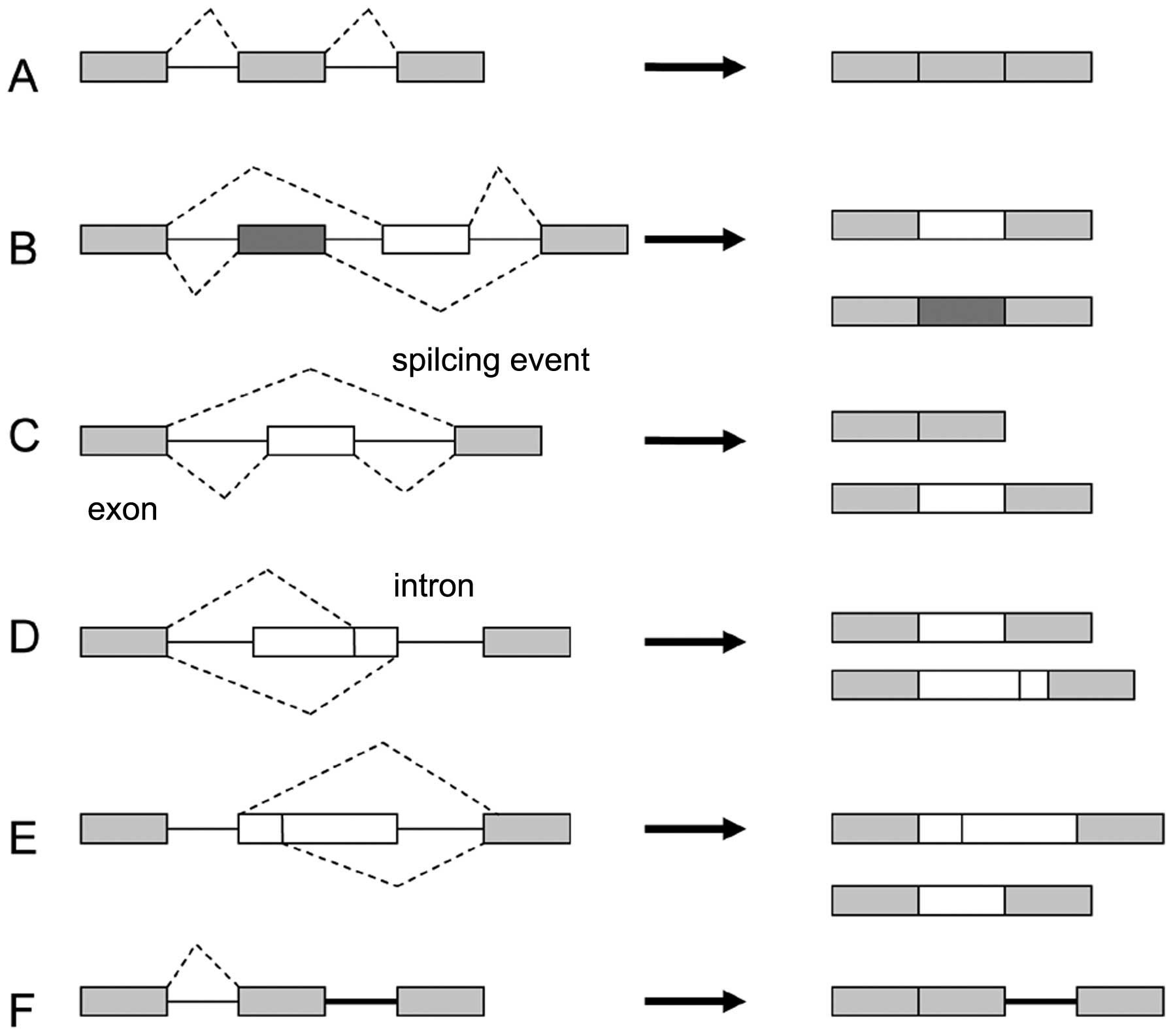
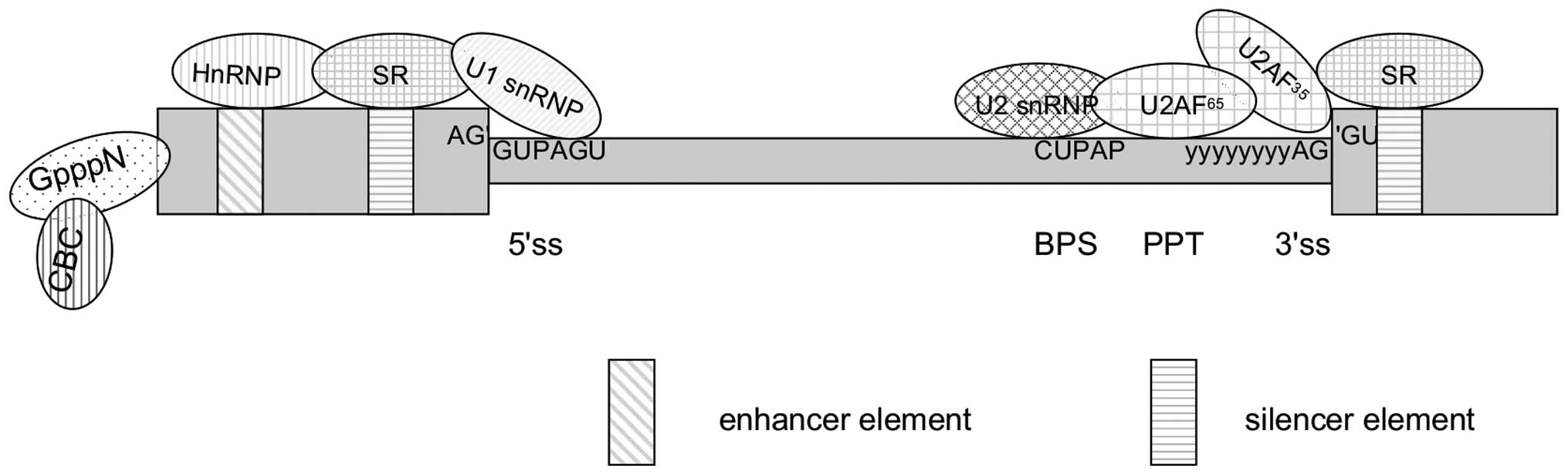
|
1
|
Gilbert W: Why genes in pieces? Nature.
271:5011978. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
C. elegans Sequencing Consortium, . Genome
sequence of the nematode C. elegans: a platform for investigating
biology. Science. 282:2012–2018. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Human Genome Sequencing Consortium I
International Human Genome Sequencing C, . Finishing the
euchromatic sequence of the human genome. Nature. 431:931–945.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Nilsen TW and Graveley BR: Expansion of
the eukaryotic proteome by alternative splicing. Nature.
463:457–463. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zheng CL, Fu XD and Gribskov M:
Characteristics and regulatory elements defining constitutive
splicing and different modes of alternative splicing in human and
mouse. RNA. 11:1777–1787. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Effenberger KA, Perriman RJ, Bray WM, et
al: A high-throughput splicing assay identifies new classes of
inhibitors of human and yeast spliceosomes. J Biomol Screen.
18:1110–1120. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wahl MC, Will CL and Luhrmann R: The
spliceosome: design principles of a dynamic RNP machine. Cell.
136:701–718. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kim E, Goren A and Ast G: Alternative
splicing: current perspectives. Bioessays. 30:38–47. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Irimia M, Penny D and Roy SW: Coevolution
of genomic intron number and splice sites. Trends Genet.
23:321–325. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Fu G, Condon KC, Epton MJ, et al:
Female-specific insect lethality engineered using alternative
splicing. Nat Biotechnol. 25:353–357. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hurt KJ, Sezen SF, Champion HC, et al:
Alternatively spliced neuronal nitric oxide synthase mediates
penile erection. Proc Natl Acad Sci USA. 103:3440–3443. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Blencowe BJ: Alternative splicing: new
insights from global analyses. Cell. 126:37–47. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Nygard AB, Cirera S, Gilchrist MJ, et al:
A study of alternative splicing in the pig. BMC Res Notes.
