Introduction
Several studies have investigated the source of
circulating microRNAs (miRNAs); however, it is an issue of debate.
Previously, it has been indicated that miRNAs are present in the
circulation as a result of the release of exosomes from cells
(1–3). A study by Esquela-Kerscher and Slack
(4) proposed that miRNAs enter the
circulation during angiogenesis following their release from tumor
cells or as a result of tumor cell death. In addition, another
study postulated that miRNAs may also be abundant in the
circulation as a result of their release from inflamed organs
(5). Changes in circulating miRNA
levels do not only result from changes within tumors, but may also
be a result of inflammatory reactions or host immune response.
Based on this fact, the expression of miRNAs in organs does not
always represent their quantity in serum. For example,
miR-195 was found to be downregulated in breast cancer
tissues (6), whereas its
circulating levels were found to be upregulated in breast cancer
patients compared to controls (7).
In support of this evidence, Waters et al (8) showed that miR-195 and
miR-497 were decreased and miR-221 was increased in
tumor tissues from murine models of breast cancer compared to
healthy tissues, with no difference in the expression of all three
miRNAs in the circulation. Additionally, miR-122 was shown
to have a low expression in liver (9,10) and
high expression in the serum of hepatocellular carcinoma (HCC)
patients compared to the controls (11). miR-181a represents another
example for this discrepancy, as upregulation of miR-181a
was reported in embryonic and epithelial cell adhesion
molecule/α-fetoprotein (EpCAM+/AFP+) liver
tissues compared to the healthy controls (12), whereas at the serum level no
difference in miR-181a expression was observed (13).
miR-181a is an immunoregulatory miRNA
(14) that is reported to have its
highest relative expression in the thymus, the primary lymphoid
organ and site of T lymphocyte maturation (15), highlighting its role in the
maturation, sensitivity and selection of T lymphocytes.
miR-181a has shown aberrant expression in viral infections,
in which it was reported to be downregulated in human
papillomavirus-positive cell lines and upregulated in HepG2 cell
lines infected with hepatitis B virus (HBV) (16,17).
In hepatitis C virus (HCV), a single study was conducted by Liu
et al (18) in which a
microarray was performed, which showed that infection of human
hepatoma cell lines (Huh 7.5.1) with JFH-1 HCV leads to
miR-181a downregulation.
From the aforementioned data, there is clear
evidence that miR-181a shows aberrant expression in liver
tissues that was not mirrored in the serum of HCC patients
(8,12,13).
Since infection with HCV is a major cause of HCC, the present study
aimed to reveal the expression profile of miR-181a in HCV
infection and correlate it to the expression reported in HCC.
Therefore, miR-181a was screened in serum, liver tissues and
peripheral mononuclear cells (PBMCs) of patients infected with
genotype 4 (GT4)-HCV, the most prevalent HCV genotype in Egypt,
which to the best of our knowledge has never been previously
investigated. The study also aimed to associate the serum
miR-181a expression with response to the standard therapy
used in Egypt, pegylated-interferon/ribavirin (PEG-IFN/RBV)
therapy.
Patients and methods
Study subjects
A total of 72 patients chronically infected with HCV
and 22 age-matched controls were included in the study. The
patients were classified as 24 naïve patients, 11 sustained
virological responders (SVRs) pre-treatment, 15 SVRs
post-treatment, 12 non-responders (NRs) pre-treatment and 10 NRs
post-treatment. All the naïve patients were candidates for
PEG-IFN/RBV therapy. The presence of HCV RNA and anti-HCV
antibodies in the serum was used to diagnose HCV infection. The
patients were determined as negative for the hepatitis B surface
antigen. The samples from post-treatment SVRs and NRs were obtained
following treatment with weekly injections of PEG-IFN-α and daily
oral doses of RBV at Al Kasr Al Ainy School of Medicine, Cairo
University (Cairo, Egypt). The controls were healthy volunteers
that were all negative for HCV, HBV and HIV infection. The patients
and healthy volunteers included in the study provided their written
informed consent. All the clinical procedures were performed in
compliance with the guidelines of the Institutional Review Board of
Al Kasr Al Ainy School of Medicine in Cairo University and in
accordance with the ethical standards of the Declaration of
Helsinki.
