Introduction
The incidence of colorectal cancer (CRC) has been on
the increase and is currently a major cause of cancer-related
morbidity and mortality worldwide, with high rates in westernized
societies and an increasing rate in developing countries (1). Although studies reported that
lifestyle, dietary habits and environmental factors may be involved
in the occurrence of CRC (2,3),
inheritance was recently recognized as an important factor
(4). Approximately one-third of
cases appeared to have inheritance as part of their pathogenesis
(5). A large twin study
demonstrated that inherited factors account for 35% of CRC cases,
whereas shared environmental factors account for 5% and non-shared
environmental factors for the remaining 60% (6).
MicroRNAs (miRNAs) are an abundant class of small
non-coding, single-stranded RNAs of 21–24 nucleotides that form
base pairs with target mRNAs and regulate their
post-transcriptional functions as tumor suppressors and oncogenes
(7–9). Several human studies provided evidence
that the presence of single-nucleotide polymorphisms (SNPs) in
miRNAs may alter miRNA processing, expression, and/or binding to
target mRNA and are another type of genetic variability that may
contribute to the susceptibility to cancer development (10–12).
A C/T polymorphism (rs11614913) was located in the
stem region opposite the mature miR-196a2 sequence. Previous
studies demonstrated that high expression levels of miR-196a2 may
promote the migration and invasion of CRC cells and the C allele of
rs11614913 polymorphism may affect miR-196a2 expression levels in
cancer (13–16). Several previous studies reported the
association between rs11614913 polymorphisms and the susceptibility
to CRC (15,17–21).
However, those studies produced controversial and inconclusive
results. Since the statistical power of an individual study may be
insufficient for the assessment of rs11614913 polymorphisms,
integration of data sets may provide improved statistical power and
detect significance.
The present meta-analysis was conducted with the aim
of addressing inconsistencies in the findings of previous studies.
The meta-analysis was based on published case-control studies, in
order to assess the association between the miR-196a2 rs11614913
polymorphism and the susceptibility to CRC.
Materials and methods
Literature search strategy
A search was conducted on PubMed, EMBASE,
ScienceDirect, the Foreign Medical Journal Service (FMJS) and the
Chinese National Knowledge Infrastructure (CNKI) databases for all
genetic association studies on the rs11614913 polymorphism of
miR-196a2 and the susceptibility to CRC, published prior to
November, 2012. The search used the following keywords and subject
terms: miR-196a2 or microRNA-196a2 or miRNA-196a2, rs11614913 or
polymorphism or SNPs and colorectal cancer/carcinoma/neoplasm. The
search was limited to English and Chinese language articles. The
reference lists were manually examined to further identify
potentially relevant studies. The corresponding authors of
conference abstracts without sufficient data were contacted via
e-mail for additional information.
Selection criteria
Any human-associated study, regardless of sample
size, was included if the following criteria were met: i) use of an
unrelated case-control design; ii) investigation of the association
between rs11614913 polymorphisms of miR-196a2 and the risk of CRC;
iii) genotype distribution of the control population in
Hardy-Weinberg equilibrium (HWE) [goodness-of-fit test, degree of
freedom (df)=1] and iv) published in English or Chinese. For
articles with the same population resource or overlapping data
sets, the publication reporting the largest or most recent data set
was included. Therefore, the data for this meta-analysis were
obtained from six case-control studies, including a total of 1,754
CRC cases and 2,430 controls.
Data extraction
Two investigators (Xiao-qing Guo and Chun-mei Wang)
independently extracted data and reached a consensus on all the
items. The following information was recorded for each study: first
author, year of publication, country of origin, cancer type,
ethnicity, number of cases and controls, study design, genotyping
methods and evidence of HWE. For subjects of different ethnicities,
data were extracted separately and classified as European or Asian
(Table I).
 | Table ICharacteristics of included studies
that investigated the association between rs11614913 polymorphisms
of miR-196a2 and colorectal cancer risk. |
Table I
Characteristics of included studies
that investigated the association between rs11614913 polymorphisms
of miR-196a2 and colorectal cancer risk.
