Introduction
Urolithiasis is a global health problem and is the
third most common urologic disease. Approximately 5% of females and
12% of males are likely to develop nephrolithiasis during their
lifetime (1). It is also notorious
for its high rate of recurrence, which is >40% in five years
(2). Various causes contribute to
the formation of urinary calculi, including genetic factors
(3,4). Efforts have been made to investigate
the causes of the disease; however, the detailed pathogenic
mechanism for the occurrence and recurrence of urolithiasis remains
intangible.
Urolithiasis formation is thought to be involved in
the progression of nucleation, growth, aggregation and retention of
mineral crystals. A number of proteins and molecules, such as
osteocalcin, osteopontin, fetuin-A, heparan sulfate and urokinase,
are likely to inhibit calcium oxalate supersaturation and
crystalization in various stages of stone formation. Urokinase was
first discovered in urine in 1947 and is a multifunctional protein
synthesized by a variety of human organs and cells, besides kidney
cells (5,6). The activity and concentration of
urokinase was verified to be lower in stone patients compared with
normal subjects (7). Urokinase,
originally known as a plasminogen activator, degrades the organic
matrix of urinary stones to prevent their complete formation and
growth (8). Thus, it is important
in averting the development of urinary stones.
Single nucleotide polymorphisms (SNPs) are
considered responsible for interindividual diversity in mediating
genetic predisposition to the complex disease (9), including cancer (10,11)
and urinary stones (12). The
urokinase gene is located on chromosome 10q24 (13). Three polymorphic sites have been
commonly reported; a T/C substitution in exon 6, a C/T change in
intron 7 (14) and a T/C
polymorphism at the +4065 nucleotide in the 3′-untranslated region
(3′-UTR). Of the three sites, the 3′-UTR T/C polymorphic site is
the most widely studied, and its relationship to complex diseases,
such as non-small cell lung, prostate and bladder cancer, has been
examined (15–17). Tsai et al(18) reported that the ‘T’ allele in 3′-UTR
increased the risk of calcium stone disease. Ozturk et
al(19) demonstrated that
3′-UTR T/C polymorphism played a role in childhood recurrence
urolithiasis. By contrast, two studies did not find any association
between 3′-UTR T/C polymorphism and nephrolithiasis (20,21).
It is assumed that the inconsistent findings may be secondary to
ethnic background or small sample size. In this study, a search was
conducted for all studies regarding the association of 3′-UTR T/C
polymorphism and urolithiasis. Results were pooled together to
obtain reliable conclusions.
Materials and methods
Identification and eligibility of
relevant studies
A search of the electronic literature was performed
via PubMed, Medline, Web of Science and the China National
Knowledge Infrastructure platform without any limit on language,
until October 2012. Search terms used included
‘urokinase/uPA/urokinase-type plasminogen activator/rs4065’ and
‘urolithiasis/nephrolithiasis/stone’.
Studies were included in the meta-analysis if they
i) focused on the relationship of urokinase gene 3′-UTR T/C
polymorphism and the risk of urolithiasis; ii) were case-control
designed; iii) provided concrete data for various genotypes for
calculating odds ratio (OR) and its corresponding 95% confidence
interval (CI). Studies not meeting the inclusion criteria, as well
as abstracts without sufficient information were excluded.
Data management
Searches and data extraction for the study were
carried out independently by two authors (D.W. Li and J.K. Liu).
The extraction information included: name of the first author, year
of publication, ratio of male/female, mean age of the subjects,
ethnicity of the population studied, number of cases and controls
with the various genotypes of urokinase gene rs4065 polymorphism,
and P-value for Hardy-Weinberg equilibrium (HWE) of the genotypes
in the patient and control groups.
Statistical analysis
The HWE was determined using the Chi-square test for
the urolithiasis and control groups, and was considered
statistically significant when P<0.05. In this meta-analysis,
RevMan 5.0 software, developed by the Cochrane Collaboration, was
used to analyze the data. Publication bias was tested by inverted
funnel plots, and subsequently assessed by the Egger’s or Begg’s
test with Stata software version 12.1. Heterogeneity between these
studies was tested using the Chi-square-based Q statistics test and
was considered statistically significant when P<0.10.
