Introduction
Malignant tumors are a complex disease relating to
an interaction effect between polygenic inheritance and
environmental factors (1).
Tumorigenesis and the development of malignant tumors are poorly
understood, and further research is required concerning their key
underlying factors and molecular abnormalities. In recent years,
the response of the immune system to malignant tumors has been the
focus of investigation (2). Studies on
the role of T lymphocytes and natural killer cells in mediating an
antitumor response have been increasing in recent years (3). For this reason, genetic polymorphisms of
the genes involved the mediate response of T lymphocytes have been
investigated for a possible association with the risk of developing
a range of malignant tumors (4,5). One of
these genes, cytotoxic T-lymphocyte-associated antigen 4 (CTLA-4),
located on chromosome 2q33, is a member of the immunoglobulin
superfamily and has an important role in the regulation of
T-lymphocyte activation and proliferation (6). CTLA-4 polymorphisms have been
demonstrated to be associated with increased susceptibility to
various malignant tumors in a number of different ethnic
populations. Previous studies have been performed to determine
whether the −318 cytosine/thymine (−318C/T) polymorphism was
involved in the etiology of various malignant tumors in Asian
population (7–26). However, the results from these studies
remain inconclusive and conflicting. The present study aimed to
achieve a more precise assessment of the association between CTLA-4
−318C/T polymorphism and malignant tumor risk in an Asian
population by performing a meta-analysis on 20 eligible
case-control studies to estimate the CTLA-4 −318C/T polymorphism
and malignant tumor.
Materials and methods
Literature search
The PubMed, Embase, Chinese Journals Full-Text
Database, Chinese Biomedical Database, and Wanfang databases were
searched for all articles published up to September 30, 2015. The
literature search used a number of keywords including, ‘genetic
polymorphism’, ‘cancer OR carcinoma’, and ‘cytotoxic T-lymphocyte
antigen-4 OR CTLA-4’. No language restrictions were applied. All of
the eligible studies included in the meta-analysis had to meet the
following inclusion criteria: i) Evaluation of the CTLA-4 −318C/T
polymorphism and malignant tumor risk; ii) independent case-control
study in design; iii) available data (distribution of alleles and
genotypes for cases and controls) estimating an odds ratio (OR)
with a 95% confidence interval (95% CI) of CTLA-4 −318C/T gene
polymorphisms in both cases and controls; iv) the studies on Asian
population; and v) genotype distributions in the controls conformed
to the Hardy-Weinberg equilibrium (HWE). The following exclusive
criteria were used: i) Repeat or overlapping publications; ii)
abstracts, reviews or letters; and iii) studies in which genotype
frequencies were not reported.
Data extraction
To enhance credibility and reduce bias in the study,
all data were abstracted by two of the authors independently, who
complied with the rigorous selection criteria and reached a
consensus on all items. The following information was extracted
from each study: First author, year of publication, ethnicity and
country of origin, type of malignant tumor, sample size, and HWE
evidence in controls (Table I).
 | Table I.Characteristics of case-controlled
studies included in the meta-analysis. |
Table I.
Characteristics of case-controlled
studies included in the meta-analysis.
|
| Sample size |
|
|---|
|
|
|
|
|---|
| First author | Year | Country | Cancer type | Genotyping
method | Cases/controls | Refs. |
|---|
| Cheng | 2006 | China | Lymphoma | PCR-RFLP |
62/250 | (7) |
| Erfani | 2006 | Iran | Breast cancer | PCR-ARMS | 283/241 | (8) |
| Wang | 2007 | China | Breast cancer | PCR-RFLP | 117/148 | (9) |
| Su | 2007 | China | Cervical cancer | PCR-RFLP | 144/378 | (10) |
| Hadinia | 2007 | Iran | Gastrointestinal
cancer | PCR-ARMS | 155/190 | (11) |
| Li | 2010 | China | Gastrointestinal
cancer | PCR-RFLP | 121/236 | (12) |
| Khaghanzadeh | 2009 | Iran | Lung cancer | PCR-ARMS | 126/122 | (13) |
| Rahimifar | 2010 | Iran | Cervical cancer | PCR-ARMS |
54/110 | (14) |
| Liu | 2011 | China | Bone sarcoma | PCR-RFLP | 267/282 | (15) |
| Jiang | 2011 | China | Cervical cancer | PCR-RFLP,
sequenom | 100/100 | (16) |
| Yang | 2012 | China | Bone sarcoma | PCR-RFLP | 223/302 | (17) |
| Gokhale | 2013 | India | Cervical
cancer | PCR-RFLP | 100/101 | (18) |
| Feng | 2013 | China | Bone sarcoma | PCR-RFLP | 308/362 | (19) |
| Bharti | 2013 | India | Oral carcinoma | PCR-RFLP | 120/180 | (20) |
| Liu | 2013 | China | Lymphoma | PCR-RFLP | 291/300 | (21) |
| Jaiswal | 2014 | India | Bladder cancer | PCR-ARMS | 200/200 | (22) |
| Xiong | 2014 | China | Cervical
cancer | RT-PCR | 365/421 | (23) |
| Hui | 2014 | China | Leukemia | RT-PCR |
86/112 | (24) |
| Cheng | 2015 | China | Lymphoma | PCR-LDR | 125/300 | (25) |
| Wang | 2015 | China | Cervical
cancer | RT-PCR | 292/355 | (26) |
Statistical analysis
The ORs with corresponding 95% CIs were calculated
for the strength of correlations between CTLA-4 −318C/T gene
polymorphisms and malignant tumor risks in an Asian population. The
significance of the pooled OR was determined by using a Z-test, and
P<0.05 was considered to indicate a statistically significant
difference. In order to assess the association between CTLA-4
−318C/T polymorphism and malignant tumor risk, five genetic models
were used to evaluate the pooled ORs of the polymorphism, as
follows: Dominant model (TT+TC vs. CC), recessive model (TT vs.
