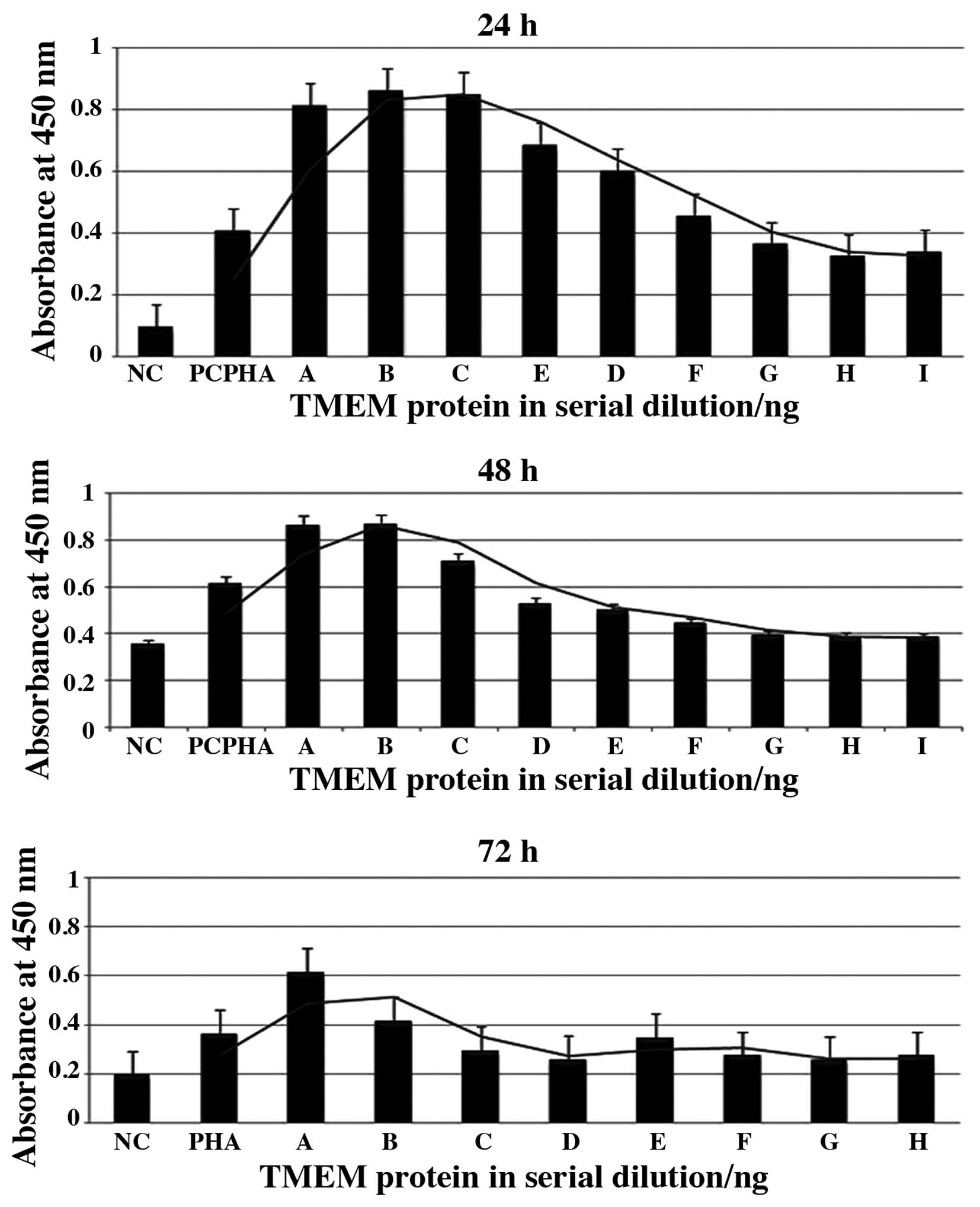
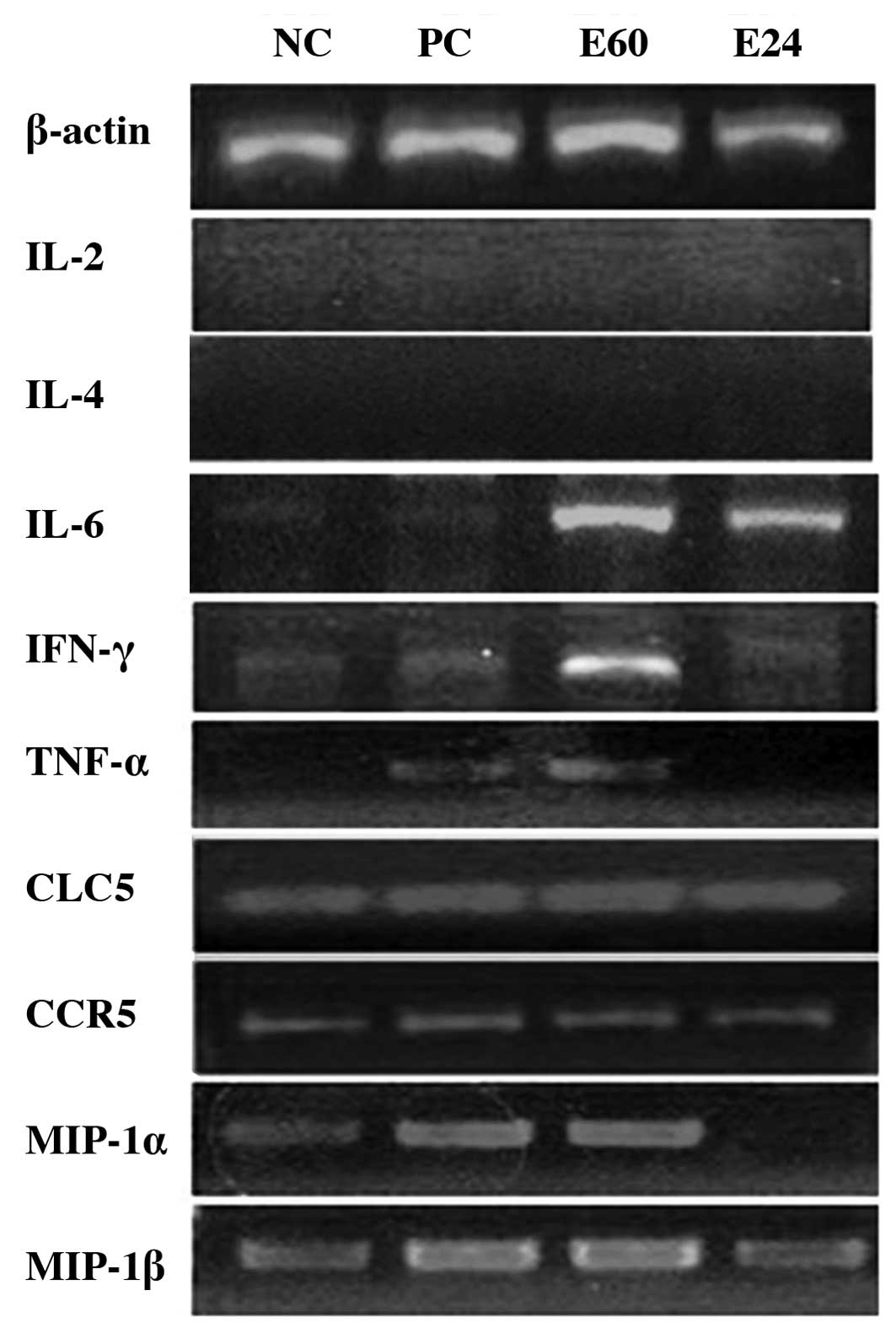
|
1
|
Broux B, Stinissen P and Hellings N: Which
immune cells matter? The immunopathogenesis of multiple sclerosis.
Crit Rev Immunol. 33:283–306. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Küçükali CI, Kürtüncü M, Coban A, Cebi M
and Tüzün E: Epigenetics of multiple sclerosis: an updated review.
Neuromolecular Med: Mar. 21:2014.(Epub ahead of print).
|
|
3
|
Ascherio A: Environmental factors in
multiple sclerosis. Expert Rev Neurother. 13 (Suppl 12):3–9. 2013.
View Article : Google Scholar
|
|
4
|
Kenealy SJ, Pericak-Vance MA and Haines
JL: The genetic epidemiology of multiple sclerosis. J Neuroimmunol.
143:7–12. 2003. View Article : Google Scholar
|
|
5
|
Noseworthy JH, Lucchinetti C, Rodriguez M
and Weinshenker BG: Multiple sclerosis. N Engl J Med. 343:938–952.
2000. View Article : Google Scholar
|
|
6
|
Alroughani RA and Al-Jumah MA: The need
for a multiple sclerosis registry in the Gulf Region. Neurosciences
(Riyadh). 19:85–86. 2014.PubMed/NCBI
|
|
7
|
Aljumah M, Alroughani R, Alsharoqi I, et
al: Future of management of multiple sclerosis in the middle East:
a consensus view from specialists in ten countries. Mult Scler Int
2013: 952321. 2013.PubMed/NCBI
|
|
8
|
Bohlega S, Inshasi J, Al Tahan AR, Madani
AB, Qahtani H and Rieckmann P: Multiple sclerosis in the Arabian
Gulf countries: a consensus statement. J Neurol. 260:2959–2963.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Al-Afasy HH, Al-Obaidan MA, Al-Ansari YA,
et al: Risk factors for multiple sclerosis in Kuwait: a
population-based case-control study. Neuroepidemiology. 40:30–35.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Prat E and Martin R: The
immunopathogenesis of multiple sclerosis. J Rehabil Res Dev.
