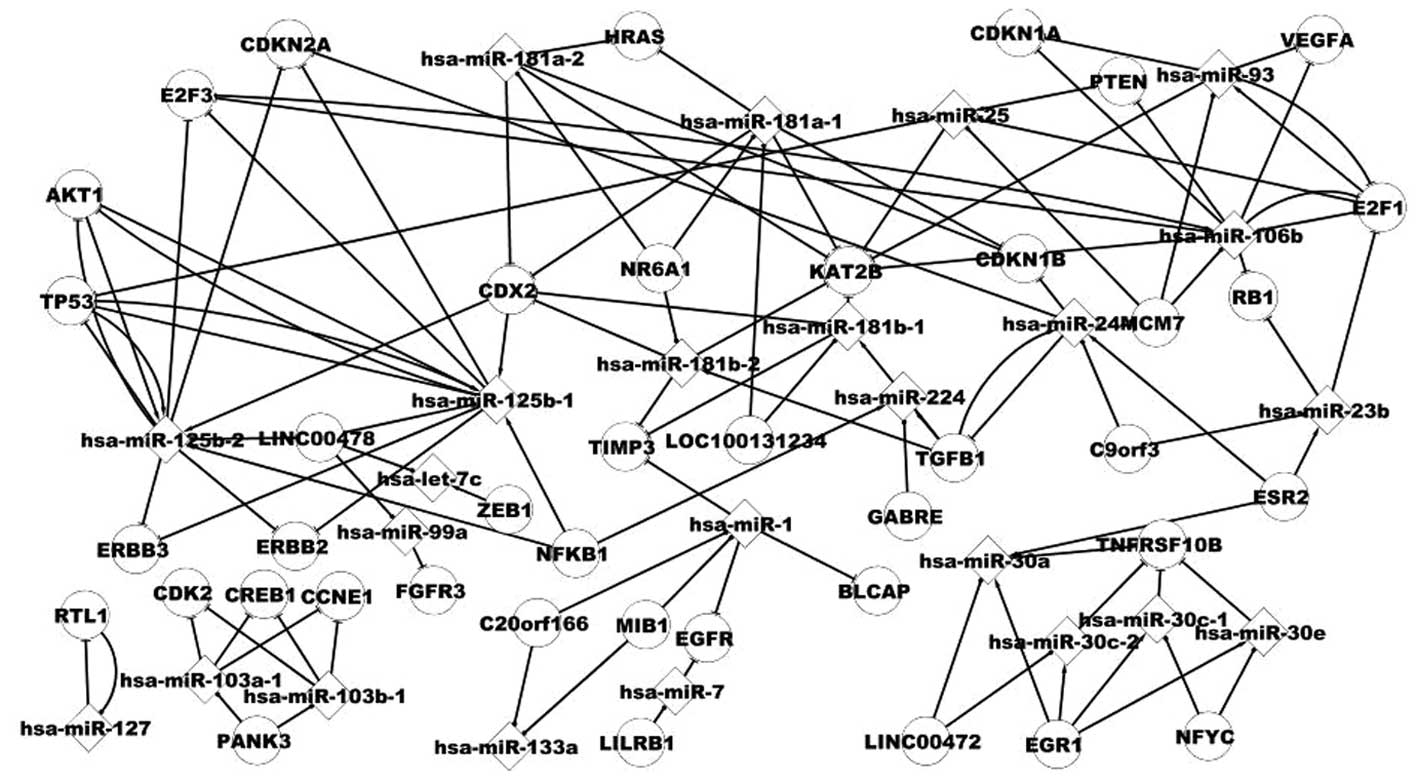
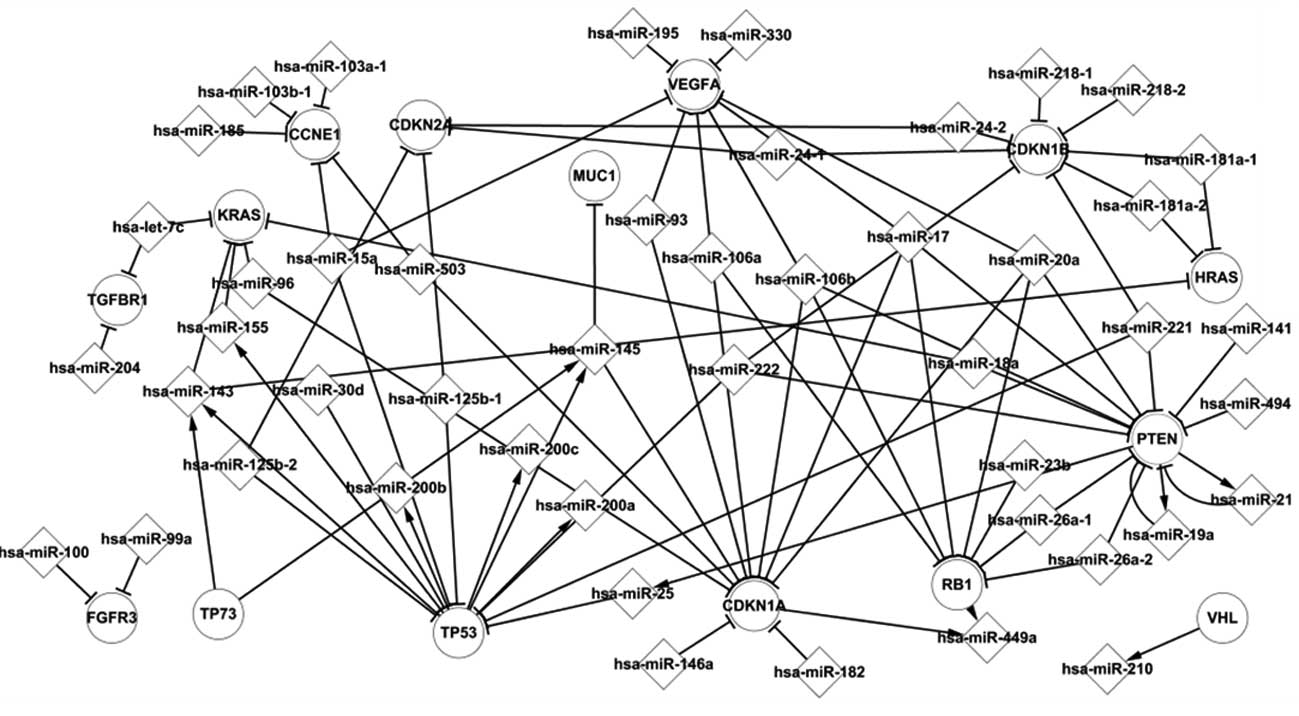
|
1
|
Jemal A, Murray T, Ward E, et al: Cancer
statistics, 2005. CA Cancer J Clin. 55:10–30. 2005. View Article : Google Scholar
|
|
2
|
Pashos CL, Botteman MF, et al: Bladder
cancer: epidemiology, diagnosis, and management. Cancer Pract.