TC+CC), homozygous model (TT vs. CC), heterozygous model (TC vs.
CC), and allele model (T vs. C) for −318C/T. The OR was calculated
by using different effect models according to the result of
heterogeneity test. The statistical heterogeneity of studies was
checked by using the Chi-square-based Q statistic: When P≥0.05, the
fixed-effects model (Mantel-Haenszel method) was used to analyze
the data, whereas the random-effects model (the DerSimonian and
Laird method) was used if P<0.05 (27). In addition, the I2 test was
used, and was considered statistically significant at an
I2 value ≥50%, and the random-effects model was
performed. An I2 value <50% indicates lack of
heterogeneity, and therefore the fixed-effects model was used.
Through an initial screen of the results, the majority of the
included populations were identified as originating from China,
hence a subgroup meta-analyses was also performed. The subgroup
meta-analyses were conducted by comparing the China population with
the population from the rest of Asia for the −318C/T polymorphism.
We checked publication bias by visual inspection of asymmetry in
funnel plots. In addition, Egger's test was investigated to further
evaluate the symmetry of the funnel plot and P<0.05 was
considered to indicate a statistically significant difference. All
statistical tests were performed using STATA version 11.2 (Stata
Corp., College Station, TX, USA).
Results
Study selection and
characteristics
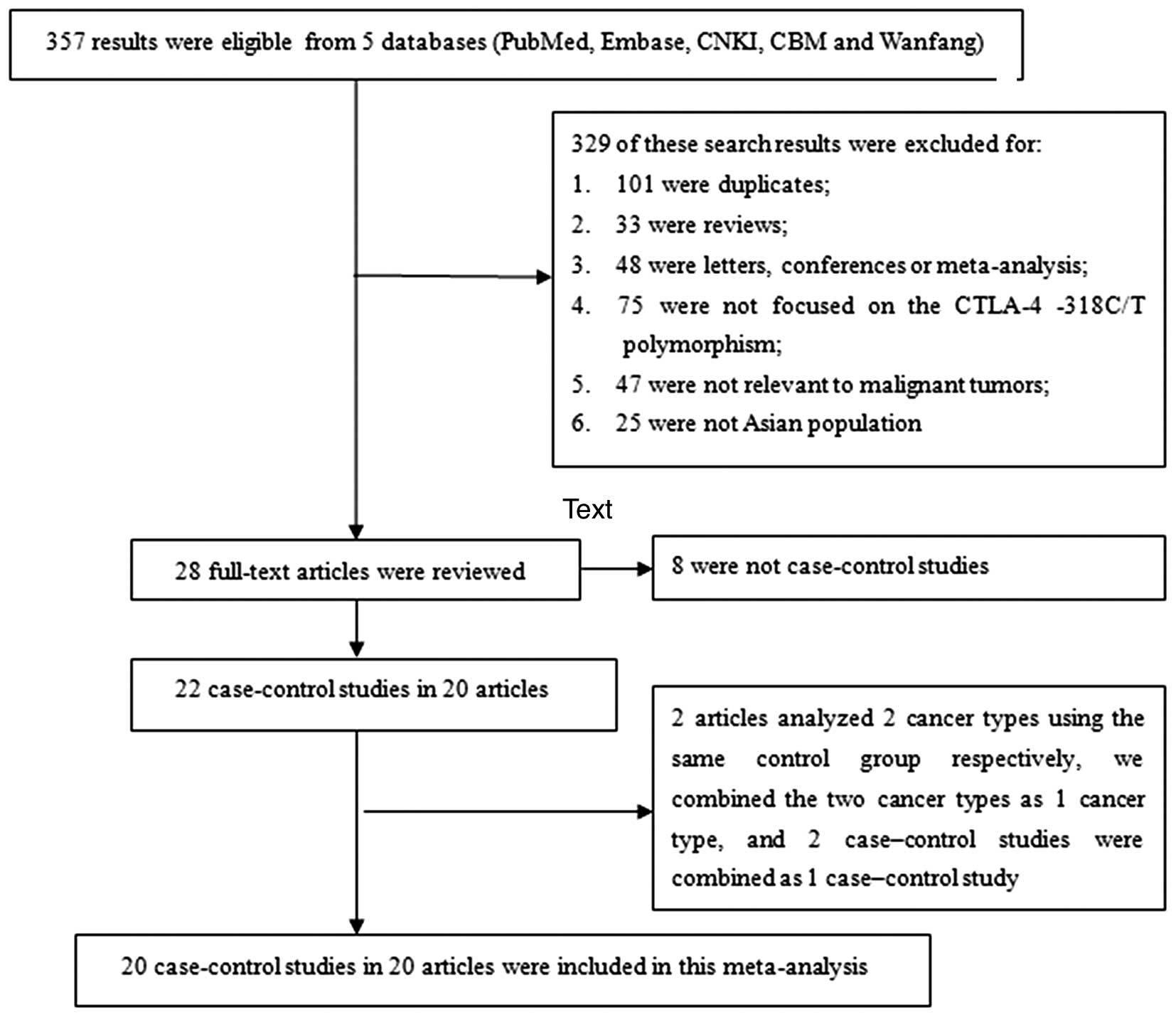
A total of 357 results were preliminarily identified
to meet the inclusion criteria of the present study. Through an
initial screening of titles, key words and abstracts, 329 of these
search results were excluded (101 were duplicates; 33 were reviews;
48 were letters, conferences or meta-analysis; 75 were not focused
on the CTLA-4 −318C/T polymorphism; 47 were not relevant to
malignant tumors, and 25 were not researching an Asian population).