Random-effects models were employed when
Pheterogenity<0.10, while the fixed-effects models
was used when Pheterogenity>0.10 (22). The allele, recessive, codominant and
dominant models of the associations between urokinase gene 3′-UTR
T/C polymorphism and urolithiasis susceptibility were
simultaneously calculated by crude ORs and 95% CIs. Sensitivity
analysis was carried out by deleting the study with the most
subjects if the meta-analysis included three or four studies. In
this study, P-value was two-tailed and was considered signficant at
0.05.
Results
Characteristics of eligible studies
The initial study search by electronic engines
supplemented with a manual search yielded 54 potential articles, of
which 49 were excluded after scanning the titles and abstracts. Of
the remaining five, two were published by the same author group and
had overlapping data, thus the article (19) with detailed information was included
following consensus. Four case-control publications (18–21)
concerning the relationship of urokinase gene 3′-UTR T/C
polymorphism and urolithiasis susceptibility were identified
(Fig. 1). In total, 1,195 subjects,
including 462 healthy controls and 733 urolithiasis patients, were
included in the meta-analysis. Of the four case-control studies,
one publication (19) involved
Caucasian populations, while the remaining three (18,20,21)
pertained to Asian populations. One (19) of the four studies meeting the
inclusion criteria focused on child patients. Control groups in two
eligible studies (19,21) deviated from the HWE (P<0.05).
Additionally, three studies (18–20)
were related to urokinase gene 3′-UTR T/C polymorphism and
urolithiasis recurrence. The detailed information of the studies
included are shown in Tables I and
II, respectively.
 | Table IDetailed characteristics of eligible
studies. |
Table I
Detailed characteristics of eligible
studies.
| Variables | Tsai et
al(18) | Mittal et
al(16) | Ozturk et
al(19) | Kim et
al(21) |
|---|
| Sample sizes, n | | | | |
| Patients | 153 | 130 | 80 | 370 |
| Controls | 105 | 150 | 40 | 167 |
| Age, mean (SD)
years | | | | |
| Controls | 54.7 | 40 (11.5) | 10.5 (0.67) | - |
| Patients | 44.2 (12.0) | 40.5 (10.5) | 10.5 (4.33) | - |
| | | 11.2 (3.8) | |
| Gender,
male/female | | | | |
| Patients | 118/35 | 106/24 | 55/25 | - |
| Controls | 65/40 | 120/30 | 25/15 | - |
| P-value for HWE | | | | |
| Patients | 0.44 | <0.05 | <0.05 | <0.05 |
| Controls | 0.84 | 0.75 | <0.05 | <0.05 |
| Ethnicity | Chinese | Indian | Turkish | Korean |
| Genotypes,
(patients/controls), n | | | | |
| TT | −/− | 32/48 | 14/12 | 158/63 |
| CT | 18/4 | 82/72 | 4/− | 212/104 |
| CC | 135/101 | 16/30 | 62/28 | −/− |
| Alleles,
(patients/controls), n | | | | |
| T | 18/4 | 146/168 | 32/24 | 528/230 |
| C | 288/206 | 114/132 | 128/56 | 212/104 |
 | Table IIDetailed results of
meta-analysis. |
Table II
Detailed results of
meta-analysis.
