Article
Detection of natural antisense non-coding RNAs transcribed from Ncam1 in mice tissues at several developmental stages
- Authors:
-
View Affiliations / Copyright
Affiliations:
Department of Biomedical Sciences, Division of Medical Life Sciences, Hirosaki University Graduate School of Health Sciences, Hirosaki, Aomori 036‑8564, Japan
-
Pages:
663-667
|
Published online on:
June 29, 2015
https://doi.org/10.3892/br.2015.482
- Expand metrics +
Metrics:
Total
Views: 0
(Spandidos Publications: | PMC Statistics:
)
Metrics:
Total PDF Downloads: 0
(Spandidos Publications: | PMC Statistics:
)
This article is mentioned in:
Abstract
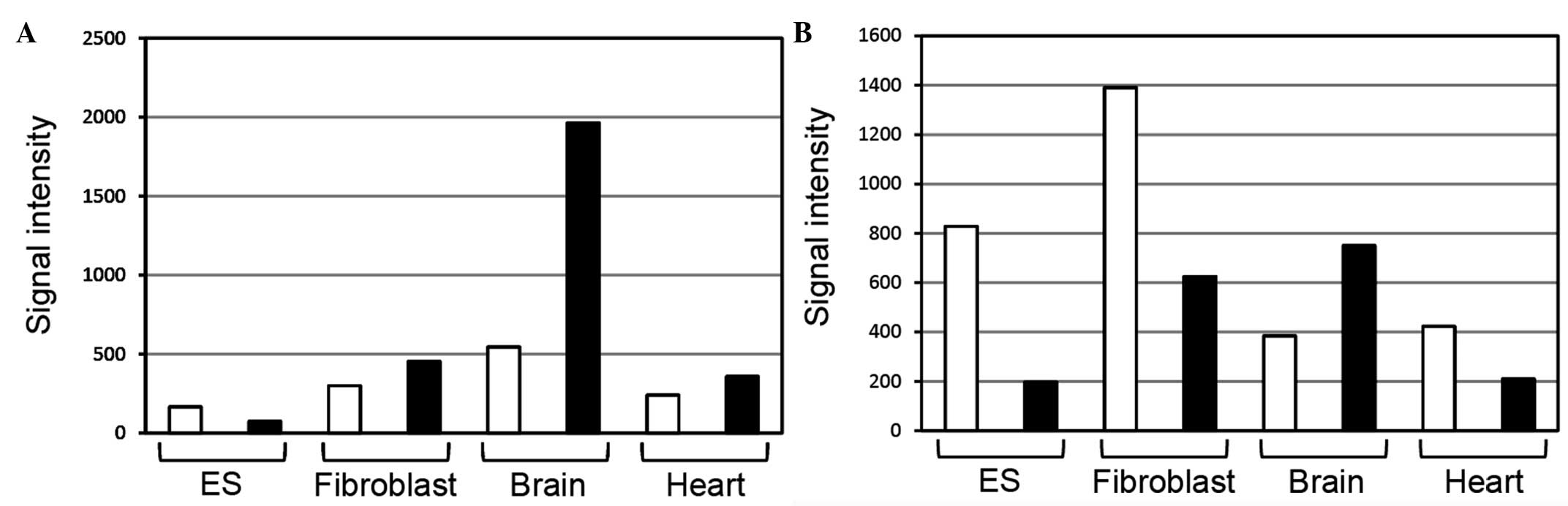
Natural antisense ribonucleic acids (RNAs) are transcribed from a large number of genes in various species, including humans and mice. The expression of neural cell adhesion molecule 1 (Ncam1) antisense non‑coding RNAs (ncRNAs) in mice has been demonstrated by functional annotation of the mammalian genome project, but the localization of Ncam1 antisense ncRNAs has not been reported in mice tissues. In the present study, the localization of Ncam1 antisense ncRNAs was examined in tissues at several developmental stages by in situ hybridization. At days 14 and 17 of embryonic development, Ncam1 antisense ncRNAs were found in the heart, liver, lung, kidney, thymus and nerve regions including the brain (cerebral cortex, olfactory bulb, hippocampus and cerebellum) and spinal cord. In newborn mice, Ncam1 antisense ncRNAs were detected in the brain, kidney and thymus, but was not detected in other tissues. In 8‑week‑old mice, Ncam1 antisense ncRNAs were detected in the lung, kidney, thymus, pancreas, cornea, stomach and nerve regions including the brain. These results indicate that Ncam1 antisense ncRNAs are expressed in mice tissues. Notably, Ncam1 messenger RNAs (mRNAs), antisense ncRNAs co‑localized in the Purkinje cells of the cerebellum and the levels of antisense ncRNAs appeared to be higher than those of mRNAs, suggesting that Ncam1 antisense ncRNAs may regulate the expression of Ncam1 mRNAs in the same cells.
View References
|
1
|
Kiyosawa H, Yamanaka I, Osato N, Kondo S
and Hayashizaki YRIKEN GER Group: Role: GSL MembersAntisense
transcripts with FANTOM2 clone set and their implications for gene
regulation. Genome Res. 13((6B)): 1324–1334. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kiyosawa H, Mise N, Iwase S, Hayashizaki Y
and Abe K: Disclosing hidden transcripts: Mouse natural
sense-antisense transcripts tend to be poly(A) negative and nuclear
localized. Genome Res. 15:463–474. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Okazaki Y, Furuno M, Kasukawa T, Adachi J,
Bono H, Kondo S, Nikaido I, Osato N, Saito R, Suzuki H, et al
FANTOM Consortium; RIKEN Genome Exploration Research Group Phase I
& II Team: Analysis of the mouse transcriptome based on
functional annotation of 60,770 full-length cDNAs. Nature.
420:563–573. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Katayama S, Tomaru Y, Kasukawa T, Waki K,
Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, et
al FANTOM Consortium: Antisense transcription in the mammalian
transcriptome. Science. 309:1564–1566. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lehner B, Williams G, Campbell RD and
Sanderson CM: Antisense transcripts in the human genome. Trends
Genet. 18:63–65. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Faghihi MA and Wahlestedt C: Regulatory
roles of natural antisense transcripts. Nat Rev Mol Cell Biol.
10:637–643. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kohno K, Chiba M, Murata S, Pak S, Nagai
K, Yamamoto M, Yanagisawa K, Kobayashi A, Yasue H and Ohkohchi N:
Identification of natural antisense transcripts involved in human
colorectal cancer development. Int J Oncol. 37:1425–1432.
2010.PubMed/NCBI
|
|
8
|
Grigoriadis A, Oliver GR, Tanney A,
Kendrick H, Smalley MJ, Jat P and Neville AM: Identification of
differentially expressed sense and antisense transcript pairs in
breast epithelial tissues. BMC Genomics. 10:3242009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Cremer H, Lange R, Christoph A, Plomann M,
Vopper G, Roes J, Brown R, Baldwin S, Kraemer P, Scheff S, et al:
Inactivation of the N-CAM gene in mice results in size reduction of
the olfactory bulb and deficits in spatial learning. Nature.
367:455–459. 1994. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Dityatev A, Dityateva G, Sytnyk V, Delling
M, Toni N, Nikonenko I, Muller D and Schachner M: Polysialylated
neural cell adhesion molecule promotes remodeling and formation of
hippocampal synapses. J Neurosci. 24:9372–9382. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lavorgna G, Dahary D, Lehner B, Sorek R,
Sanderson CM and Casari G: In search of antisense. Trends Biochem
Sci. 29:88–94. 2004. View Article : Google Scholar : PubMed/NCBI
|