A total of 28 full-text studies were then reviewed and an
additional 8 studies were excluded for not being a case-control
study. Thus, 20 articles with a total of 3,539 cases and 4,690
controls were included in the final data extraction (7–26). There
were two articles that analyzed two cancer types (colorectal cancer
and gastric cancer) using the same control group (11,12). We
combined the two cancer types into one (gastrointestinal cancer),
and therefore, the two case-controlled studies were combined.
Finally, there were 20 case-controlled studies included in the
meta-analysis, Fig. 1 shows the
detailed procedure for screening eligible studies. The publication
years ranged from 2006 to 2015. Among the 20 case-controlled
studies, 13 evaluated the relevant correlation in a Chinese
population (7,9,10,12,15–17,19,21,23–26), 4 studies in an Iranian population
(8,11,13,14), and 3 researched an Indian population
(18,20,22). All
enrolled studies showed that the genotypes in the healthy control
group did not deviate from the HWE, (all P>0.05). The
characteristics of each study are listed in Table I, and the distributions of genotypes
and alleles in CTLA-4 −318C/T polymorphisms are listed in Table II.
 | Table II.Distributions of genotypes and
alleles in cytotoxic T-lymphocyte antigen 4 −318C/T
polymorphisms. |
Table II.
Distributions of genotypes and
alleles in cytotoxic T-lymphocyte antigen 4 −318C/T
polymorphisms.
|
| Case | Control |
|---|
|
|
|
|
|---|