Collection of samples
Peripheral venous blood (8 ml) was collected from
each patient and control in the presence of an anticoagulant, EDTA,
for isolation of PBMCs. All the samples were processed on the same
day and within a few hours after collection. In addition, 2 ml
blood samples were collected from patients and healthy controls for
serum separation. The liver tissue samples were collected from
patients by fine-needle aspiration and healthy liver tissues were
obtained during liver transplantation. The samples were directly
cryopreserved following biopsy collection until required for
use.
Serum separation and isolation of
PBMCs
The serum samples were collected in serum separator
tubes, centrifuged at 1,500 × g for 10 min and immediately frozen
at −80°C until required for use.
PBMCs were isolated using the Ficoll (Axis-Shield
PoC AS, Kjelsåsveien, Oslo, Norway) density gradient centrifugation
method as previously described (19), and frozen at −80°C until required
for use. Cell counting and viability testing were performed using
trypan blue.
Genotyping
Genotyping was performed using the
Versant® HCV Genotype 2.0 assay (LiPA; Bayer HealthCare,
Tarrytown, NY, USA), at the National Cancer Institute, according to
the manufacturer’s instructions.
Cell culture
Huh7 cells were maintained in Dulbecco’s Modified
Eagle’s medium supplemented with L-glutamine,
penicillin/streptomycin and fetal bovine serum.
In vitro transcription and transfection
of GT4-HCV full length genome
The intergenotypic recombinant
pED435′UTR-NS2/JFH1T8271,T977S encompassing
the 5′ untranslated region (UTR) to NS2 region of the GT4a-HCV
genome (provided by Professor Jens Bukh) (20) was linerized using the Xba1
restriction enzyme (Thermo Scientific, Waltham, MA, USA) and
purified using phenol-chloroform. Confirmation of linearization was
performed using gel electrophoresis. Linearized plasmid was in
vitro transcribed using the MEGAscript T7 in vitro
transcription kit (Ambion, Life Technologies, Carlsbad, CA, USA).
The transcribed viral RNA was transfected into the Huh7 cells by
lipofection (Superfect; Qiagen, Hilden, Germany). The supernatant
containing HCV particles was collected and filtered through 0.45-μm
pore size syringe filters for further use.
Infection
Naïve Huh7 cells were inoculated overnight with the
filtered supernatant harvested from the HCV RNA-transfected
cells.
Total RNA extraction and reverse
transcription
Total cellular RNA was extracted from the Huh7
cells, liver biopsies, PBMCs and serum samples under sterile
conditions using Biozol (Bioer Technology Co., Ltd., Binjiang
Hanchuan, China) according to the manufacturer’s instructions, as
first demonstrated by Chomczynski and Sacchi (21). The TaqMan MicroRNA Reverse
Transcription kit (Applied Biosystems, Foster City, CA, USA) was
subsequently used to reverse transcribe the total extracted RNA
into single-stranded complementary DNA according to the
manufacturer’s instructions.
Quantification of miRNAs expression
The relative expression of miR-181a (TaqMan
miRNA; ID: 000480) was quantified using StepOne Real-Time PCR
(Applied Biosystems) using the TaqMan MicroRNA assay (Applied
Biosystems). The comparative cycle threshold (CT) method, which
involves comparing the CT values of the samples of interest to that
of the healthy controls, was used to calculate the amount of
miR-181a. In the Huh7 cell lines, liver biopsies and PBMCs,
the amount of miRNA was calculated relative to the amount of the
reference gene, RNU6B, in the same sample. Reactions,
including the controls, were run in duplicates.
Statistical analysis
miRNA expression is represented as relative
quantification (RQ). For the Huh7 cells, liver tissues and PBMCs
samples, RQ=2−ΔΔCT, whereas for the serum samples,
RQ=2−ΔCT, as described previously (22). Mann-Whitney U test was employed to
compare miRNA expression and the results are expressed as median.
Correlation studies were analyzed using Spearman test. P<0.05
was considered to indicate a statistically significant difference.
Analysis and calculations were performed using GraphPad Prism 5.00
software (GraphPad Software, San Diego, CA, USA).