3:1232010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kelemen O, Convertini P, Zhang Z, et al:
Function of alternative splicing. Gene. 514:1–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kim E, Magen A and Ast G: Different levels
of alternative splicing among eukaryotes. Nucleic Acids Res.
35:125–131. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Galante PA, Sakabe NJ, Kirschbaum-Slager N
and de Souza SJ: Detection and evaluation of intron retention
events in the human transcriptome. RNA. 10:757–765. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sakabe NJ and de Souza SJ: Sequence
features responsible for intron retention in human. BMC Genomics.
8:592007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Goldstrohm AC, Greenleaf AL and
Garcia-Blanco MA: Co-transcriptional splicing of pre-messenger
RNAs: considerations for the mechanism of alternative splicing.
Gene. 277:31–47. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Di Giammartino DC, Nishida K and Manley
JL: Mechanisms and consequences of alternative polyadenylation. Mol
Cell. 43:853–866. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mastrangelo AM, Marone D, Laido G, et al:
Alternative splicing: enhancing ability to cope with stress via
transcriptome plasticity. Plant Sci. 185–186:40–49. 2012.
View Article : Google Scholar
|
|
21
|
Hoskins AA and Moore MJ: The spliceosome:
a flexible, reversible macromolecular machine. Trends Biochem Sci.
37:179–188. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang Z and Burge CB: Splicing regulation:
from a parts list of regulatory elements to an integrated splicing
code. RNA. 14:802–813. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang Z, Xiao X, Van Nostrand E and Burge
CB: General and specific functions of exonic splicing silencers in
splicing control. Mol Cell. 23:61–70. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Jin Y, Yang Y and Zhang P: New insights
into RNA secondary structure in the alternative splicing of
pre-mRNAs. RNA Biol. 8:450–457. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu W, Zhou Y, Hu Z, et al: Regulation of
splicing enhancer activities by RNA secondary structures. FEBS
Lett. 584:4401–4407. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
McManus CJ and Graveley BR: RNA structure
and the mechanisms of alternative splicing. Curr Opin Genet Dev.
21:373–379. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Nasim FU, Hutchison S, Cordeau M and
Chabot B: High-affinity hnRNP A1 binding sites and duplex-forming
inverted repeats have similar effects on 5′ splice site selection
in support of a common looping out and repression mechanism. RNA.
8:1078–1089. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Han SP, Kassahn KS, Skarshewski A, Ragan
MA, Rothnagel JA and Smith R: Functional implications of the
emergence of alternative splicing in hnRNP A/B transcripts. RNA.
16:1760–1768. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang E and Cambi F: Heterogeneous nuclear
ribonucleoproteins H and F regulate the proteolipid protein/DM20
ratio by recruiting U1 small nuclear ribonucleoprotein through a
complex array of G runs. J Biol Chem. 284:11194–11204. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Park E, Iaccarino C, Lee J, et al:
Regulatory roles of heterogeneous nuclear ribonucleoprotein M and
Nova-1 protein in alternative splicing of dopamine D2 receptor
pre-mRNA. J Biol Chem. 286:25301–25308. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liu G, Razanau A, Hai Y, et al: A
conserved serine of heterogeneous nuclear ribonucleoprotein L
(hnRNP L) mediates depolarization-regulated alternative splicing of
potassium channels. J Biol Chem. 287:22709–22716. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yu J, Hai Y, Liu G, Fang T, Kung SK and
Xie J: The heterogeneous nuclear ribonucleoprotein L is an
essential component in the Ca2+/calmodulin-dependent protein kinase
IV-regulated alternative splicing through cytidine-adenosine
repeats. J Biol Chem. 284:1505–1513. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ghosh G and Adams JA: Phosphorylation
mechanism and structure of serine-arginine protein kinases. FEBS J.
278:587–597. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Twyffels L, Gueydan C and Kruys V:
Shuttling SR proteins: more than splicing factors. FEBS J.