| First author | CC | TC | TT | C | T | CC | TC | TT | C | T | Refs. |
| Cheng | 59 | 3 | 0 | 121 | 3 | 209 | 40 | 1 | 458 | 42 |
(7) |
| Erfani | 244 | 38 | 1 | 526 | 40 | 206 | 31 | 4 | 443 | 39 |
(8) |
| Wang | 84 | 33 | 0 | 201 | 33 | 129 | 19 | 0 | 277 | 19 |
(9) |
| Su | 105 | 38 | 1 | 248 | 40 | 306 | 67 | 5 | 679 | 77 | (10) |
| Hadinia | 129 | 24 | 1 | 282 | 26 | 159 | 24 | 4 | 342 | 32 | (11) |
| Li | 99 | 17 | 5 | 215 | 27 | 206 | 27 | 3 | 439 | 33 | (12) |
| Khaghanzadeh | 107 | 17 | 2 | 231 | 21 | 105 | 16 | 1 | 226 | 18 | (13) |
| Rahimifar | 51 | 3 | 0 | 105 | 3 | 89 | 20 | 1 | 198 | 22 | (14) |
| Liu | 175 | 77 | 15 | 427 | 107 | 195 | 80 | 7 | 470 | 94 | (15) |
| Jiang | 75 | 24 | 1 | 174 | 26 | 92 | 8 | 0 | 192 | 8 | (16) |
| Yang | 149 | 65 | 9 | 363 | 83 | 210 | 85 | 7 | 505 | 99 | (17) |
| Gokhale | 93 | 7 | 0 | 193 | 7 | 94 | 7 | 0 | 195 | 7 | (18) |
| Feng | 213 | 89 | 6 | 515 | 101 | 249 | 102 | 11 | 600 | 124 | (19) |
| Bharti | 112 | 8 | 0 | 232 | 8 | 170 | 10 | 0 | 350 | 10 | (20) |
| Liu | 222 | 64 | 5 | 508 | 74 | 222 | 73 | 5 | 517 | 83 | (21) |
| Jaiswal | 106 | 89 | 5 | 301 | 99 | 112 | 81 | 7 | 305 | 95 | (22) |
| Xiong | 232 | 127 | 6 | 591 | 139 | 316 | 104 | 1 | 736 | 106 | (23) |
| Hui | 48 | 31 | 7 | 127 | 45 | 84 | 25 | 3 | 193 | 31 | (24) |
| Cheng | 88 | 36 | 1 | 212 | 38 | 222 | 73 | 5 | 517 | 83 | (25) |
| Wang | 183 | 97 | 12 | 463 | 121 | 266 | 85 | 4 | 617 | 93 | (26) |
Quantitative synthesis
The summary of results from the meta-analysis of the