| Category | Patients, n |
Pheterogenity | I2
(%) | Statistical
method | OR | 95% CI | P-value |
|---|
| T vs. C | | | | | | | |
| Total | 733 | 0.05 | 62 | R | 1.05 | 0.72–1.53 | 0.82 |
| Asian | 653 | 0.14 | 49 | F | 1.13 | 0.92–1.40 | 0.24 |
| Male | 224 | 0.03 | 78 | R | 2.14 | 0.25–18.51 | 0.49 |
| Female | 59 | 0.82 | 0 | F | 1.54 | 0.77–3.08 | 0.22 |
| HWE | 283 | 0.97 | 0 | F | 3.29 | 2.45–4.41 | 0.000 |
| Recurrence | 323 | 0.12 | 54 | F | 1.09 | 0.82–1.45 | 0.54 |
| TT+TC vs. CC | | | | | | | |
| Total | 363 | 0.06 | 64 | R | 1.53 | 0.66–3.51 | 0.32 |
| Asian | 283 | 0.33 | 0 | F | 2.51 | 1.23–3.77 | 0.007 |
| Male | 224 | 0.14 | 55 | F | 2.46 | 1.21–4.99 | 0.01 |
| Female | 59 | 0.73 | 0 | F | 1.67 | 0.62–4.50 | 0.31 |
| HWE | 283 | 0.33 | 0 | F | 2.51 | 1.23–3.77 | 0.007 |
| Recurrence | 323 | 0.34 | 8 | F | 1.83 | 1.14–2.95 | 0.01 |
| TT vs. TC+CC | | | | | | | |
| Total | 580 | 0.07 | 62 | R | 0.83 | 0.49–1.40 | 0.48 |
| Asian | 500 | 0.08 | 67 | R | 0.95 | 0.55–1.67 | 0.87 |
| Recurrence | 170 | 0.97 | 0 | F | 0.69 | 0.43–1.10 | 0.12 |
| TC vs. CC | | | | | | | |
| Total | 317 | 0.74 | 0 | F | 2.53 | 1.43–4.46 | 0.001 |
| Asian | 251 | 0.49 | 0 | F | 2.46 | 1.38–4.40 | 0.002 |
| Male | 201 | 0.21 | 37 | F | 2.98 | 1.43–6.21 | 0.004 |
| Female | 50 | 0.83 | 0 | F | 1.52 | 0.53–4.34 | 0.44 |
| HWE | 251 | 0.49 | 0 | F | 2.46 | 1.38–4.40 | 0.002 |
| Recurrence | 282 | 0.53 | 0 | F | 2.66 | 1.51–4.67 | 0.000 |
| TT vs. CC | | | | | | | |
| Total | 124 | 0.15 | 53 | F | 0.88 | 0.50–1.56 | 0.66 |
| Recurrence | 84 | 0.46 | 0 | F | 1.06 | 0.58–1.93 | 0.86 |