278:3246–3255. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Long JC and Caceres JF: The SR protein
family of splicing factors: master regulators of gene expression.
Biochem J. 417:15–27. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kalnina Z, Zayakin P, Silina K and Linē A:
Alterations of pre-mRNA splicing in cancer. Genes Chromosomes
Cancer. 42:342–357. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Beyer AL, Bouton AH and Miller OL Jr:
Correlation of hnRNP structure and nascent transcript cleavage.
Cell. 26:155–165. 1981. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kornblihtt AR, de la Mata M, Fededa JP, et
al: Multiple links between transcription and splicing. RNA.
10:1489–1498. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Montes M, Becerra S, Sanchez-Alvarez M and
Sune C: Functional coupling of transcription and splicing. Gene.
501:104–117. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ryman K, Fong N, Bratt E, Bentley DL and
Ohman M: The C-terminal domain of RNA Pol II helps ensure that
editing precedes splicing of the GluR-B transcript. RNA.
13:1071–1078. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Bentley DL: Rules of engagement:
co-transcriptional recruitment of pre-mRNA processing factors. Curr
Opin Cell Biol. 17:251–256. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Bartkowiak B, Liu P, Phatnani HP, et al:
CDK12 is a transcription elongation-associated CTD kinase, the
metazoan ortholog of yeast Ctk1. Genes Dev. 24:2303–2316. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Kim H, Erickson B, Luo W, et al:
Gene-specific RNA polymerase II phosphorylation and the CTD code.
Nat Struct Mol Biol. 17:1279–1286. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Phatnani HP and Greenleaf AL:
Phosphorylation and functions of the RNA polymerase II CTD. Genes
Dev. 20:2922–2936. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Rosonina E and Blencowe BJ: Analysis of
the requirement for RNA polymerase II CTD heptapeptide repeats in
pre-mRNA splicing and 3′-end cleavage. RNA. 10:581–589. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Kalsotra A and Cooper TA: Functional
consequences of developmentally regulated alternative splicing. Nat
Rev Genet. 12:715–729. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Schor IE, Gómez Acuña LI and Kornblihtt
AR: Coupling between transcription and alternative splicing. Cancer
Treat Res. 158:1–24. 2013.PubMed/NCBI
|
|
48
|
Dujardin G, Lafaille C, Petrillo E, et al:
Transcriptional elongation and alternative splicing. Biochim
Biophys Acta. 1829:134–140. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Gómez Acuña LI, Fiszbein A, Alló M, et al:
Connections between chromatin signatures and splicing. Wiley
Interdiscip Rev RNA. 4:77–91. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Iannone C and Valcárcel J: Chromatin's
thread to alternative splicing regulation. Chromosoma. 122:465–474.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Shukla S and Oberdoerffer S:
Co-transcriptional regulation of alternative pre-mRNA splicing.
Biochim Biophys Acta. 1819:673–683. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Ma X, Li-Ling J, Huang Q, et al:
Systematic analysis of alternative promoters correlated with
alternative splicing in human genes. Genomics. 93:420–425. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Xin D, Hu L and Kong X: Alternative
promoters influence alternative splicing at the genomic level. PLoS
One. 3:e23772008. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Shabalina SA, Spiridonov AN, Spiridonov NA
and Koonin EV: Connections between alternative transcription and
alternative splicing in mammals. Genome Biol Evol. 2:791–799. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Roy B, Haupt LM and Griffiths LR: Review:
Alternative splicing (AS) of genes as an approach for generating
protein complexity. Curr Genomics. 14:182–194. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Lareau LF, Brooks AN, Soergel DA, Meng Q
and Brenner SE: The coupling of alternative splicing and
nonsense-mediated mRNA decay. Adv Exp Med Biol. 623:190–211.
2007.PubMed/NCBI
|
|
57
|
Lewis BP, Green RE and Brenner SE:
Evidence for the widespread coupling of alternative splicing and
nonsense-mediated mRNA decay in humans. Proc Natl Acad Sci USA.