39:187–199. 2002.
|
|
11
|
Achiron A, Gurevich M, Magalashvili D,
Kishner I, Dolev M and Mandel M: Understanding autoimmune
mechanisms in multiple sclerosis using gene expression microarrays:
treatment effect and cytokine-related pathways. Clin Dev Immunol.
11:299–305. 2004. View Article : Google Scholar
|
|
12
|
Martins TB, Rose JW, Jaskowski TD, et al:
Analysis of proinflammatory and anti-inflammatory cytokine serum
concentrations in patients with multiple sclerosis by using a
multiplexed immunoassay. Am J Clin Pathol. 136:696–704. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kolbert CP, Taylor WR, Krajnik KL and
O'Kane DJ: Gene expression microarrays. Methods Mol Med.
85:239–255. 2003.
|
|
14
|
Watson A, Mazumder A, Stewart M and
Balasubramanian S: Technology for microarray analysis of gene
expression. Curr Opin Biotechnol. 9:609–614. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Baranzini SE and Hauser SL: Large-scale
gene-expression studies and the challenge of multiple sclerosis.
Genome Biol. 3:10272002. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Polman CH, Reingold SC, Banwell B, et al:
Diagnostic criteria for multiple sclerosis: 2010 revisions to the
McDonald criteria. Ann Neurol. 69:292–302. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zanzinger K, Schellack C, Nausch N and
Cerwenka A: Regulation of triggering receptor expressed on myeloid
cells 1 expression on mouse inflammatory monocytes. Immunology.
128:185–195. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Rasmussen R: Quantification on the
LightCyclerRapid Cycle Real-Time PCR, Methods and Applications.
Meuer S, Wittwer C and Nakagawara K: Springer Press; Heidelberg:
pp. 22–34. 2001
|
|
19
|
Ausubel FM, Brent R, Kingston RE, Moore
DD, Seidman JG, Smith JA and Struhl K: Current Protocols in
Molecular Biology. John Wiley & Sons; New York, NY: 1994
|
|
20
|
Decker T and Lohmann-Matthes ML: A quick
and simple method for the quantitation of lactate dehydrogenase
release in measurements of cellular cytotoxicity and tumor necrosis
factor (TNF) activity. J Immunol Methods. 115:61–69. 1988.
View Article : Google Scholar
|
|
21
|
Korzeniewski C and Callewaert DM: An
enzyme-release assay for natural cytotoxicity. J Immunol Methods.
64:313–320. 1983. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chomczynski P: A reagent for the
single-step simultaneous isolation of RNA, DNA and proteins from
cell and tissue samples. Biotechniques. 15:532–534, 536–537.
1993.PubMed/NCBI
|
|
23
|
Chomczynski P and Sacchi N: Single-step
method of RNA isolation by acid guanidinium
thiocyanate-phenol-chloroform extraction. Anal Biochem.
162:156–159. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hummon AB, Lim SR, Difilippantonio MJ and
Ried T: Isolation and solubilization of proteins after TRIzol
extraction of RNA and DNA from patient material following prolonged
storage. Biotechniques. 42:467–470, 472. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Irizarry RA, Hobbs B, Collin F, et al:
Exploration, normalization, and summaries of high density
oligonucleotide array probe level data. Biostatistics. 4:249–264.
2003. View Article : Google Scholar
|
|
26
|
Wu Z, Irizarry R, Gentlemen R, et al: A
model-based background adjustment for oligonucleotide expression
arrays. J Am Statist Assoc. 99:909–917. 2004. View Article : Google Scholar
|
|
27
|
Palty R, Raveh A, Kaminsky I, Meller R and
Reuveny E: SARAF inactivates the store operated calcium entry
machinery to prevent excess calcium refilling. Cell. 149:425–438.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Su AI, Wiltshire T, Batalov S, et al: A
gene atlas of the mouse and human protein-encoding transcriptomes.
Proc Natl Acad Sci USA. 101:6062–6067. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Romanuik TL, Wang G, Holt RA, Jones SJ,
Marra MA and Sadar MD: Identification of novel androgen-responsive
genes by sequencing of LongSAGE libraries. BMC Genomics.
10:4762009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Romanuik TL, Ueda T, Le N, et al: Novel
biomarkers for prostate cancer including noncoding transcripts. Am
J Pathol. 175:2264–2276. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Hartung HP, Aktas O, Menge T and Kieseier
BC: Immune regulation of multiple sclerosis. Handb Clin Neurol.
122:3–14. 2014. View Article : Google Scholar
|