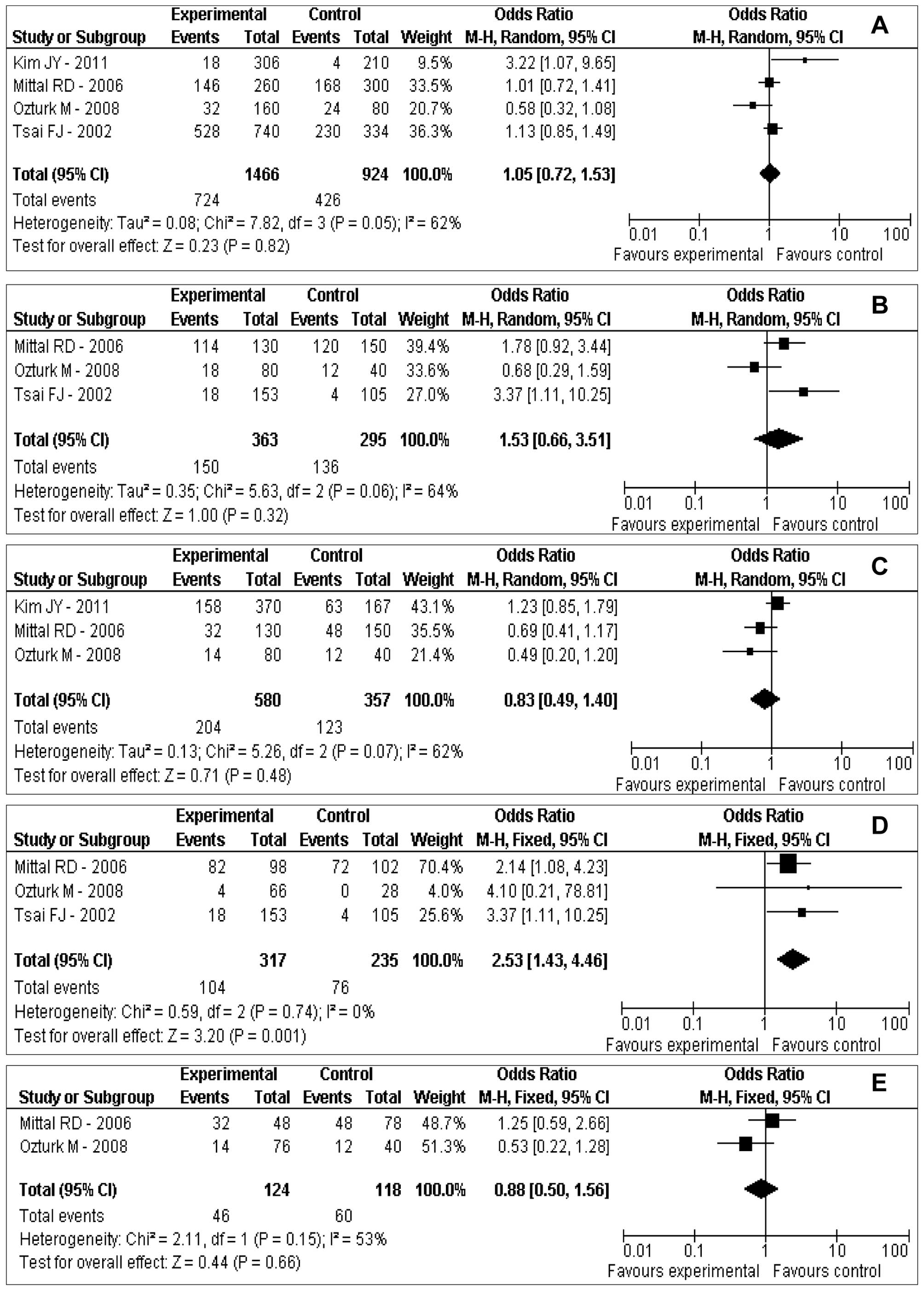
Main meta-analysis results
Table II and
Fig. 2 show the results of the
association between urokinase gene 3′-UTR T/C polymorphism and risk
of urolithiasis in the total population. A significant association
was noted in the codominant model (TC vs. CC: OR=2.53; 95% CI,
1.43–4.46; P=0.001). However, no significant associations between
urokinase gene 3′-UTR T/C polymorphism and risk of urolithiasis
were observed in the allele model (T vs. C: OR=1.05; 95% CI,
0.72–1.53; P=0.82), the dominant model (TT+TC vs. CC: OR=1.53; 95%
CI, 0.66–3.51; P= 0.32), the recessive model (TT vs. TC+CC: OR=
0.83; 95% CI, 0.49–1.40; P= 0.48) and the codominant model (TT vs.
CC: OR=0.88; 95% CI, 0.50–1.56; P=0.66) (Table II and Fig. 2).
As for the subgroup meta-analysis, we stratified the
analysis in terms of gender (male/female), ethnicity (Asian), HWE
and urolithiasis status (recurrence) (Table II). The association of urokinase
gene 3′-UTR T/C polymorphism and urolithiasis risk was evident in
some genetic models, such as: allele model (T vs. C: HWE OR=3.29;
95% CI, 2.45–4.41; P=0.000), the dominant model (TT+TC vs. CC:
Asian OR=2.51; 95% CI, 1.23–3.77; P=0.007; male OR=2.46; 95% CI,
1.21–4.99; P=0.01; HWE OR=2.51; 95% CI, 1.23–3.77; P= 0.007;
recurrence OR=1.83; 95% CI, 1.14–2.95; P= 0.01) and the codominant
model (TC vs. CC: Asian OR=2.46; 95% CI, 1.38–4.40; P=0.002; male
OR=2.98; 95% CI, 1.43–6.21; P=0.004; HWE OR=2.46; 95% CI,
1.38–4.40; P=0.002; recurrence OR=2.66; 95% CI, 1.51–4.67; P=0.000)
(Table II).