| First author | Year | Country | Ethnic descent | Genotyping | Source of
control | Sample size
(case/control) | P-valuea | Case | Control | Refs. |
|---|
|
|
|---|
| CC | CT | TT | CC | CT | TT |
|---|
| Chen et
al | 2012 | China | Asian | PCR-LDR | HB | 126/407 | 0.788 | 27 | 64 | 35 | 94 | 206 | 107 | (17) |
| Hezova et
al | 2012 | Czech | European | Taqman | HB | 197/212 | 0.291 | 82 | 89 | 26 | 87 | 103 | 22 | (18) |
| Min et
al | 2012 | Korea | Asian | PCR-RFLP | PB | 446/502 | 0.633 | 120 | 201 | 125 | 100 | 254 | 148 | (19) |
| Vinci et
al | 2013 | Italy | European | HRMA | HB | 160/178 | 0.087 | 62 | 86 | 12 | 83 | 84 | 11 | (20) |
| Zhan et
al | 2011 | China | Asian | PCR-RFLP | HB | 252/543 | 0.849 | 68 | 128 | 56 | 113 | 267 | 163 | (15) |
| Zhu et
al | 2012 | China | Asian | Taqman | HB | 573/588 | 0.790 | 140 | 303 | 130 | 121 | 295 | 172 | (21) |
Statistical analysis
Observed genotype frequencies for rs11614913
polymorphisms in controls were assessed for deviation from HWE
using a goodness-of-fit Chi-square test with df=1. P<0.05 was
considered representative of departure from HWE. Summary odds
ratios (ORs) and 95% confidence intervals (CIs) were calculated for
the allelic (C allele vs. T allele) and genotypic comparisons,
following the co-dominant (CT vs. TT and CC vs. TT), dominant
(CT+CC vs. TT) and recessive (CC vs. TT+CT) genetic models. For age
comparison, the pooled weighted mean difference (WMD) was also
performed. The significance of pooled ORs was determined by the
Z-test and P<0.05 was considered to indicate a statistically
significant difference.
Statistical heterogeneity among the studies was
assessed by the Chi-square-based Q-test. A Q-test P>0.05
indicated no significant heterogeneity among the studies (22) and the pooled OR was estimated by the
fixed-effects model (Mantel-Haenszel method) (23). If the heterogeneity was significant,
the random- effects model (inverse variance method) was employed
(24).
Publication bias was investigated with a funnel
plot, which was used as the main graphical method. To supplement
the funnel plot approach, the Begg and Mazumdar’s adjusted rank
correlation test (25) and the
Egger’s regression asymmetry test (26) were utilized.
Analyses were performed with Review Manager software
(RevMan, version 5.0; The Cochrane Collaboration, Oxford, England)
and Stata software, version 10.0 (StataCorp LP, College Station,
TX, USA). All the P-values were two-sided. The statistical tests
performed in the present analysis were considered to indicate a
statistically significant difference whenever the corresponding
null-hypothesis probability was P<0.05.
Results
Characteristics of studies
Overall, six studies including 1,754 cases and 2,430
controls were available for this analysis. The study
characteristics are provided in Table
I. The sample size in these case-control studies varied
considerably (338–1,161 individuals). There were four studies on
Asian descendants and two on European descendants. Four genotyping
methods were used, including Taqman, polymerase chain
reaction-restriction fragment length polymorphism (PCR-RFLP),
high-resolution melting analysis (HRMA) and polymerase chain
reaction-ligation detection reaction (PCR-LDR). Furthermore, ~83%
(5/6) of these studies included described genotyping quality
control measures, such as a different genotyping assay to confirm
the data and random repetition of a portion of samples. The
genotype distributions among the controls of all studies were
consistent with HWE.
In addition, the cases and controls of all the
studies included in our analysis were matched by gender and age [no
specific data were presented in the study by Vinci et
al(20)]. The meta-analysis for
age and gender detected no significant difference between the cases
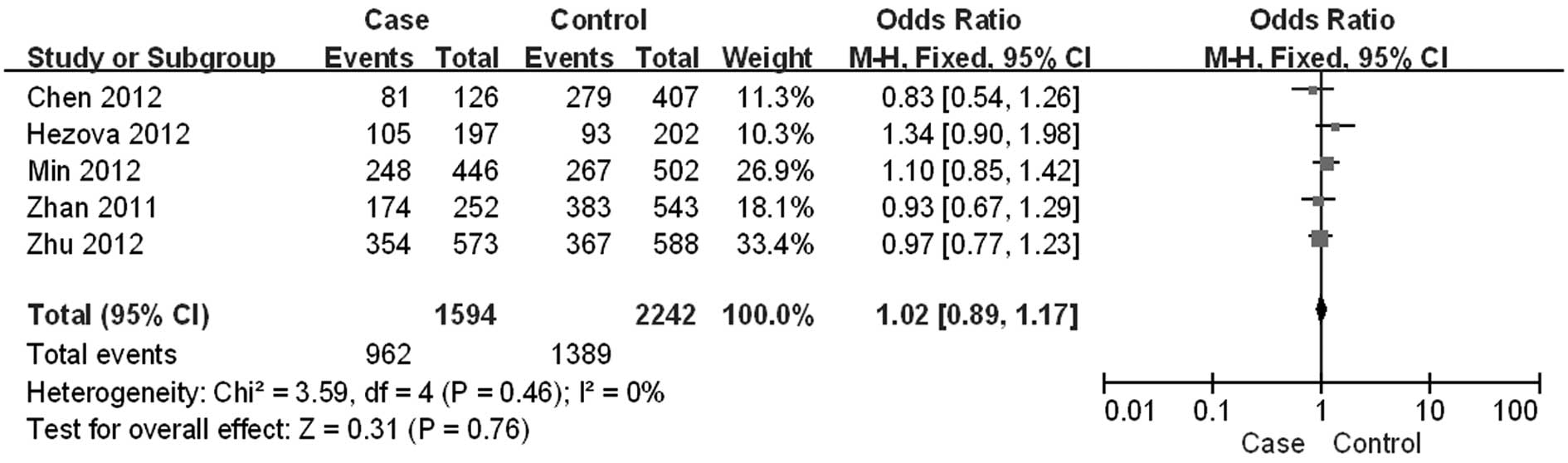
and the controls (for gender: OR=1.02; 95% CI: 0.89–1.17;
heterogeneity test, P=0.46, Fig. 1;
for age: WMD=0.22; 95% CI: 0.69–1.14; heterogeneity test, P=0.08),
which suggested that age and gender were adequately matched in this
meta-analysis.
