metabolism related gene polymorphisms interact with
alcohol drinking to influence the risk of colorectal cancer in
Japan. Carcinogenesis. 26:2164–2171. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Jiang Q, Chen K, Ma X, Li Q, Yu W, Shu G
and Yao K: Diets, polymorphisms of methylenetetrahydrofolate
reductase, and the susceptibility of colon cancer and rectal
cancer. Cancer Detect Prev. 29:146–154. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Miao XP, Yang S, Tan W, Zhang XM, et al:
Association between genetic variations in methylenetetrahydrofolate
reductase and risk of colorectal cancer in a Chinese population.
Chin J Prev Med. 39:409–411. 2005.(In Chinese).
|
|
49
|
Otani T, Iwasaki M, Hanaoka T, Kobayashi
M, Ishihara J, Natsukawa S, Shaura K, Koizumi Y, Kasuga Y,
Yoshimura K, Yoshida T and Tsugane S: Folate, vitamin B6, vitamin
B12, and vitamin B2 intake, genetic polymorphisms of related
enzymes, and risk of colorectal cancer in a hospital-based
case-control study in Japan. Nutr Cancer. 53:42–50. 2005.
View Article : Google Scholar
|
|
50
|
Wang J, Gajalakshmi V, Jiang J, Kuriki K,
Suzuki S, Nagaya T, Nakamura S, Akasaka S, Ishikawa H and Tokudome
S: Associations between 5,10-methylenetetrahydrofolate reductase
codon 677 and 1298 genetic polymorphisms and environmental factors
with reference to susceptibility to colorectal cancer: a
case-control study in an Indian population. Int J Cancer.
118:991–997. 2006. View Article : Google Scholar
|
|
51
|
Chang SC, Lin PC, Lin JK, Yang SH, Wang HS
and Li AF: Role of MTHFR polymorphisms and folate levels in
different phenotypes of sporadic colorectal cancers. Int J
Colorectal Dis. 22:483–489. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Zeybek U, Yaylim I, Yilmaz H, Agaçhan B,
Ergen A, Arikan S, Bayrak S and Isbir T: Methylenetetrahydrofolate
reductase C677T polymorphism in patients with gastric and
colorectal cancer. Cell Biochem Funct. 25:419–422. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Jin XX, Zhu ZZ, Wang AZ and Jia HR:
Association of methylenetetrahydrofolate reductase C677T
polymorphism with genetic susceptibility to colorectal cancer.
World Chin J Digestol. 15:2754–2757. 2007.(In Chinese).
|
|
54
|
Mokarram P, Naghibalhossaini F, Saberi
Firoozi M, Hosseini SV, Izadpanah A, Salahi H, Malek-Hosseini SA,
Talei A and Mojallal M: Methylenetetrahydrofolate reductase C677T
genotype affects promoter methylation of tumor-specific genes in
sporadic colorectal cancer through an interaction with
folate/vitamin B12 status. World J Gastroenterol. 14:3662–3671.
2008. View Article : Google Scholar
|
|
55
|
Haghighi MM, Radpour R, Mahmoudi T,
Mohebbi SR, Vahedi M and Zali MR: Association between MTHFR
polymorphism (C677T) with nonfamilial colorectal cancer. Oncol Res.
18:57–63. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Zhang YL, Yuan XY, Zhang Z, Yang H, Zhou
YH, Pan YM, Zhou ZY, Liang HJ and Cao J: Relationship of genetic
polymorphisms in methylenetetrahydrofolate reductase and alcohol
drinking with the risk of colorectal cancer. Chin J Cancer Prev
Treat. 15:1298–1301. 2008.(In Chinese).
|
|
57
|
Promthet SS, Pientong C, Ekalaksananan T,
Wiangnon S, Poomphakwaen K, Songserm N, Chopjitt P, Moore MA and
Tokudome S: Risk factors for colon cancer in Northeastern Thailand:
interaction of MTHFR codon 677 and 1298 genotypes with
environmental factors. J Epidemiol. 20:329–338. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Naghibalhossaini F, Mokarram P, Khalili I,
Vasei M, Hosseini SV, Ashktorab H, Rasti M and Abdollahi K: MTHFR
C677T and A1298C variant genotypes and the risk of microsatellite
instability among Iranian colorectal cancer patients. Cancer Genet
Cytogenet. 197:142–151. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Kim JW, Park HM, Choi YK, Chong SY, Oh D
and Kim NK: Polymorphisms in genes involved in folate metabolism
and plasma DNA methylation in colorectal cancer patients. Oncol
Rep. 25:167–172. 2011.PubMed/NCBI
|
|
60
|
Chandy S, Sadananda Adiga MN, Ramachandra
N, Krishnamoorthy S, Ramaswamy G, Savithri HS and Krishnamoorthy L:
Association of methylenetetrahydrofolate reductase gene
polymorphisms and colorectal cancer in India. Indian J Med Res.