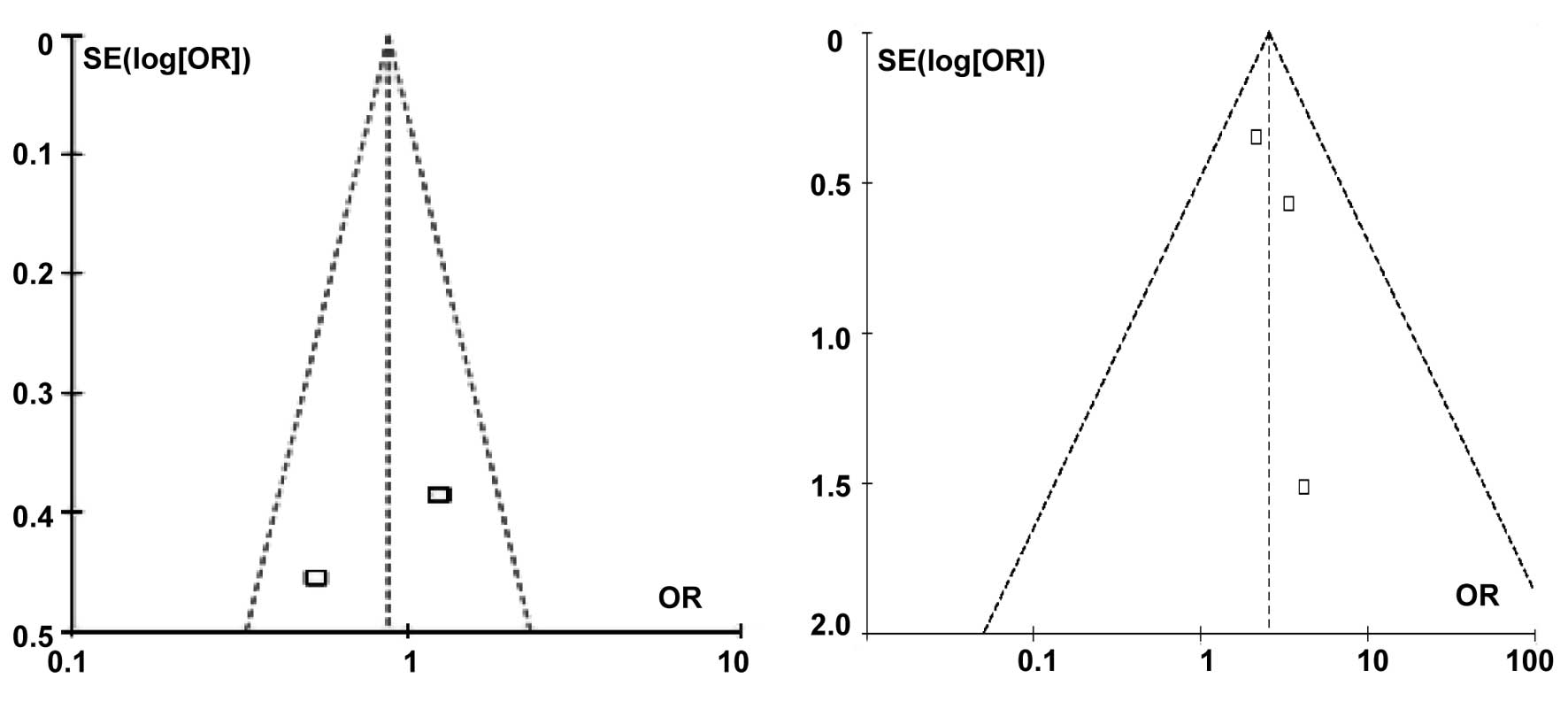
The publication bias was assessed by the inverted
funnel plot. Funnel plots of the codominant models (TC vs. CC and
TT vs. CC) for the total population are shown in Fig. 3. No existing publication bias was
evident and neither the Egger’s nor Begg’s tests identified
publication bias. A significant difference was observed in the
subgroup analysis of recurrence urolithiasis for the dominant model
when the study by Mittal et al(20), with the largest number of subjects,
was deleted.