Results
Expression pattern of miR-181a in
HCV-infected Huh7 cells
Huh7 cells infected with
pED435′UTR-NS2/JFH1T8271,T977S encompassing
the GT4a-HCV genome for 24 h showed lower miR-181a
expression when compared to uninfected cells (n=4) (P=0.0286)
(Fig. 1).
Expression pattern of miR-181a in liver
tissues of naïve HCV-infected patients and healthy control
subjects
All the samples were found to be infected with
GT4-HCV. The miR-181a expression in HCV-infected (n=10) and
healthy (n=10) liver tissues was examined using quantitative
reverse transcription-polymerase chain reaction. miR-181a
showed similar expression in liver tissues of patients and healthy
controls (P=0.1431) (Fig. 2).
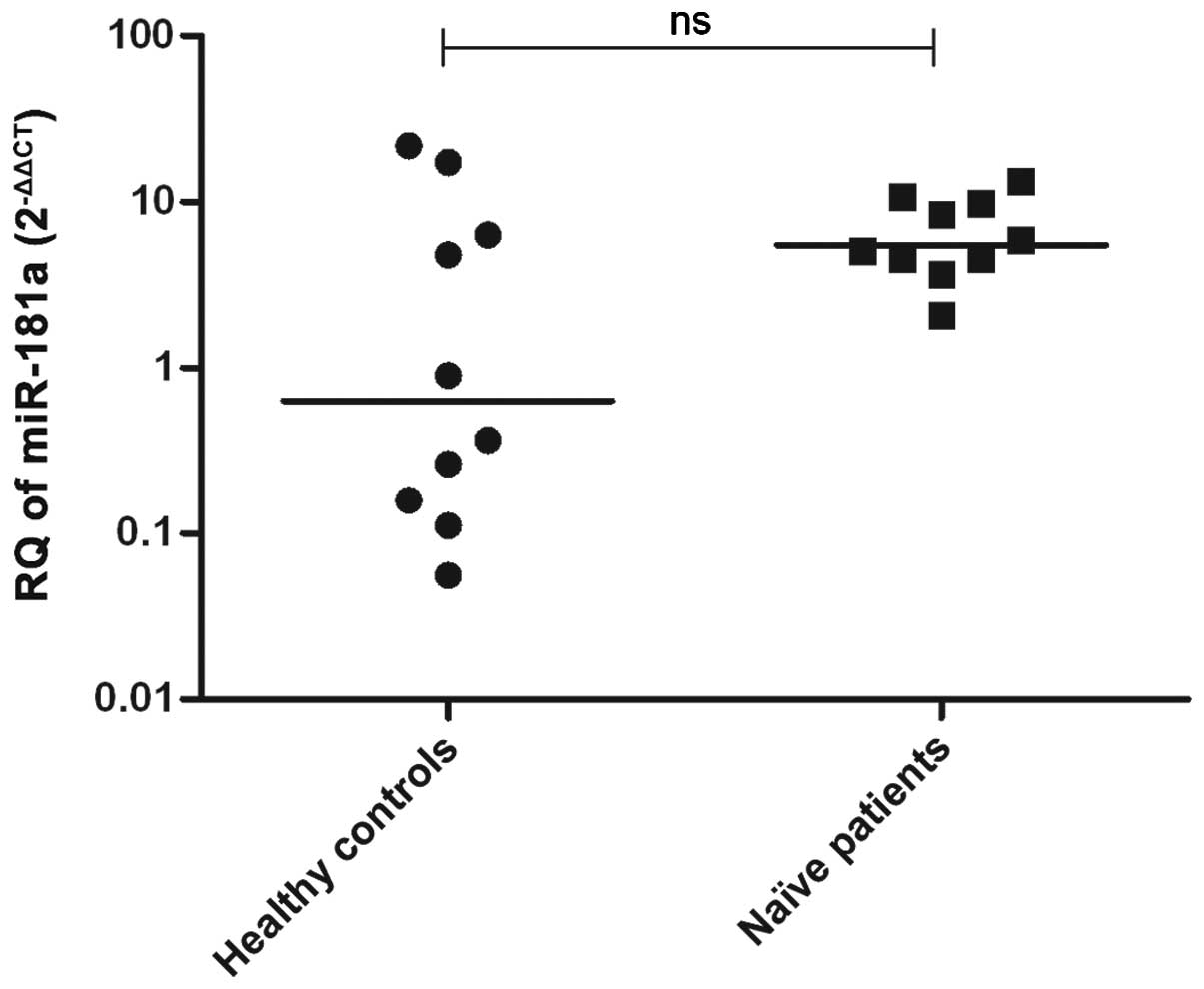
Expression pattern of miR-181a in PBMCs
of naïve HCV-infected patients and healthy control subjects
miR-181a expression was measured in the PBMCs
isolated from naïve GT4-HCV-infected patients (n=14) and healthy
controls (n=12). No significant difference was observed in
miR-181a expression in patients compared to the controls
(P=0.4714) (Fig. 3).