Meta-analysis results
Six studies involving a total of 1,754 cases and
2,430 controls were assessed for the association between miR-196a2
rs11614913 polymorphism and CRC risk. There were no significant
statistical heterogeneities in any of the comparison models;
therefore, the fixed-effects model was used. Overall, the C allele
were associated with a significantly increased risk when compared
to the T allele (OR=1.14; 95% CI: 1.04–1.24). Similarly, moderately
elevated risks were also observed in overall analyses in the
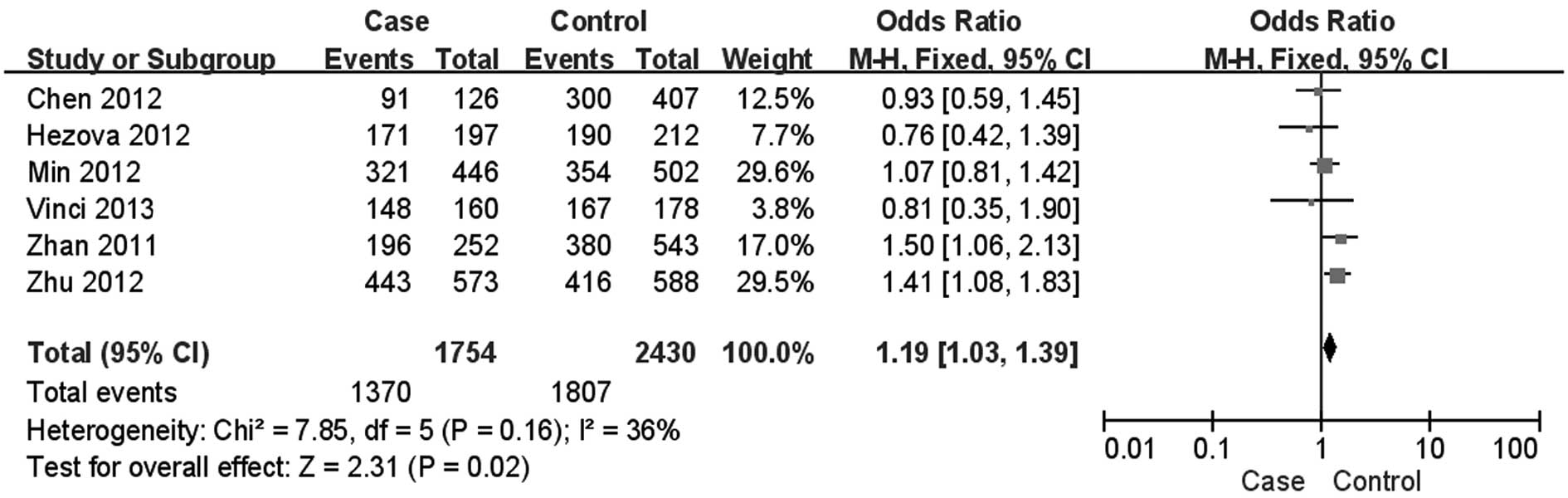
dominant (OR=1.19; 95% CI: 1.03–1.39; Fig. 2) and the recessive model (OR=1.18;
95% CI: 1.02–1.36). Moreover, the subgroup analysis in an Asian
population also demonstrated similar results. Almost all the
genetic models indicated a significant association between
rs11614913 polymorphism and CRC risk (C allele vs. T allele:
OR=1.20; 95% CI: 1.08–1.32; CC vs. TT: OR=1.44; 95% CI: 1.18–1.75;
dominant model: OR=1.25; 95% CI: 1.06–1.46; recessive model:
OR=1.30; 95% CI: 1.10–1.53) except for the model of CT vs. TT. For
CT vs. TT, there was no significant association in the overall
analysis and the subgroup analysis in an Asian population. The
results are shown in Table II.