Discussion
Urolithiasis is a global problem and affects almost
all populations with a prevalence rate of 4–20% (23) and a recurrence rate of >40% in
five years (2). Due to such
distinguishing features, extensive efforts have mainly focused on
three aspects: i) screening useful markers to prevent the
occurrence of urolithiasis; ii) perfecting the means of clearing
stones; and iii) finding effective methods, to avoid the recurrence
of nephrolithiasis.
Stone formation in the kidney is a multifactor and
complex process. According to the matrix theory, proteins, such as
uromucoid, promote precipitation of calcium crystals to initiate
the crystallization process. Therefore, factors that affect this
procedure may be important in precluding stone development.
Urokinase, originally isolated from human urine, is a plasminogen
activator synthesized by the kidney, which can cleave plasminogen
to plasmin and then stimulate fibrinolysis. Urokinase then becomes
a natural inhibitor of abnormal initiation and growth of stone. The
quantity and activity of urokinase were evidently downregulated in
in vivo(8) and in
vitro(24) studies. These
findings suggest that a higher urinary excretion of urokinase may
play a protective role in preventing calcium urolithiasis
formation.
Single nucleotide polymorphisms (SNP) have been
identified as a powerful tool for predicting complex diseases,
including cancers (10,11) and urolithiasis (18–21,25).
Determination of SNPs in these genes is useful in the
identification of available markers and in the clarification of the
role of urokinase in the development of stones. Three polymorphic
sites exist within the urokinase gene, of which 3′-UTR T/C
polymorphism is the most widely studied (18–21).
Several case-control studies (18–21)
have investigated the possibility that urokinase gene 3′-UTR T/C
polymorphism is associated with the formation of urolithiasis.
However, the results of these studies were inconsistent. To the
best of our knowledge, the present study is the first meta-analysis
to assess the association of 3′-UTR T/C polymorphism and
urolithiasis susceptibility by synthesizing individual studies.
In the final analysis, a significant association was
noted in the ‘T vs. C’ allele model (HWE OR=3.29; 95% CI,
2.45–4.41; P=0.000). Significant results were also observed in the
‘TC vs. CC’ codominant model (total population OR=2.53; 95% CI,
1.43–4.46; P=0.001; Asian OR=2.46, 95% CI, 1.38–4.40; P=0.002; male
OR=2.98; 95% CI, 1.43–6.21; P=0.004; HWE OR=2.46; 95% CI,
1.38–4.40; P=0.002; recurrence OR=2.66; 95% CI, 1.51–4.67; P=
0.000). A significant association of urokinase gene 3′-UTR T/C
polymorphism and urolithiasis risk was also evident in the ‘TT+TC
vs. CC’ dominant model (Asian OR=2.51; 95% CI, 1.23–3.77; P=0.007;
male OR=2.46; 95% CI, 1.21–4.99; P=0.01; HWE OR=2.51; 95% CI,
1.23–3.77; P= 0.007; recurrence OR=1.83; 95% CI, 1.14–2.95; P=
0.01). It is likely that the rare variant ‘T’ increases
susceptibility to urolithiasis, particularly in Asian populations.
A population with a ‘T’ allele has a 2.51-fold risk of stone
susceptibility compared to population groups without ‘T’ allele.
Males with ‘T’ allele have a 2.46-fold risk of stone susceptibility
compared to those without ‘T’ allele, while for females with or
without ‘T’ allele there is no difference with regard to the risk
of urolithiasis. Nevertheless, in the present study, ‘T’ allele was
closely associated with the recurrence of urinary stone.
The specific mechanism regarding whether T/C
polymorphism localized in 3′-UTR affects susceptibility to
urolithiasis remains to be determined. The 3′-UTR is a particular
section of messenger RNA (mRNA) located between the stop codon and
the polyA tail. It is well documented that sequences in the 3′-UTR
may effectively regulate gene expression. One feasible model is
that 3′-UTR contains binding sites for regulatory proteins.