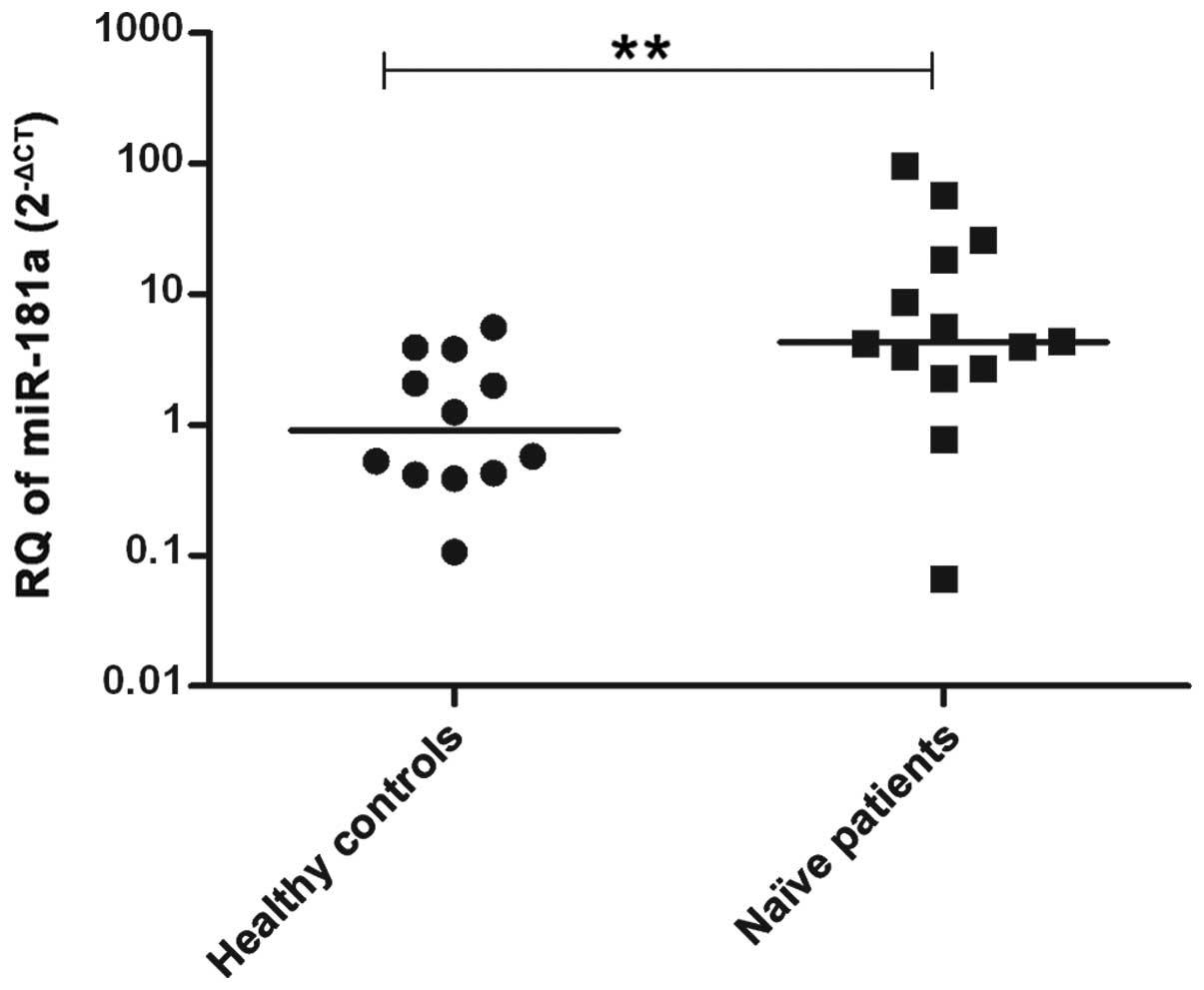
Expression pattern of miR-181a in serum
of naïve HCV-infected patients compared to healthy control
subjects
The miR-181a expression was measured in the
serum samples of naïve GT4-HCV-infected patients (n=14) and was
compared to the expression in the healthy controls (n=12).