 | Table IISummary of ORs in the meta-analysis
of SNP rs11614913 with the fixed-effects model. |
Table II
Summary of ORs in the meta-analysis
of SNP rs11614913 with the fixed-effects model.
| Genetic model | Population | No. of
case/controls | OR (95% CI) | P-value | Ph |
|---|
| C allele vs. T
allele | Overall | 3508/4860 | 1.14
(1.04–1.24) | 0.005 | 0.06 |
| Asian | 2794/4080 | 1.20
(1.08–1.32) | 0.0004 | 0.28 |
| CT vs. TT | Overall | 1255/1832 | 1.12
(0.96–1.32) | 0.15 | 0.23 |
| Asian | 1042/1612 | 1.17
(0.99–1.38) | 0.07 | 0.19 |
| CC vs. TT | Overall | 883/1221 | 1.33
(1.10–1.59) | 0.003 | 0.11 |
| Asian | 701/1018 | 1.44
(1.18–1.75) | 0.0003 | 0.28 |
| Dominant model
(CT+CC vs. TT) | Overall | 1754/2430 | 1.19
(1.03–1.39) | 0.02 | 0.16 |
| Asian | 1397/2040 | 1.25
(1.06–1.46) | 0.006 | 0.20 |
| Recessive model (CC
vs. TT+CT) | Overall | 1754/2430 | 1.18
(1.02–1.36) | 0.02 | 0.08 |
| Asian | 1397/2040 | 1.30
(1.10–1.53) | 0.002 | 0.38 |
The effect of miR-196a2 rs11614913 polymorphism was
then evaluated based on the clinical characteristics of patients
with CRC in three studies on the Han Chinese ethnic group (15,17,21).
The clinical CRC stage in two of those studies (15,17)
was evaluated on the basis of the TNM classification system;
however, in the third study (21)
stage was classified into Dukes’ A, B, C and D. It is well known
that Dukes’ A+B is equivalent to clinical stage I+II and Dukes’ C+D
is equivalent to clinical stage III+IV [Ling-jun Zhu, the author of
article (21), was contacted via
e-mail and confirmed this classification]. Therefore, tumor stage
was classified into two groups, III+IV vs. I+II. The tumor grade
was also divided into two groups, intermediate/high vs. low. When
we evaluated the associations between rs11614913 polymorphism and
tumor stage or grade separately, no significant association was
observed in any comparison model (Table III), which suggested that
miR-196a2 rs11614913 polymorphism may not be associated with CRC
stage or grade.
 | Table IIIMeta-analyses of miR-196a2 rs11614913
polymorphism and clinical characteristics of colorectal cancer