Sequence alterations in the 3′-UTR may change the binding patterns
of regulatory proteins and then the stability of mRNA. One example
of this model involves the manner in which the translation of the
transferring receptor gene is regulated by its 3′-UTR (26). Another feasible model involves miRNA
(27,28) recognizing and combining the site
located in the 3′-UTR of the gene due to the minor allele. In such
a hypothesis, some miRNAs may bind to the sequence containing
pathogenic ‘T’ allele, leading to transcript degradation or
translation repression. However, how this polymorphism localizing
in 3′-UTR affects the expression and function of urokinase remains
to be elucidated.
In the present meta-analysis, we collected published
case-control studies to obtain a more reliable association between
urokinase gene 3′-UTR T/C polymorphism and urolithiasis. The
urokinase gene 3′-UTR ‘T’ is a potential genetic marker for
urolithiasis. Of note, however, is that the final conclusion was
based on a moderate sample size. Thus, these results should be
confirmed in further studies. Screening genetic markers with a
significant association with the risk of urolithiasis presents an
important challenge as genotyping of gene-gene and gene-enviroment
factor interaction remains to be clarified.
In conclusion, urokinase gene 3′-UTR T/C
polymorphism contributes to the susceptibility of nephrolithiasis,
particularly in Asian populations. Additionally, the minor allele
‘T’ correlates with the recurrence of urolithiasis. Well-designed
studies with a large sample size and different population
characteristics should be performed to confirm our findings,
thereby providing clinical and/or biological support for our
results.
Acknowledgements
This study was partly supported by the
grant Y2003C10 from the Science Foundation of Shandong (Hainan
Liu). Dawei Li received a scholarship (no. 2011622132) from the
China Scholarship Council.
References
|
1
|
Johnson CM, Wilson DM, O’Fallon WM, Malek
RS and Kurland LT: Renal stone epidemiology: a 25-year study in
Rochester, Minnesota. Kidney Int. 16:624–631. 1979.PubMed/NCBI
|
|
2
|
Caudarella R, Tonello L, Rizzoli E and
Vescini F: Predicting five-year recurrence rates of kidney stones:
an artificial neural network model. Arch Ital Urol Androl.
83:14–19. 2011.PubMed/NCBI
|
|
3
|
Coe FL, Parks JH and Asplin JR: The
pathogenesis and treatment of kidney stones. N Engl J Med.
327:1141–1152. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Goodman HO, Brommage R, Assimos DG and
Holmes RP: Genes in idiopathic calcium oxalate stone disease. World
J Urol. 15:186–194. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wagner SN, Atkinson MJ, Wagner C, Höfler
H, Schmitt M and Wilhelm O: Sites of urokinase-type plasminogen
activator expression and distribution of its receptor in the normal
human kidney. Histochem Cell Biol. 105:53–60. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kitching AR, Holdsworth SR, Ploplis VA, et
al: Plasminogen and plasminogen activators protect against renal
injury in crescentic glomerulonephritis. J Exp Med. 185:963–968.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
du Toit PJ, van Aswegen CH, Steyn PL, Pols
A and du Plessis DJ: Effects of bacteria involved with the
pathogenesis of infection-induced urolithiasis on the urokinase and
sialidase (neuraminidase) activity. Urol Res. 20:393–397. 1992.