miR-181a was found to be significantly upregulated in
HCV-infected patients compared to the controls (P=0.0069) (Fig. 4).
Correlation between miR-181a and clinical
parameters
The miR-181a expression measured in the serum
samples of naïve GT4-HCV-infected patients (n=14) was correlated
with viral load and liver transaminases [alanine aminotransferase
(ALT) and aspartate aminotransferase (AST)]. A significant negative
correlation was observed between miR-181a expression and
viral load (P=0.0304) (r=−0.5780), ALT (P=0.0162) (r=−0.6278), as
well as AST (P=0.0314) (r=−0.5752) (Fig. 5).
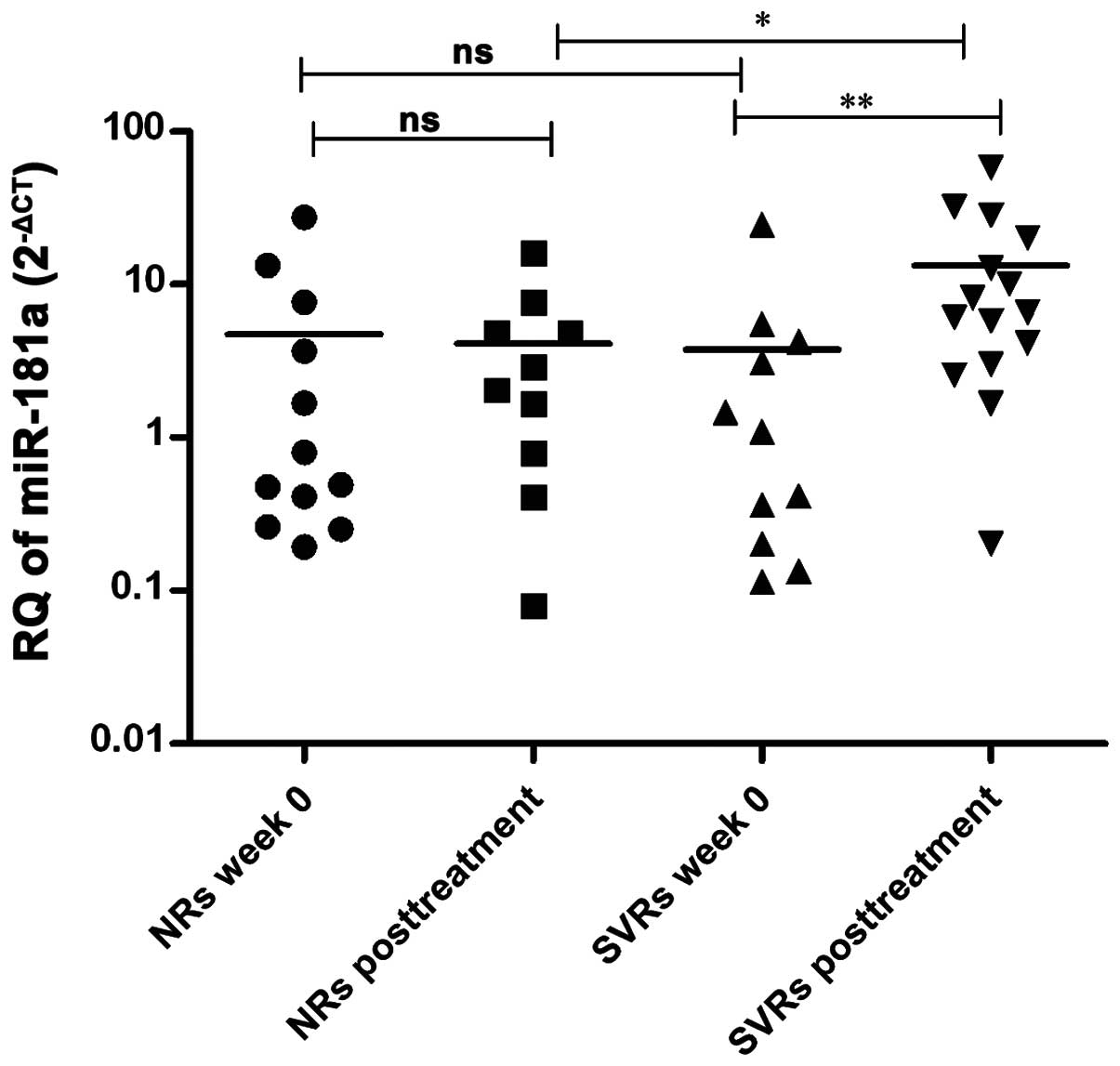
Variation in miR-181a expression in the
serum of HCV-infected patients of different response prior and
subsequent to IFN therapy
miR-181a expression was assessed in the serum
of SVR and NR HCV-infected patients prior (n=11 and 12,
respectively) and subsequent to therapy (n=15 and 10,
respectively).