131:659–664. 2010.PubMed/NCBI
|
|
61
|
Zhu F, Wang YM and QY Z: A case-control
study of plasma homocysteine, serum folate, the polymorphism of
methylenetetrahydrofolate reductase in colorectal cancer. J
Southeast Univ Med Sci Edi. 29:88–92. 2010.
|
|
62
|
Kang BS, Ahn DH, Kim NK and Kim JW:
Relationship between metabolic syndrome and MTHFR polymorphism in
colorectal cancer. J Korean Soc Coloproctol. 27:78–82. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Prasad VV and Wilkhoo H: Association of
the functional polymorphism C677T in the methylenetetrahydrofolate
reductase gene with colorectal, thyroid, breast, ovarian, and
cervical cancers. Onkologie. 34:422–426. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Sameer AS, Shah ZA, Nissar S, Mudassar S
and Siddiqi MA: Risk of colorectal cancer associated with the
methylenetetrahydrofolate reductase (MTHFR) C677T polymorphism in
the Kashmiri population. Genet Mol Res. 10:1200–1210. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Li H, Xu WL, Shen HL, Chen QY, Hui LL,
Long LL and Zhu XL: Methylenetetrahydrofolate reductase genotypes
and haplotypes associated with susceptibility to colorectal cancer
in an eastern Chinese Han population. Genet Mol Res. 10:3738–3746.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Yin G, Ming H, Zheng X, Xuan Y, Liang J
and Jin X: Methylenetetrahydrofolate reductase C677T gene
polymorphism and colorectal cancer risk: A case-control study.
Oncol Lett. 4:365–369. 2012.PubMed/NCBI
|
|
67
|
Kim J, Cho YA, Kim DH, Lee BH, Hwang DY,
Jeong J, Lee HJ, Matsuo K, Tajima K and Ahn YO: Dietary intake of
folate and alcohol, MTHFR C677T polymorphism, and colorectal cancer
risk in Korea. Am J Clin Nutr. 95:405–412. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
El Awady MK, Karim AM, Hanna LS, El
Husseiny LA, El Sahar M, Menem HA and Meguid NA:
Methylenetetrahydrofolate reductase gene polymorphisms and the risk
of colorectal carcinoma in a sample of Egyptian individuals. Cancer
Biomark. 5:233–240. 2009.PubMed/NCBI
|
|
69
|
Guimarães JL, de Ayrizono ML, Coy CS and
Lima CS: Gene polymorphisms involved in folate and methionine
metabolism and increased risk of sporadic colorectal
adenocarcinoma. Tumour Biol. 32:853–861. 2011.PubMed/NCBI
|
|
70
|
Slattery ML, Potter JD, Samowitz W,
Schaffer D and Leppert M: Methylenetetrahydrofolate reductase,
diet, and risk of colon cancer. Cancer Epidemiol Biomarkers Prev.
8:513–518. 1999.PubMed/NCBI
|
|
71
|
Curtin K, Bigler J, Slattery ML, Caan B,
Potter JD and Ulrich CM: MTHFR C677T and A1298C polymorphisms:
diet, estrogen, and risk of colon cancer. Cancer Epidemiol
Biomarkers Prev. 13:285–292. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Murtaugh MA, Curtin K, Sweeney C, Wolff
RK, Holubkov R, Caan BJ and Slattery ML: Dietary intake of folate
and co-factors in folate metabolism, MTHFR polymorphisms, and
reduced rectal cancer. Cancer Causes Control. 18:153–163. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Iacopetta B, Heyworth J, Girschik J, Grieu
F, Clayforth C and Fritschi L: The MTHFR C677T and DeltaDNMT3B
C-149T polymorphisms confer different risks for right- and
left-sided colorectal cancer. Int J Cancer. 125:84–90. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Reeves SG, Meldrum C, Groombridge C,
Spigelman AD, Suchy J, Kurzawski G, Lubinski J, McElduff P and
Scott RJ: MTHFR 677 C>T and 1298 A>C polymorphisms and the
age of onset of colorectal cancer in hereditary nonpolyposis
colorectal cancer. Eur J Hum Genet. 17:629–635. 2009.
|
|
75
|
Yang XX, Li FX, Yi J, Li X, Sun JZ and Hu
NY: An association of methylenetetrahydrofolate reductase reductase
C677T and gastric cancer, colorectal cancer and lung cancer
susceptibility. Guangdong Med J. 31:2375–2377. 2010.(In
Chinese).
|
|
76
|
Zhu Q, Jin Z, Yuan Y, Lu Q, Ge D and Zong
M: Impact of MTHFR gene C677T polymorphism on Bcl-2 gene
methylation and protein expression in colorectal cancer. Scand J
Gastroenterol. 46:436–445. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Le Marchand L, Wilkens LR, Kolonel LN and
Henderson BE: The MTHFR C677T polymorphism and colorectal cancer:
the multiethnic cohort study. Cancer Epidemiol Biomarkers Prev.
14:1198–1203. 2005.PubMed/NCBI
|