association between CTLA-4 −318C/T polymorphisms and malignant
tumor in Asian populations is shown in Table III. The heterogeneity of the dominant
model (TT+TC vs. CC), heterozygous model (TC vs. CC) and allele
model (T vs. C) were performed for the 20 case-controlled studies
and there was statistical heterogeneity among these models
(Table III). However, the
heterogeneity of the recessive model (TT vs. TC+CC) and homozygous
model (TT vs. CC) were performed for 17 case-control studies (there
were three case-control studies excluded by the STATA software
automatically for lack of homozygote genotype TT both in cases and
controls) and there was no statistical heterogeneity in the two
models (Table III). Data from the
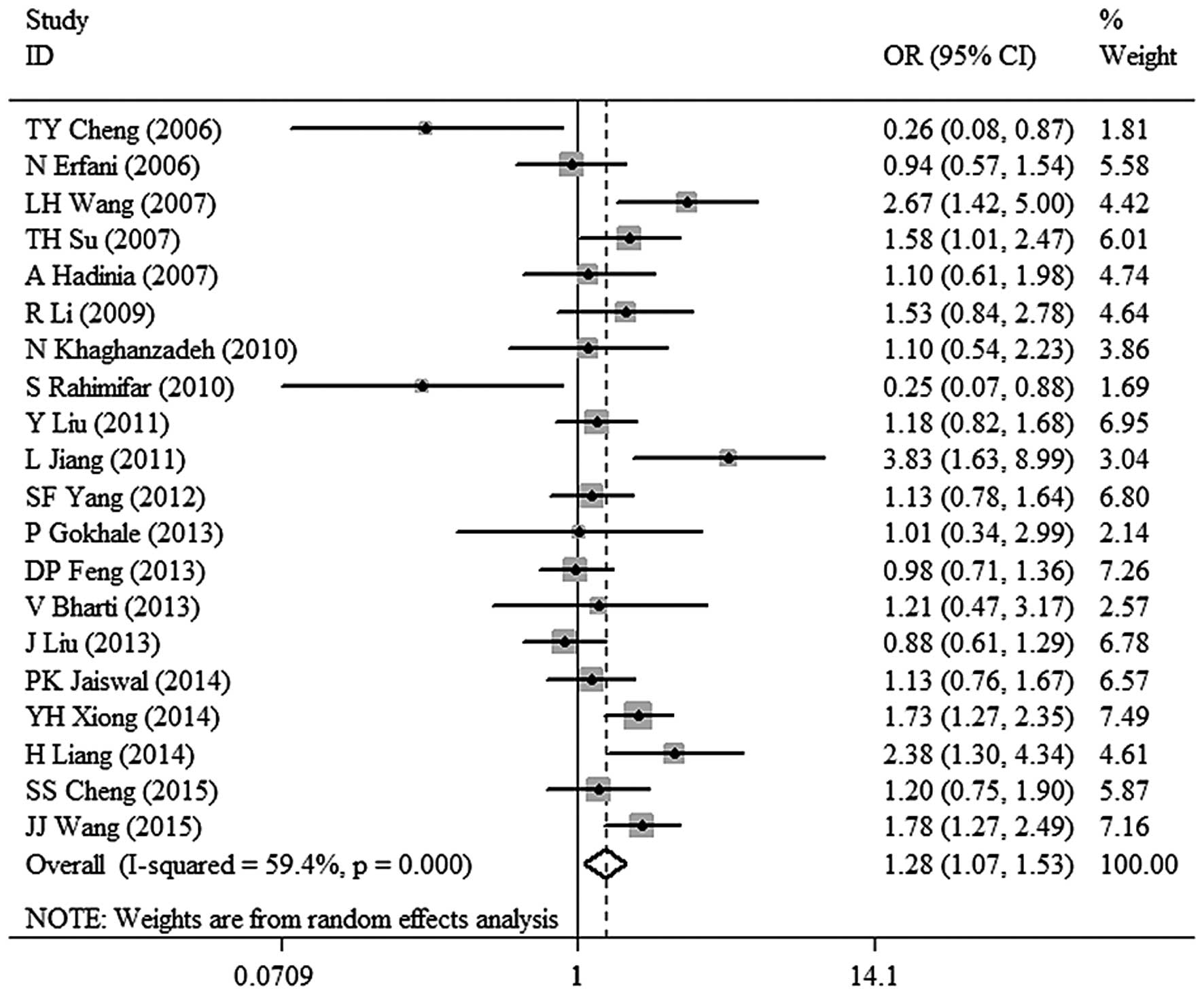
meta-analysis for TT+TC vs. CC was as follows: χ2,
46.84; df=19; P=0.0004 and I2=59.4%. A random-effects
model was used to analyze the data. A significant association
between CTLA-4 −318C/T polymorphism and malignant tumor risk (OR,
1.28; 95% CI, 1.07–1.53; P=0.008) was observed in dominant model
(Fig. 2). The results also revealed an
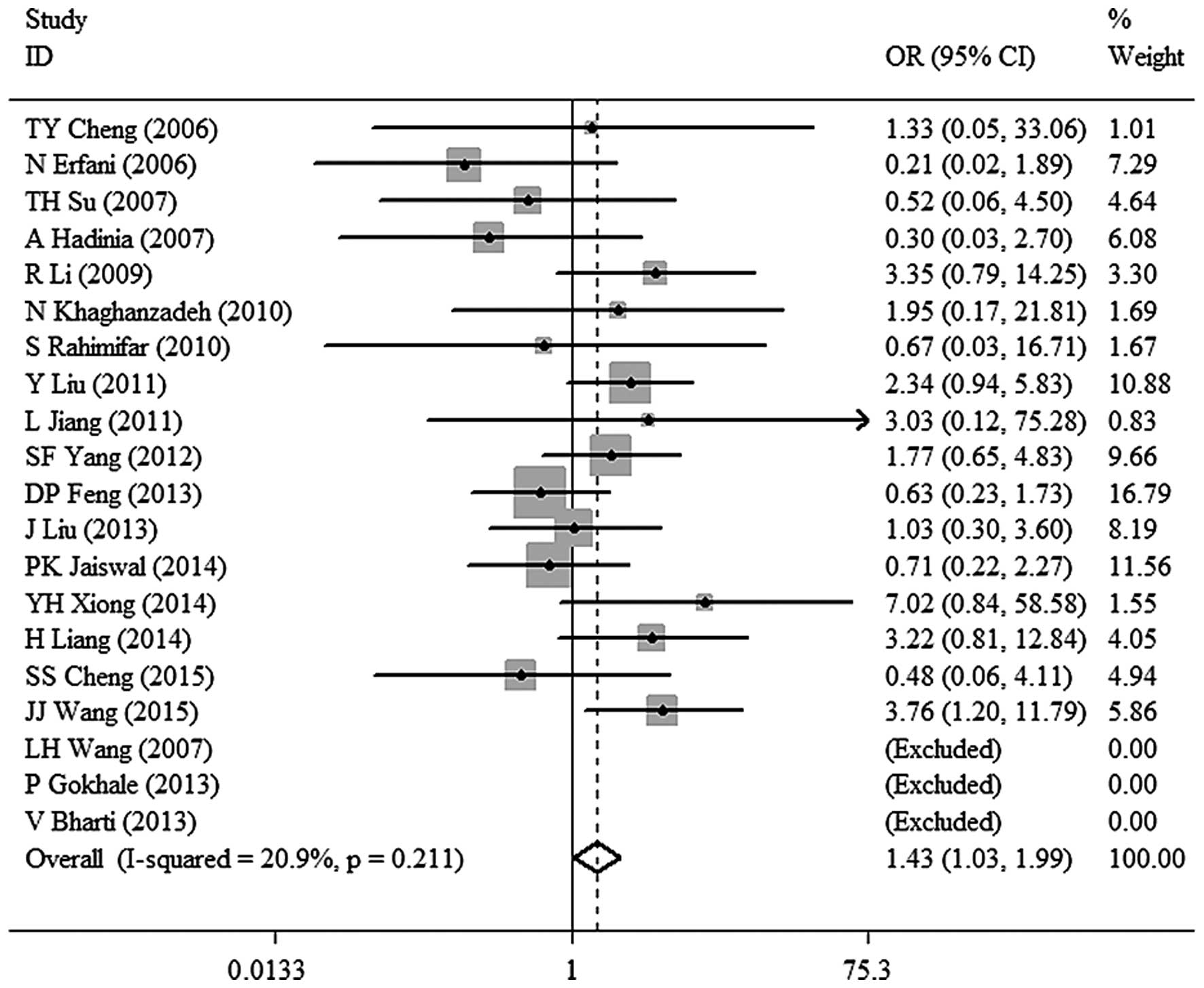
association between CTLA-4 −318C/T polymorphism and malignant tumor
risk in the Asian population under other models [TT vs. TC+CC: OR,
1.43; 95% CI, 1.03–1.99; P=0.032 (Fig.
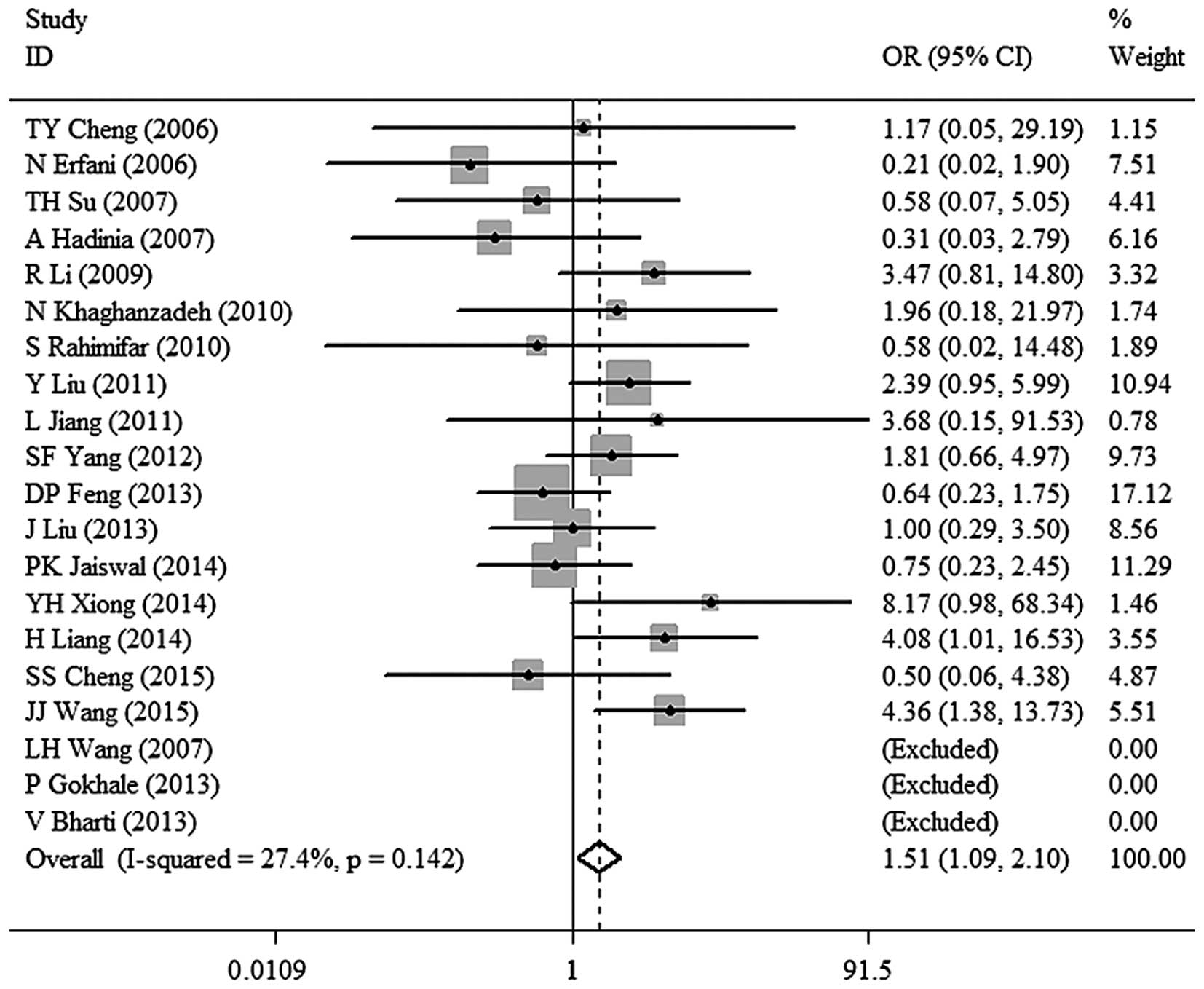
3). TT vs. CC: OR, 1.51; 95% CI, 1.09–2.10; P=0.014 (Fig. 4). TC vs. CC: OR, 1.26; 95% CI,
1.06–1.50; P=0.008 (Fig. 5). T vs. C:
OR, 1.25; 95% CI, 1.05–1.47; P=0.010 (Fig.