risk. |
Table III
Meta-analyses of miR-196a2 rs11614913
polymorphism and clinical characteristics of colorectal cancer
risk.
| Genetic model | Variables | No. of
case/controls | OR (95% CI) | P-value | Ph |
|---|
| C allele vs. T
allele | Tumor stage | 910/968 | 1.10
(0.92–1.32) | 0.29 | 0.23 |
| Tumor grade | 1172/610 | 1.14
(0.83–1.56) | 0.43 | 0.88 |
| CT vs. TT | Tumor stage | 341/366 | 1.28
(0.93–1.76) | 0.13 | 0.16 |
| Tumor grade | 439/241 | 0.90
(0.52–1.58) | 0.72 | 0.59 |
| CC vs. TT | Tumor stage | 208/238 | 1.24
(0.85–1.79) | 0.27 | 0.20 |
| Tumor grade | 281/141 | 1.28
(0.69–2.38) | 0.44 | 0.89 |
| CT+CC vs. TT | Tumor stage | 455/484 | 1.26
(0.93–1.72) | 0.13 | 0.12 |
| Tumor grade | 586/305 | 1.02
(0.60–1.71) | 0.95 | 0.84 |
| CC vs. TT+CT | Tumor stage | 455/484 | 1.04
(0.77–1.40) | 0.80 | 0.72 |
| Tumor grade | 586/305 | 1.38
(0.82–2.31) | 0.23 | 0.52 |
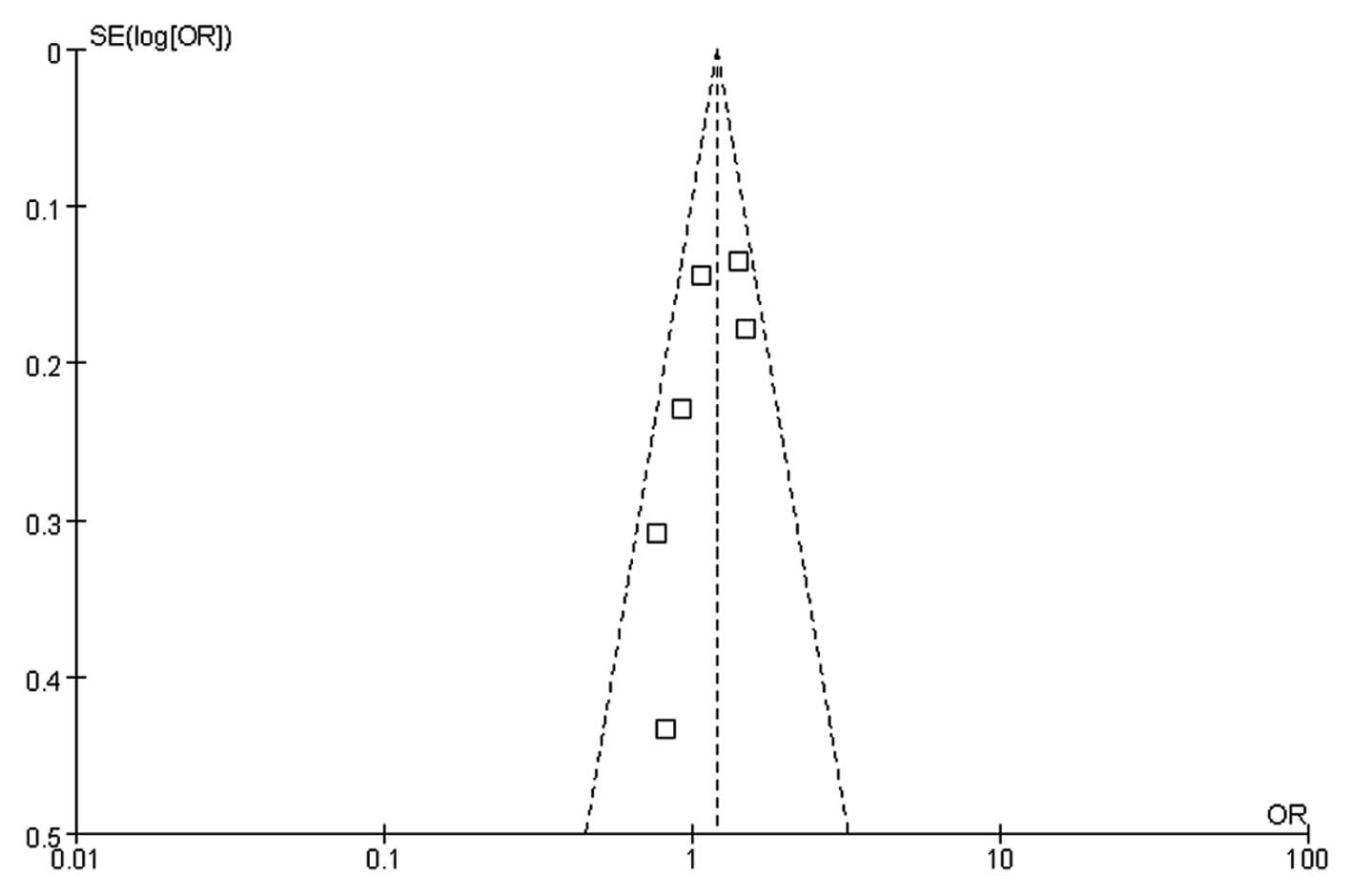
Publication bias
Funnel plots, Egger’s and Begg’s tests were used to
evaluate the publication bias of the literature on CRC. Symmetrical
funnel plots were obtained in all the models. Egger’s and Begg’s
tests further confirmed the absence of publication bias in this
meta-analysis (P>0.05). Fig. 3
displays a funnel plot that assessed the miR-196a2 rs11614913
polymorphism and CRC risk in the dominant model.
Discussion
The association between miR-196a2 rs11614913
polymorphisms and the risk of CRC has been previously investigated
(15,17–21).
However, due to the limited sample size and the potential bias of
case selection, these results were controversial and inconclusive.
In this meta-analysis, we systematically summarized six eligible
case-control studies on the association between SNP rs11614913 and
the susceptibility to CRC.
The common genetic variant (rs11614913) was located
in the 3p mature miRNA region of hsa-miR-196a2. This C>T
polymorphism results in a change from G:C to G:T in the stem region
of the miR-196a2 precursor. Several studies investigated the
association between miR-196a2 rs11614913 polymorphism and the
miR-196a2 expression level in various types of cancer (13–15).