No significant difference was observed in the
pre-treatment expression of miR-181a in the serum of NR
patients compared to post-treatment levels in NRs (P=0.5095) and
pre-treatment levels in SVRs (P=0.6891). However, SVR patients
showed higher post-treatment levels of miR-181a compared to
pre-treatment levels in SVRs (P=0.0075) and post-treatment levels
in NRs (P=0.0375) (Fig. 6).
Discussion
miR-181a was found to be highly expressed in
HCC embryonic livers and EpCAM+/AFP+ HCC
cells isolated from fetal livers compared to adult livers or
freshly isolated mature hepatocytes (12), however, its level was similar in the
serum of HCC patients and healthy controls (13). Thus far, it is not known whether
miR-181a expression in HCV infection, as a major cause of
HCC, shows the same discrepancy as reported in HCC patients.
Therefore, the present study was interested in investigating the
expression of miR-181a in GT4-HCV infection with the aim to
examine whether it depicts a correlation to its profile in HCC and
to the treatment response with PEG-IFN/RBV therapy, the standard
therapy used in Egypt.
In order to examine miR-181a expression in
GT4-HCV, Huh7 cells were infected with HCVcc (infectious cell
culture HCV model) derived from ED43/JFH1 (provided by Professor
Jens Buch). The expression of miR-181a was decreased in the
infected Huh7 cells (Fig. 1). This
finding exhibits a similarity with a study performed by Liu et
al (18), which showed that
miR-181a is downregulated in Huh7 cells infected with
GT2-HCV (JFH1-HCV). The finding of this study, as well as the
present study, confers that miR-181a is downregulated in
HCV-infected cell lines irrespective of the genotype. To the best
of our knowledge, for the first time the expression pattern of
miR-181a in liver biopsies, PBMCs and serum of naïve
GT4-HCV-infected patients was investigated. miR-181a
expression did not demonstrate variation in liver tissues and PBMCs
of the patients compared to the controls (Figs. 2 and 3, respectively). By contrast, there was a
significant increase in the serum of patients compared to the
controls (Fig. 4). This elevated
expression of miR-181a could be a result of released
exosomes in the circulation (1–3).
Notably, the miR-181a expression pattern in the liver
tissues and serum of GT4-HCV-infected patients showed an inverse
correlation to its expression pattern in HCC, in which HCC
miR-181a was found to be upregulated in liver tissues and
normally expressed in the serum of patients (12,13),
in contrast to no expression variation in the liver tissues and a
significantly increased expression in the serum of HCV-infected
patients. Subsequently, whether the expression of miR-181a
correlates to the patient clinical parameters [viral load and liver
enzymes (ALT and AST)] was examined. A clear finding that serum
miR-181a expression of HCV patients is inversely correlated
with the level of viremia, as well as liver enzymes (ALT, AST)
(Fig. 5), was found. To compare the
miR-181a expression pattern among different groups of
responders to standard PEG-IFN/RBV treatment, miR-181a was
quantified in the serum of pre- and post-treatment SVRs and NRs.