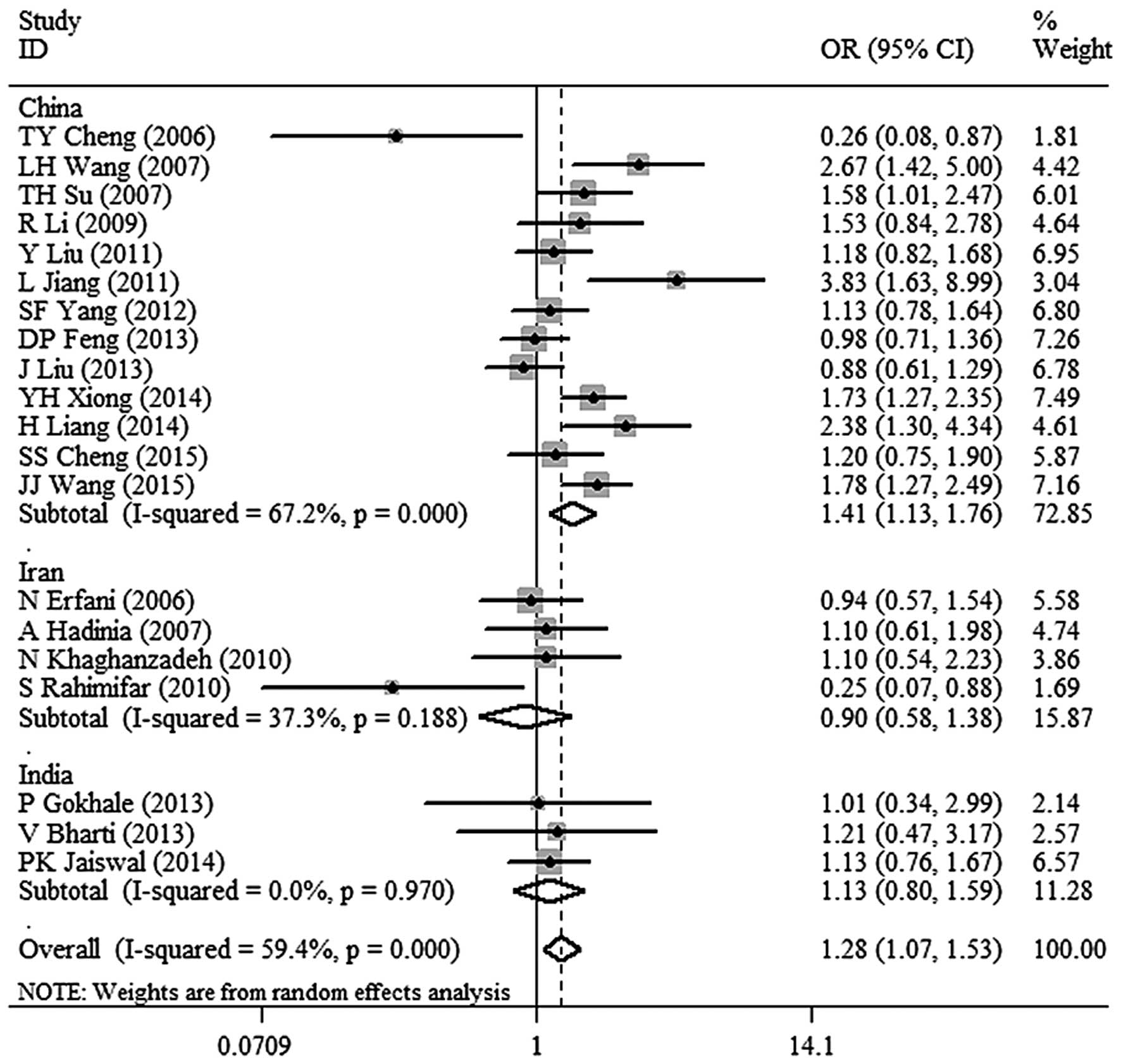
6)]. In the subgroup analysis, a significant association
between CTLA-4 −318C/T polymorphism and malignant tumor risk in the
Chinese population was identified (OR, 1.41; 95% CI, 1.13–1.76;
P=0.002 for TT+TC vs. CC; Fig. 7). In
addition, there was no association between CTLA-4 −318C/T
polymorphism and malignant tumor risk in the populations of other
countries (Fig. 7).
 | Table III.Summary of total pooled results from
different comparative genetic models. |
Table III.
Summary of total pooled results from
different comparative genetic models.
|
|
|
| Test of
heterogeneity |
|---|
|
|
|
|
|
|---|
| Genetic models | OR (95% CI) |
P-valuea | I2
(%)b | P-value | Model |
|---|
| TT+TC vs. CC | 1.28
(1.07–1.53) | 0.008 | 59.4 |
0.0004 | Random |
| TT vs. TC+CC | 1.43
(1.03–1.99) | 0.032 | 20.9 | 0.211 | Fixed |
| TT vs. CC | 1.51
(1.09–2.10) | 0.014 | 27.4 | 0.142 | Fixed |
| TC vs. CC | 1.26
(1.06–1.50) | 0.008 | 53.6 | 0.002 | Random |
| T vs. C | 1.25
(1.05–1.47) | 0.010 | 62.3 |
0.0004 | Random |
Publication bias
No publication bias was detected in either the
Begg's funnel plot (data not shown) or Egger's test for TT+TC vs.