Hoffman et al(13)
demonstrated that mature miR-196a2 levels were increased 9.3-fold
in cells transfected with pre-miR-196a2-C but only 4.4-fold with
pre-miR-196a2-T. In the genotype-phenotype correlation analysis of
cancer tissues, the C allele of rs11614913 increased the expression
of mature miR-196a2 in lung cancer (14) and CRC tissues (15). These results indicated that
rs11614913 polymorphism may affect the processing of the pre-miRNA
to its mature form. It was previously reported that a high
expression level of miR-196a may promote the migration and invasion
of CRC cells (16) and recent
studies reported the association of miR-196a2 rs11614913
polymorphism with CRC risk. For example, Zhan et al(15) first observed the association between
SNP miR-196a2 rs11614913 and susceptibility to CRC. However, the
results from other studies reported a lack of association of
miR-196a2 rs11614913 and the risk for CRC (17,18) or
only associations between miR-196a2 SNP and the non-diabetic or
rectal cancer groups (19).
In this meta-analysis, six case-control studies were
analyzed to provide a comprehensive assessment of the association
between the miR-196a2 rs11614913 polymorphism and CRC. Our results
supported a genetic association between rs11614913 and
susceptibility to CRC. It was observed that the hsa-miR-196a2
rs11614913 polymorphism was associated with an increased CRC risk
in almost all the genetic models, except the model of CT vs. TT,
indicating that the hsa-miR-196a2 rs11614913 polymorphism may be
important in the development of CRC. Since the incidence of gene
polymorphisms may vary between different ethnic groups and this
variation may interfere with the detection of minor effects of SNPs
on CRC risk, a subgroup analysis in an Asian population was
performed to further investigate the potential association between
rs11614913 and the risk of CRC. The subgroup analysis also
demonstrated a significant association of miR-196a2 rs11614913
polymorphism with susceptibility to CRC. Moreover, since the
clinical characteristics of patients were well-established risk
factors for the development of CRC, we also evaluated the effect of
rs11614913 polymorphism on CRC by a stratified analysis of tumor
stage and grade. However, there was no association identified
between rs11614913 polymorphism and the risk for clinical
characteristics of patients in any of the genetic models,
suggesting that tumor stage and grade may not be the main factors
affecting the stability of these comparisons. It also indicated
that rs11614913 polymorphism may be involved in the occurrence of
CRC, but not in its progression. The sensitivity analysis did not
detect a significant effect of any single study on pooled ORs,
indicating that the stability of this meta-analysis was
acceptable.
Ten previous meta-analyses reviewed the potential
role of polymorphism rs11614913 in the development of cancer, seven
of which reported a statistically significant association between
this polymorphism and susceptibility to cancer without
pre-specified tissue origin (27–33)
and three focused on a pre-specified cancer type, such as breast
cancer (34), hepatocellular
carcinoma (35) or digestive system
cancer (36). However, clinical
heterogeneity due to inherent differences between cancers of
distinct tissue origins may limit the reliability of the
conclusions of those meta-analyses (35). Therefore, a meta-analysis focusing
on one specific type of cancer may increase the stability of the
conclusions. Thus far, only two meta-analyses investigated the
association between rs11614913 and CRC by means of subgroup
analysis (32,36). Those results were consistent with
ours in the allele frequency comparison (C allele vs. T allele),
co-dominant model (CC vs. TT) and recessive model (CC vs. TT+CT).
However, the meta-analysis of Wang et al(32) analyzed three case-control studies
and reported a moderately elevated risk in the dominant model,
which was consistent with our findings but inconsistent with those
of Guo et al(36).
Similarly, the two subgroup results (32,36)
indicated that rs11614913 C/T heterozygosity is significantly
associated with the risk of CRC, which was inconsistent with our
results. These inconsistencies may be partly due to the fact that
none of the meta-analyses included all the available studies on the
association between rs11614913 and susceptibility to CRC.
To the best of our knowledge, this is the first
meta-analysis evaluating the potential association between
rs11614913 in miR-196a2 and susceptibility to CRC. It has been well
established that environmental factors, dietary habits and
lifestyle habits, such as drinking and smoking, are associated with
increased risk of CRC (2,3,37). In
addition, a positive family history may play a key role in CRC risk
(38,39). However, lack of available data
prevented an adjustment for subgroup factors, such as family
history of CRC, alcohol consumption and smoking status. However,
our meta-analysis held some key benefits. First, no heterogeneity
was detected in any comparison and the sensitivity analysis
demonstrated that none of the studies exerted a significant effect
on the evaluation of potential association, which indicated that
the results were more reliable. Second, the evaluation of
rs11614913 polymorphism and the risk of patient clinical
characteristics were evaluated in our study, but not in those of
Wang et al(32) and Guo
et al(36). The stratified
analysis in our study may have enhanced the stability of the
results and the reliability of the conclusions. Thirdly, the cases
and controls of all the studies included in our analysis were
adequately matched by gender and age, which ensured the stability
of our results and conclusions.