|
|
8
|
du Toit PJ, Van Aswegen CH, Steinmann CM,
Klue L and Du Plessis DJ: Does urokinase play a role in renal stone
formation? Med Hypotheses. 49:57–59. 1997.PubMed/NCBI
|
|
9
|
Loder N: Genetic variations can point the
way to disease genes. Nature. 401:7341999. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li D, Liu H, Yan L, Tang Y, Ren J and Xu
Z: Lack of association between hOGG1 Ser326Cys polymorphism and the
risk of bladder cancer: a meta-analysis. Urol Int. 88:88–94. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li D, Tian T, Guo C, et al: No association
of the MTHFR gene A1298C polymorphism with the risk of prostate
cancer: a meta-analysis. Exp Ther Med. 3:493–498. 2012.PubMed/NCBI
|
|
12
|
Gögebakan B, Igci YZ, Arslan A, et al:
Association between the T-593A and C6982T polymorphisms of the
osteopontin gene and risk of developing nephrolithiasis. Arch Med
Res. 41:442–448. 2010.PubMed/NCBI
|
|
13
|
Tripputi P, Blasi F, Verde P, Cannizzaro
LA, Emanuel BS and Croce CM: Human urokinase gene is located on the
long arm of chromosome 10. Proc Natl Acad Sci USA. 82:4448–4452.
1985. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Conne B, Berczy M and Belin D: Detection
of polymorphisms in the human urokinase-type plasminogen activator
gene. Thromb Haemost. 77:434–435. 1997.PubMed/NCBI
|
|
15
|
Shih CM, Kuo WH, Lin CW, et al:
Association of polymorphisms in the genes of the urokinase
plasminogen activation system with susceptibility to and severity
of non-small cell lung cancer. Clin Chim Acta. 412:194–198. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mittal RD, Srivastava DS and Mishra DK: Is
urokinase gene 3′-UTR polymorphism associated with prostate cancer?
Cancer Biomark. 1:287–292. 2005.
|
|
17
|
Manchanda PK, Bid HK and Mittal RD:
Association of urokinase gene 3′-UTR T/C polymorphism with bladder
cancer. Urol Int. 77:81–84. 2006.
|
|
18
|
Tsai FJ, Lin CC, Lu HF, Chen HY and Chen
WC: Urokinase gene 3′-UTR T/C polymorphism is associated with
urolithiasis. Urology. 59:458–461. 2002.
|
|
19
|
Ozturk M, Kordan Y, Cangul H, et al:
Association of urokinase gene 3′-UTR T/C polymorphism with calcium
oxalate urolithiasis in children. Int Urol Nephrol. 40:563–568.
2008.
|
|
20
|
Mittal RD, Bid HK, Kumar A and Bhandari M:
Association of urokinase gene 3′-UTR polymorphism with calcium
oxalate nephrolithiasis. J Endourol. 20:157–160. 2006.
|
|
21
|
Kim JY, Kim YS, Chang IH, Kim TH and Kim
HR: Interleukin-1β, calcium-sensing receptor, and urokinase gene
polymorphisms in korean patients with urolithiasis. Korean J Urol.
52:340–344. 2011.
|
|
22
|
Higgins JP, Thompson SG, Deeks JJ and
Altman DG: Measuring inconsistency in meta-analyses. BMJ.
327:557–560. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lee YH, Huang WC, Tsai JY, et al:
Epidemiological studies on the prevalence of upper urinary calculi
in Taiwan. Urol Int. 68:172–177. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
van Aswegen CH, Dirksen van Sckalckwyk JC,
du Toit PJ, Verster L, Franz RC and du Plessis DJ: The effect of
calcium and magnesium ions on urinary urokinase and sialidase
activity. Urol Res. 20:41–44. 1992.PubMed/NCBI
|
|
25
|
Wang S, Wang X, Wu J, et al: Association
of vitamin D receptor gene polymorphism and calcium urolithiasis in
the Chinese Han population. Urol Res. 40:277–284. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Klausner RD, Rouault TA and Harford JB:
Regulating the fate of mRNA: the control of cellular iron
metabolism. Cell. 72:19–28. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chen K, Song F, Calin GA, Wei Q, Hao X and
Zhang W: Polymorphisms in microRNA targets: a gold mine for
molecular epidemiology. Carcinogenesis. 29:1306–1311. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ha M, Pang M, Agarwal V and Chen ZJ:
Interspecies regulation of microRNAs and their targets. Biochim
Biophys Acta. 1779:735–742. 2008. View Article : Google Scholar : PubMed/NCBI
|