10:311–322. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wang DS, Rieger-Christ K, Latini JM, et
al: Molecular analysis of PTEN and MXI1 in primary bladder
carcinoma. Int J Cancer. 88:620–625. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hobert O: Gene regulation by transcription
factors and microRNAs. Science. 319:1785–1786. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Calin GA, Dumitru CD, Shimizu M, et al:
Frequent deletions and down-regulation of micro-RNA genes miR15 and
miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci
USA. 99:15524–15529. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Rodriguez A, Griffiths-Jones S, Ashurst JL
and Bradley A: Identification of mammalian microRNA host genes and
transcription units. Genome Res. 14:1902–1910. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Baskerville S and Bartel DP: Microarray
profiling of microRNAs reveals frequent coexpression with
neighboring miRNAs and host genes. RNA. 11:241–247. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cao GJ, Huang BC, Liu ZH, et al: Intronic
miR-301 feedback regulates its host gene, ska2, in A549 cells by
targeting MEOX2 to affect ERK/CREB pathways. Biochem Biophys Res
Commun. 396:978–982. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sethupathy P, Corda B and Hatzigeorgiou
AG: TarBase: a comprehensive database of experimentally supported
animal microRNA targets. RNA. 12:192–197. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hsu SD, Lin FM, Wu WY, et al: miRTarBase:
a database curates experimentally validated microRNA-target
interactions. Nucleic Acids Res. 39:D163–D169. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang J, Lu M, Qiu CX and Cui QH: TransmiR:
a transcription factor-microRNA regulation database. Nucleic Acids
Res. 38:D119–D122. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Griffiths-Jones S, Saini HK, van Dongen S
and Enright AJ: miRBase: tools for microRNA genomics. Nucleic Acids
Res. 36:D154–D158. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kanehisa M, Goto S, Furumichi M, Tanabe M
and Hirakawa M: KEGG for representation and analysis of molecular
networks involving diseases and drugs. Nucleic Acids Res.
38:D355–D360. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chekmenev DS, Haid C and Kel AE: P-Match:
transcription factor binding site search by combining patterns and
weight matrices. Nucleic Acids Res. 33:W432–W437. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Dreszer TR, Karolchik D, Zweig AS, et al:
The UCSC Genome Browser database: extensions and updates 2011.
Nucleic Acids Res. 40:D918–D923. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kao A, Poteet SR, Jones DH, et al:
P-MATCH: Identifying Part Name in Noisy Text Data.
Cross-Disciplinary Advances in Applied Natural Language Processing:
Issues and Approaches. Boonthum-Denecke C, McCarthy PM and Lamkin
T: IGI Global; Hershey: pp. 172–184. 2011
|
|
17
|
Jiang QH, Wang YD, Hao YY, et al:
miR2Disease: a manually curated database for microRNA deregulation
in human disease. Nucleic Acids Res. 37:D98–D104. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Swallows DM, Gendler S, Gribths B, et al:
The hypervariable gene locus PUM, which encodes for the tumour
associated epithelial mucins, is located on chromosome 1, within
the region 1q21–24. Ann Hum Genet. 51:289–294. 1987.PubMed/NCBI
|
|
19
|
Zotter S, Hageman PC, Lossnitzer A, Mooi
WJ and Hilgers J: Tissue and tumour distribution of human
polymorphicepithelial mucin. Cancer Rev. 11:55–79. 1988.
|
|
20
|
Girling A, Bartkova J, Burchell J, et al:
A core protein epitope of the polymorphic epithelial mucin detected
by the monoclonal antibody SM-3 is selectively exposed in a range
of primary carcinomas. Int J Cancer. 43:1072–1076. 1989. View Article : Google Scholar
|
|
21
|
Simms MS, Hughes ODM, Limb M, Price MR and
Bishop MC: MUC1 mucin as a tumour marker in bladder cancer. BJU
Int. 84:350–352. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kompier LC, Lurkin I, van der Aa MN, van
Rhijn BW, van der Kwast TH and Zwarthoff EC: FGFR3, HRAS, KRAS,
NRAS and PIK3CA mutations in bladder cancer and their potential as
biomarkers for surveillance and therapy. PloS One. 5:e138212010.
View Article : Google Scholar : PubMed/NCBI
|