In summary, miR-196a2 rs11614913 polymorphisms may
be associated with the risk of CRC. Well-designed studies including
larger sample sizes and more ethnic groups are required to further
elucidate this association. Other factors, such as gender, age,
smoking status, tumor stage and grade, lymph node status and tumor
invasiveness should also be considered in future studies.
Acknowledgements
We would like to thank Guicheng Zhang (School of
Paediatrics and Child Health, University of Western Australia,
Australia) for his helpful comments. This study was supported by
the Natural Science Foundation of China (grant no. 81172333).
References
|
1
|
Schnekenburger M and Diederich M:
Epigenetics offer new horizons for colorectal cancer prevention.
Curr Colorectal Cancer Rep. 8:66–81. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Glade MJ: Food, nutrition, and the
prevention of cancer: a global perspective. American Institute for
Cancer Research/World Cancer Research Fund, American Institute for
Cancer Research, 1997. Nutrition. 15:523–526. 1999.PubMed/NCBI
|
|
3
|
Potter JD: Colorectal cancer: molecules
and populations. J Natl Cancer Inst. 91:916–932. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jasperson KW, Tuohy TM, Neklason DW and
Burt RW: Hereditary and familial colon cancer. Gastroenterology.
138:2044–2058. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Burt RW: Colon cancer screening.
Gastroenterology. 119:837–853. 2000. View Article : Google Scholar
|
|
6
|
Lichtenstein P, Holm NV, Verkasalo PK,
Iliadou A, Kaprio J, Koskenvuo M, Pukkala E, Skytthe A and Hemminki
K: Environmental and heritable factors in the causation of cancer -
analyses of cohorts of twins from Sweden, Denmark, and Finland. N
Engl J Med. 343:78–85. 2000. View Article : Google Scholar
|
|
7
|
Bartel DP: MicroRNAs: genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Esquela-Kerscher A and Slack FJ: Oncomirs
- microRNAs with a role in cancer. Nat Rev Cancer. 6:259–269. 2006.
View Article : Google Scholar
|
|
9
|
Vasudevan S, Tong Y and Steitz JA:
Switching from repression to activation: microRNAs can up-regulate
translation. Science. 318:1931–1934. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Loktionov A: Common gene polymorphisms,
cancer progression and prognosis. Cancer Lett. 208:1–33. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Duan R, Pak C and Jin P: Single nucleotide
polymorphism associated with mature miR-125a alters the processing
of pri-miRNA. Hum Mol Genet. 16:1124–1131. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zeng Y and Cullen BR: Sequence
requirements for micro RNA processing and function in human cells.
RNA. 9:112–123. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hoffman AE, Zheng T, Yi C, Leaderer D,
Weidhaas J, Slack F, Zhang Y, Paranjape T and Zhu Y: microRNA
miR-196a-2 and breast cancer: a genetic and epigenetic association
study and functional analysis. Cancer Res. 69:5970–5977. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hu Z, Chen J, Tian T, Zhou X, Gu H, Xu L,
Zeng Y, Miao R, Jin G, Ma H, et al: Genetic variants of miRNA
sequences and non-small cell lung cancer survival. J Clin Invest.
118:2600–2608. 2008.PubMed/NCBI
|
|
15
|
Zhan JF, Chen LH, Chen ZX, Yuan YW, Xie
GZ, Sun AM and Liu Y: A functional variant in microRNA-196a2 is
associated with susceptibility of colorectal cancer in a Chinese
population. Arch Med Res. 42:144–148. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Schimanski CC: High miR-196a levels
promote the oncogenic phenotype of colorectal cancer cells. World J
Gastroenterol. 15:2089–2096. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chen H, Sun LY, Chen LL, Zheng HQ and
Zhang QF: A variant in microRNA-196a2 is not associated with
susceptibility to and progression of colorectal cancer in Chinese.
Intern Med J. 42:e115–e119. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hezova R, Kovarikova A, Bienertova-Vasku
J, Sachlova M, Redova M, Vasku A, Svoboda M, Radova L, Kiss I,
Vyzula R and Slaby O: Evaluation of SNPs in miR-196-a2, miR-27a and
miR-146a as risk factors of colorectal cancer. World J
Gastroenterol. 18:2827–2831. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Min KT, Kim JW, Jeon YJ, Jang MJ, Chong
SY, Oh D and Kim NK: Association of the miR-146aC>G, 149C>T,
196a2C>T, and 499A>G polymorphisms with colorectal cancer in
the Korean population. Mol Carcinog. 51(Suppl 1): E65–E73.