Serum pre- and post-treatment expression of miR-181a did not
differ in NRs (Fig. 6). Notably, a
significant upregulation of miR-181a was found in SVR
patients following treatment compared to NR patients and
treatment-naïve SVRs, with no difference shown between the groups
(SVRs and NRs) prior to therapy (Fig.
6). This is in accordance with a previous study reporting
comparable pre-treatment levels of miR-122 in SVRs and NR
patients infected with GT1-HCV (23). The expression of miR-181a
observed in pre-treatment SVRs and NRs opposes that of several
miRNAs depicted to show higher levels in responders compared to in
NRs. For example, serum pre-treatment levels of miR-122, the
most extensively studied miRNA in HCV, were found to be
significantly higher in SVR compared to NR GT2-HCV-infected
patients (24). Similarly,
pre-treatment levels of miR-122 were reported to be higher
in liver tissues of responders compared to NR GT1, 2, 3 and 4
HCV-infected patients (25).
Furthermore, GT1, 2 and 3 SVR patients showed high pre-treatment
levels of miR-155 in liver tissues and PBMCs, which
decreased following viral clearance (26,27).
In conclusion, to the best of our knowledge, the
present study demonstrates for the first time, a disparity in the
expression of miR-181a in the liver tissues and serum of
GT4-HCV-infected patients compared to controls, which is
conflicting to the expression pattern of miR-181a reported
in HCC (12,13). Additionally, although pre-treatment
miR-181a expression did not differentiate between SVRs and
NRs, the serum of SVR patients post-treatment was shown to exhibit
a significant upregulation of miR-181a compared to NR
patients and treatment-naïve SVRs, which indicates viral
eradication. Thus, the data show that the upregulation of
miR-181a in the serum of HCV patients is an indication of
good prognosis and any decrease during follow-up may be an early
marker for progression to HCC.
Acknowledgements
The authors would like to acknowledge Professor Jens
Bukh (Copenhagen University Hospital, Hvidovre, Denmark) and
Professor Takaji Wakita (Department of Virology II, National
Institute of Infectious Diseases, Tokyo, Japan) for the
pED435′UTR-NS2/JFH1T8271,T977S viral
vector.
References
|
1
|
Lima LG, Chammas R, Monteiro RQ, Moreira
ME and Barcinski MA: Tumor-derived microvesicles modulate the
establishment of metastatic melanoma in a
phosphatidylserine-dependent manner. Cancer Lett. 283:168–175.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cocucci E, Racchetti G and Meldolesi J:
Shedding microvesicles: artefacts no more. Trends Cell Biol.
19:43–51. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ghosh AK, Secreto CR, Knox TR, Ding W,
Mukhopadhyay D and Kay NE: Circulating microvesicles in B-cell
chronic lymphocytic leukemia can stimulate marrow stromal cells:
implications for disease progression. Blood. 115:1755–1764. 2010.
View Article : Google Scholar
|
|
4
|
Esquela-Kerscher A and Slack FJ: Oncomirs
- microRNAs with a role in cancer. Nat Rev Cancer. 6:259–269. 2006.
View Article : Google Scholar
|
|
5
|
Zhang Q, Pu R, Du Y, Han Y, Su T, Wang H
and Cao G: Non-coding RNAs in hepatitis B or C-associated
hepatocellular carcinoma: potential diagnostic and prognostic
markers and therapeutic targets. Cancer Lett. 321:1–12. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Li D, Zhao Y, Liu C, et al: Analysis of
miR-195 and miR-497 expression, regulation and role in breast
cancer. Clin Cancer Res. 17:1722–1730. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Heneghan HM, Miller N, Lowery AJ, Sweeney
KJ, Newell J and Kerin MJ: Circulating microRNAs as novel minimally
invasive biomarkers for breast cancer. Ann Surg. 251:499–505. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Waters PS, McDermott AM, Wall D, et al:
Relationship between circulating and tissue microRNAs in a murine
model of breast cancer. PLoS One. 7:e504592012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kutay H, Bai S, Datta J, et al:
Downregulation of miR-122 in the rodent and human hepatocellular
carcinomas. J Cell Biochem. 99:671–678. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gramantieri L, Ferracin M, Fornari F, et
al: Cyclin G1 is a target of miR-122a, a microRNA frequently
down-regulated in human hepatocellular carcinoma. Cancer Res.