CC (t=−0.64; P>|=0.533). The relative symmetrical distribution
for the Begg's funnel plot (data not shown) or Egger's test
(t=−0.58; P>|=0.570) indicated there was no publication bias for
allele model (T vs. C).
Discussion
In recent years, an increasing number of studies
have revealed the effect of gene polymorphisms in tumorigenesis or
the susceptibility to malignant tumors. Tumor immunity is
increasing as a focus in malignant tumors research. CTLA-4, an
important mediator in T cell proliferation and activation, is
important in cancer immunosurveillance (28). Previous studies demonstrated that
variants of CTLA-4 −318C/T may increase susceptibility to malignant
tumors in Asian populations (7,14,29). However, negative results have been
reported in a previous study (16). To
date, no meta-analysis has been conducted to comprehensively
evaluate the association between the CTLA-4 −318C/T polymorphism
and malignant tumor risk in Asian populations. Thus, the current
study performed a meta-analysis to explore the exact associations
between the CTLA-4 −318C/T polymorphism and malignant tumor risk in
an Asian population.
The results of our meta-analysis indicated that
CTLA-4 −318C/T polymorphism was significantly association with the
risk of developing a malignant tumor. The TT+TC genotype and the TC
heterozygote carriers exhibited higher malignant tumor incident
risks when compared to the CC genotype carriers (TT+TC vs. CC: OR,
1.28; 95% CI, 1.07–1.53; P=0.008. TC vs. CC: OR, 1.26; 95% CI,
1.06–1.50; P=0.008). When the comparisons were conducted between
the TT and CC homozygotes or between the T and C alleles, there
remained a significant effect from the TT homozygote or the T
allele on the risk of malignant tumor, respectively (TT vs. CC: OR,
1.51; 95% CI, 1.09–2.10; P=0.014. T vs. C: OR, 1.25; 95% CI,
1.05–1.47; P=0.010). The analysis reveals that CTLA-4 −318C/T
genetic variants may be an important factor in the development and
progression of malignant tumors in Asian populations, and
individuals with the variant T allele have a ~25% higher risk of
developing malignant tumors. In the subgroup analysis, we found
that the dominant models (TT+TC vs. CC) had significantly increased
the risk of malignant tumor in China (OR, 1.41; 95% CI, 1.13–1.76).
In addition, there were no such correlation in Iran and India,
indicating that country differences may be a potential factor of
heterogeneity for this correlation.
However, the results from current meta-analysis must
be interpreted with caution, as some limitations should be
considered. First, for the Iran and India study populations, only 5
and 3 case-control studies were included in each, respectively.
Second, only published studies in the selected databases were
included, and some unpublished studies with null results were
ignored. Third, the studies included in the meta-analysis were
lacking in original information concerning environmental and
lifestyle factors, which prevented us from further performing
subgroup analysis according to these variables. All of these
limitations may have limited power to reveal a credible association
and distort the results. Hence, we addressed the heterogeneity and
publication bias through rigorously selecting case-control studies,
using a statistical approach to analyze the inconsistent data of
selected studies, contributing to a more reliable association
between the CTL-4 −318C/T polymorphism and malignant tumor risk in
Asian populations. Significant publication bias was not detected
for the gene polymorphisms, indicating the reliability of the
results from this meta-analysis.
In conclusion, the results from the meta-analysis
suggest that the CTL-4 −318 C/T polymorphisms, may be a risk factor
for the development of malignant tumors in Asian populations.
However, this result should be interpreted with caution, because
the eligible studies are only from 3 Asian countries, and the
studies contained no more than five individuals, except for those
originating from China. A subgroup analysis was performed by
country, and the dominant model (TT+TC vs. CC) revealed a
significantly increased risk of malignant tumors in the Chinese
population (OR, 1.41; 95% CI, 1.13–1.76; P=0.002). However, the
analysis did not reveal any association between CTL-4 −318 C/T
polymorphisms and the risk of malignant tumor in the Iranian and
Indian populations. The fact that there were not sufficient numbers
of studies from other Asian nations may distort our conclusion.
More intensive studies based on individuals from additional
countries in Asia are required to further reveal the precise
association between CTL-4 −318C/T polymorphisms and malignant tumor
risk in Asian populations.
References
|
1
|
Bredberg A: Cancer: More of polygenic
disease and less of multiple mutations? A quantitative viewpoint.
Cancer. 117:440–445. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wong YK, Chang KW, Cheng CY and Liu CJ:
Association of CTLA-4 gene polymorphism with oral squamous cell
carcinoma. J Oral Pathol Med. 35:51–54. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ramakrishnan R and Gabrilovich DI: Novel
mechanism of synergistic effects of conventional chemotherapy and
immune therapy of cancer. Cancer Immunol Immunother. 62:405–410.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhang Y, Zhang J, Deng Y, Tian C, Li X,
Huang J and Fan H: Polymorphisms in the cytotoxic T-lymphocyte
antigen 4 gene and cancer risk: A meta-analysis. Cancer.
117:4312–4324. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ghaderi A: CTLA4 gene variants in
autoimmunity and cancer: A comparative review. Iran J Immunol.
8:127–149. 2011.PubMed/NCBI
|
|
6
|
Sun T, Zhou Y, Yang M, Hu Z, Tan W, Han X,
Shi Y, Yao J, Guo Y, Yu D, et al: Functional genetic variations in
cytotoxic T-lymphocyte antigen 4 and susceptibility to multiple
types of cancer. Cancer Res. 68:7025–7034. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Cheng TY, Lin JT, Chen LT, Shun CT, Wang
HP, Lin MT, Wang TE, Cheng AL and Wu MS: Association of T-cell
regulatory gene polymorphisms with susceptibility to gastric
mucosa-associated lymphoid tissue lymphoma. J Clin Oncol.