100:189–192. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Nyiko T, Kerenyi F, Szabadkai L, et al:
Plant nonsense-mediated mRNA decay is controlled by different
autoregulatory circuits and can be induced by an EJC-like complex.
Nucleic Acids Res. 41:6715–6728. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Kalyna M, Simpson CG, Syed NH, et al:
Alternative splicing and nonsense-mediated decay modulate
expression of important regulatory genes in Arabidopsis. Nucleic
Acids Res. 40:2454–2469. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Saltzman AL, Kim YK, Pan Q, et al:
Regulation of multiple core spliceosomal proteins by alternative
splicing-coupled nonsense-mediated mRNA decay. Mol Cell Biol.
28:4320–4330. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Ge Y and Porse BT: The functional
consequences of intron retention: alternative splicing coupled to
NMD as a regulator of gene expression. Bioessays. 36:236–243. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Herai RH and Yamagishi ME: Detection of
human interchromosomal trans-splicing in sequence databanks. Brief
Bioinform. 11:198–209. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Horiuchi T and Aigaki T: Alternative
trans-splicing: a novel mode of pre-mRNA processing. Biol Cell.
98:135–140. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Lee SW and Jeong JS: Use of
tumor-targeting trans-splicing ribozyme for cancer treatment.
Methods Mol Biol. 1103:83–95. 2014.PubMed/NCBI
|
|
65
|
Jung HS and Lee SW: Ribozyme-mediated
selective killing of cancer cells expressing carcinoembryonic
antigen RNA by targeted trans-splicing. Biochem Biophys Res Commun.
349:556–563. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Avale ME, Rodriguez-Martin T and Gallo JM:
Trans-splicing correction of tau isoform imbalance in a mouse model
of tau mis-splicing. Hum Mol Genet. 22:2603–2611. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Luco RF and Misteli T: More than a
splicing code: integrating the role of RNA, chromatin and
non-coding RNA in alternative splicing regulation. Curr Opin Genet
Dev. 21:366–372. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Singh RK and Cooper TA: Pre-mRNA splicing
in disease and therapeutics. Trends Mol Med. 18:472–482. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Zhang J and Manley JL: Misregulation of
pre-mRNA alternative splicing in cancer. Cancer Discov.
3:1228–1237. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Shkreta L, Bell B, Revil T, et al:
Cancer-associated perturbations in alternative pre-messenger RNA
splicing. Cancer Treat Res. 158:41–94. 2013.PubMed/NCBI
|
|
71
|
Kim YJ and Kim HS: Alternative splicing
and its impact as a cancer diagnostic marker. Genomics Inform.
10:74–80. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Lapuk AV, Volik SV, Wang Y and Collins CC:
The role of mRNA splicing in prostate cancer. Asian J Androl.
16:515–521. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Pajares MJ, Ezponda T, Catena R, et al:
Alternative splicing: an emerging topic in molecular and clinical
oncology. Lancet Oncol. 8:349–357. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Sitohy B, Nagy JA and Dvorak HF:
Anti-VEGF/VEGFR therapy for cancer: reassessing the target. Cancer
Res. 72:1909–1914. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Pal S, Gupta R and Davuluri RV:
Alternative transcription and alternative splicing in cancer.
Pharmacol Ther. 136:283–294. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Pan Q, Shai O, Lee LJ, et al: Deep
surveying of alternative splicing complexity in the human
transcriptome by high-throughput sequencing. Nat Genet.
40:1413–1415. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
77
|
Calarco JA, Xing Y, Caceres M, et al:
Global analysis of alternative splicing differences between humans
and chimpanzees. Genes Dev. 21:2963–2975. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Reddy AS, Rogers MF, Richardson DN,
Hamilton M and Ben-Hur A: Deciphering the plant splicing code:
experimental and computational approaches for predicting
alternative splicing and splicing regulatory elements. Front Plant
Sci. 3:182012. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Tang W, Fei Y and Page M: Biological
significance of RNA editing in cells. Mol Biotechnol. 52:91–100.
2012. View Article : Google Scholar : PubMed/NCBI
|