67:6092–6099. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Xu J, Wu C, Che X, et al: Circulating
microRNAs, miR-21, miR-122, and miR-223, in patients with
hepatocellular carcinoma or chronic hepatitis. Mol Carcinog.
50:136–142. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ji J, Yamashita T, Budhu A, et al:
Identification of microRNA-181 by genome-wide screening as a
critical player in EpCAM-positive hepatic cancer stem cells.
Hepatology. 50:472–480. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhan MX, Li Y, Hu BS, et al: Expression of
serum microRNAs (miR-222, miR-181, miR-216) in human hepatocellular
carcinoma and its clinical significance. Zhonghua Yi Xue Za Zhi.
93:1830–1832. 2013.(In Chinese).
|
|
14
|
Li QJ, Chau J, Ebert PJ, et al: miR-181a
is an intrinsic modulator of T cell sensitivity and selection.
Cell. 129:147–161. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chen CZ, Li L, Lodish HF and Bartel DP:
MicroRNAs modulate hematopoietic lineage differentiation. Science.
303:83–86. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liu Y, Zhao JJ, Wang CM, et al: Altered
expression profiles of microRNAs in a stable hepatitis B
virus-expressing cell line. Chin Med J (Engl). 122:10–14.
2009.PubMed/NCBI
|
|
17
|
Wald AI, Hoskins EE, Wells SI, Ferris RL
and Khan SA: Alteration of microRNA profiles in squamous cell
carcinoma of the head and neck cell lines by human papillomavirus.
Head Neck. 33:504–512. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Liu X, Wang T, Wakita T and Yang W:
Systematic identification of microRNA and messenger RNA profiles in
hepatitis C virus-infected human hepatoma cells. Virology.
398:57–67. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
El-Ekiaby, Hamdi N, Negm M, et al:
Repressed induction of interferon-related microRNAs miR-146a and
miR-155 in peripheral blood mononuclear cells infected with HCV
genotype 4. FEBS Open Bio. 2:179–186. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li YP, Gottwein JM, Scheel TK, Jensen TB
and Bukh J: MicroRNA-122 antagonism against hepatitis C virus
genotypes 1–6 and reduced efficacy by host RNA insertion or
mutations in the HCV 5′ UTR. Proc Natl Acad Sci USA. 108:4991–4996.
2011.PubMed/NCBI
|
|
21
|
Chomczynski P and Sacchi N: Single-step
method of RNA isolation by acid guanidinium
thiocyanate-phenol-chloroform extraction. Anal Biochem.
162:156–159. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hu Z, Chen X, Zhao Y, et al: Serum
microRNA signatures identified in a genome-wide serum microRNA
expression profiling predict survival of non-small-cell lung
cancer. J Clin Oncol. 28:1721–1726. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Köberle V, Waidmann O, Kronenberger B, et
al: Serum microRNA-122 kinetics in patients with chronic hepatitis
C virus infection during antiviral therapy. J Viral Hepat.
20:530–535. 2013.PubMed/NCBI
|
|
24
|
Su TH, Liu CH, Liu CJ, et al: Serum
microRNA-122 level correlates with virologic responses to pegylated
interferon therapy in chronic hepatitis C. Proc Natl Acad Sci USA.
110:7844–7849. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Sarasin-Filipowicz M, Krol J, Markiewicz
I, Heim MH and Filipowicz W: Decreased levels of microRNA miR-122
in individuals with hepatitis C responding poorly to interferon
therapy. Nat Med. 15:31–33. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bala S, Tilahun Y, Taha O, Alao H, Kodys
K, Catalano D and Szabo G: Increased microRNA-155 expression in the
serum and peripheral monocytes in chronic HCV infection. J Transl
Med. 10:1512012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang Y, Wei W, Cheng N, Wang K, Li B,
Jiang X and Sun S: Hepatitis C virus-induced up-regulation of
microRNA-155 promotes hepatocarcinogenesis by activating Wnt
signaling. Hepatology. 56:1631–1640. 2012. View Article : Google Scholar : PubMed/NCBI
|