24:3483–3489. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Erfani N, Razmkhah M, Talei AR, Pezeshki
AM, Doroudchi M, Monabati A and Ghaderi A: Cytotoxic T lymphocyte
antigen-4 promoter variants in breast cancer. Cancer Genet
Cytogenet. 165:114–120. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang L and Li D, Fu Z, Li H, Jiang W and
Li D: Association of CTLA-4 gene polymorphisms with sporadic breast
cancer in Chinese Han population. BMC Cancer. 7:1732007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Su TH, Chang TY, Lee YJ, Chen CK, Liu HF,
Chu CC, Lin M, Wang PT, Huang WC, Chen TC, et al: CTLA-4 gene and
susceptibility to human papillomavirus-16-associated cervical
squamous cell carcinoma in Taiwanese women. Carcinogenesis.
28:1237–1240. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hadinia A, Hossieni SV, Erfani N,
Saberi-Firozi M, Fattahi MJ and Ghaderi A: CTLA-4 gene promoter and
exon 1 polymorphisms in Iranian patients with gastric and
colorectal cancers. J Gastroenterol Hepatol. 22:2283–2287. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li R, Xia B, Xiao H, Jiang Y and Zhou F:
Association between CTLA-4 gene promoter region polymorphisms at
positions −1661 and −318 and gastric cancer. Clin J Gastroenterol.
14:332–336. 2009.
|
|
13
|
Khaghanzadeh N, Erfani N, Ghayumi MA and
Ghaderi A: CTLA4 gene variations and haplotypes in patients with
lung cancer. Cancer Genet Cytogenet. 196:171–174. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Rahimifar S, Erfani N, Sarraf Z and
Ghaderi A: ctla-4 gene variations may influence cervical cancer
susceptibility. Gynecol Oncol. 119:136–139. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu Y, He Z, Feng D, Shi G, Gao R, Wu X,
Song W and Yuan W: Cytotoxic T-lymphocyte antigen-4 polymorphisms
and susceptibility to osteosarcoma. DNA Cell Biol. 30:1051–1055.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Jiang L, Luo RY, Zhang W, Wang LR, Wang F
and Cheng YX: Single nucleotide polymorphisms of CTLA4 gene and
their association with human cervical cancer. Zhonghua Yi Xue Yi
Chuan Xue Za Zhi. 28:313–317. 2011.(In Chinese). PubMed/NCBI
|
|
17
|
Yang S, Wang C, Zhou Y, Sun G, Zhu D and
Gao S: Cytotoxic T-lymphocyte antigen-4 polymorphisms and
susceptibility to Ewing's sarcoma. Genet Test Mol Biomarkers.
16:1236–1240. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gokhale P, Kerkar S, Tongaonkar H, Salvi V
and Mania-Pramanik J: CTLA-4 gene polymorphism at position +49
A>G in exon 1: A risk factor for cervical cancer in Indian
women. Cancer Genet. 206:154–161. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Feng D, Yang X, Li S, Liu T, Wu Z, Song Y,
Wang J, Gao W, Huang Q, Huang W, et al: Cytotoxic T-lymphocyte
antigen-4 genetic variants and risk of Ewing's sarcoma. Genet Test
Mol Biomarkers. 17:458–463. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bharti V, Mohanti BK and Das SN:
Functional genetic variants of CTLA-4 and risk of tobacco-related
oral carcinoma in high-risk North Indian population. Hum Immunol.
74:348–352. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liu J, Liu J, Song B, Wang T, Liu Y, Hao J
and Yu J: Genetic variations in CTLA-4, TNF-α, and LTA and
susceptibility to T-cell lymphoma in a Chinese population. Cancer
Epidemiol. 37:930–934. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Jaiswal PK, Singh V and Mittal RD:
Cytotoxic T lymphocyte antigen 4 (CTLA4) gene polymorphism with
bladder cancer risk in North Indian population. Mol Biol Rep.
41:799–807. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xiong YH, He L and Fei J: Genetic
variations in cytotoxic T-lymphocyte antigen-4 and susceptibility
to cervical cancer. Int Immunopharmacol. 18:71–76. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hui L, Lei Z, Peng Z, Ruobing S and
Fenghua Z: Polymorphism analysis of CTLA-4 in childhood acute
lymphoblastic leukemia. Pak J Pharm Sci. 27(Suppl 4): 1005–1013.
2014.PubMed/NCBI
|
|
25
|
Cheng S, Li J, Liu W, Liu C, Su L, Liu X,
Guo L, Ma Y, Song B and Liu J: LTA + 252A > G polymorphism is
associated with risk of nasal NK/T-cell lymphoma in a Chinese
population: A case-control study. BMC Cancer. 15:4802015.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang JJ, Wang ZB and Huang WF: The
correlation of cytotoxic T lymphocyte antigen 4 polymorphisms with
occurrence of cervical cancer. Pract J Clin Med. 12:77–80.
2015.
|
|
27
|
Jackson D, White IR and Riley RD:
Quantifying the impact of between-study heterogeneity in
multivariate meta-analyses. Stat Med. 31:3805–3820. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Leach DR, Krummel MF and Allison JP:
Enhancement of antitumor immunity by CTLA-4 blockade. Science.
271:1734–1736. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Xu HB, Yang H, Liu T and Chen H:
Association of CTLA4 gene polymorphism (rs5742909) with cervical
cancer: A meta-analysis. Tumour Biol. 35:1605–1608. 2014.
View Article : Google Scholar : PubMed/NCBI
|