2012.
|
|
20
|
Vinci S, Gelmini S, Mancini I, Malentacchi
F, Pazzagli M, Beltrami C, Pinzani P and Orlando C: Genetic and
epigenetic factors in regulation of microRNA in colorectal cancers.
Methods. 59:138–146. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhu L, Chu H, Gu D, Ma L, Shi D, Zhong D,
Tong N, Zhang Z and Wang M: A functional polymorphism in
miRNA-196a2 is associated with colorectal cancer risk in a Chinese
population. DNA Cell Biol. 31:350–354. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lau J, Ioannidis JP and Schmid CH:
Quantitative synthesis in systematic reviews. Ann Intern Med.
127:820–826. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Mantel N and Haenszel W: Statistical
aspects of the analysis of data from retrospective studies of
disease. J Natl Cancer Inst. 22:719–748. 1959.PubMed/NCBI
|
|
24
|
DerSimonian R and Laird N: Meta-analysis
in clinical trials. Control Clin Trials. 7:177–188. 1986.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Begg CB and Mazumdar M: Operating
characteristics of a rank correlation test for publication bias.
Biometrics. 50:1088–1101. 1994. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Egger M, Davey Smith G, Schneider M and
Minder C: Bias in meta-analysis detected by a simple, graphical
test. BMJ. 315:629–634. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang P, Xie S, Cui A, Zhang Y and Jiang B:
miR-196a2 polymorphisms and susceptibility to cancer: A
meta-analysis involving 24,697 subjects. Exp Ther Med. 3:324–330.
2012.PubMed/NCBI
|
|
28
|
Wang F, Ma YL, Zhang P, Yang JJ, Chen HQ,
Liu ZH, Peng JY, Zhou YK and Qin HL: A genetic variant in
microRNA-196a2 is associated with increased cancer risk: a
meta-analysis. Mol Biol Rep. 39:269–275. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chu H, Wang M, Shi D, Ma L and Zhang Z,
Tong N, Huo X, Wang W, Luo D, Gao Y and Zhang Z: Hsa-miR-196a2
Rs11614913 polymorphism contributes to cancer susceptibility:
evidence from 15 case-control studies. PLoS One. 6:e181082011.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Xu W, Xu J, Liu S, Chen B, Wang X, Li Y,
Qian Y, Zhao W and Wu J: Effects of common polymorphisms rs11614913
in miR-196a2 and rs2910164 in miR-146a on cancer susceptibility: a
meta-analysis. PLoS One. 6:e204712011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Qiu LX, Wang Y, Xia ZG, Xi B, Mao C, Wang
JL, Wang BY, Lv FF, Wu XH and Hu LQ: miR-196a2 C allele is a
low-penetrant risk factor for cancer development. Cytokine.
56:589–592. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang J, Wang Q, Liu H, Shao N, Tan B,
Zhang G, Wang K, Jia Y, Ma W, Wang N and Cheng Y: The association
of miR-146a rs2910164 and miR-196a2 rs11614913 polymorphisms with
cancer risk: a meta-analysis of 32 studies. Mutagenesis.
27:779–788. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Tian T, Xu Y, Dai J, Wu J, Shen H and Hu
Z: Functional polymorphisms in two pre-microRNAs and cancer risk: a
meta-analysis. Int J Mol Epidemiol Genet. 1:358–366.
2010.PubMed/NCBI
|
|
34
|
Gao LB, Bai P, Pan XM, Jia J, Li LJ, Liang
WB, Tang M, Zhang LS, Wei YG and Zhang L: The association between
two polymorphisms in pre-miRNAs and breast cancer risk: a
meta-analysis. Breast Cancer Res Treat. 125:571–574. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wang Z, Cao Y, Jiang C, Yang G, Wu J and
Ding Y: Lack of association of two common polymorphisms rs2910164
and rs11614913 with susceptibility to hepatocellular carcinoma: a
meta-analysis. PLoS One. 7:e400392012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Guo J, Jin M, Zhang M and Chen K: A
genetic variant in miR-196a2 increased digestive system cancer
risks: a meta-analysis of 15 case-control studies. PLoS One.
7:e305852012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zambirinis CP, Theodoropoulos G and
Gazouli M: Undefined familial colorectal cancer. World J
Gastrointest Oncol. 1:12–20. 2009. View Article : Google Scholar
|
|
38
|
Burt R: Inheritance of colorectal cancer.
Drug Discov Today Dis Mech. 4:293–300. 2007. View Article : Google Scholar
|
|
39
|
Patel SG and Ahnen DJ: Familial colon
cancer syndromes: an update of a rapidly evolving field. Curr
Gastroenterol Rep. 14:428–438. 2012. View Article : Google Scholar : PubMed/NCBI
|