Introduction
Tuberculosis (TB), caused by Mycobacterium
tuberculosis (MTB) infection, is a public health problem
globally (1,2). According to the WHO report on the
worldwide control of TB, approximately 8.6 million new cases
occurred in 2012 (3). Although almost
33% of the population is infected with TB, only 5–10% of infected
cases develop active TB (3), which
suggests a major role of genetic factors in host immunity.
Interferon-γ (IFNγ) is produced and released by host
cells in response to the presence of numerous pathogens (4). It plays a key role in macrophage
activation during MTB infection (5).
Individuals defective in the genes for IFNγ or IFNγ
receptor (IFNγR) have been indicated to be susceptible for
mycobacterial infections including MTB (6). Previously, we showed an association
between IFNγ and IFNγR variants and risk of pulmonary
TB (PTB) (7,8).
Interferon-induced transmembrane protein-3 (IFITM3)
is a double transmembrane protein that can be upregulated by IFNs
and participates in INF-triggered processes, such as homotypic cell
adhesion, anti-proliferative activities in tumor pathogenesis, and
the innate immune response to virus infections (9–13). The
IFITM3 gene is mapped to an IFITM gene cluster on
chromosome 11p15.5 (14). In a genome
wide scan, Stein et al (15)
identified that one of the TB-linked loci was located in this
chromosome region. To the best of our knowledge, there is only one
report regarding the impact of IFITM gene polymorphisms on
the risk of TB (16). Therefore, the
present study aimed to examine the possible associations between
polymorphisms of IFITM3 gene and susceptibility to PTB in a
sample of Iranian population.
Materials and methods
Patients
This case-control study was performed on 188 PTB
patients and 169 age- and gender-matched healthy individuals. The
enrollment process and study design are described elsewhere
(17–23). Briefly, the cases were chosen from PTB
patients admitted to a University-Affiliated Hospital (Bou-Ali
Hospital, Zahedan, Iran, referral center for TB) with no clinical
symptoms or family history of TB. TB was diagnosed by clinical
symptoms, posterior-anterior chest radiography, presence of
acid-fast-bacilli on a sputum smear, and culturing MTB organisms
from a specimen taken from the patient and response to therapy, as
described previously (20–23). The project was approved by the local
Ethics Committee of the Zahedan University of Medical Sciences and
informed consent was obtained from all subjects. DNA was extracted
from whole blood samples using the salting out method (24).
Genotyping
Genotyping of IFTIM3 rs7478728 and rs3888188
polymorphisms was performed using the polymerase chain reaction
(PCR)-restriction fragment length polymorphism method. The primer
sequences are shown in Table I. In
each 0.20 ml PCR reaction tube, 1 µl of genomic DNA (~100 ng/ml), 1
µl of each primer (10 µM), 10 µl of 2X Prime Taq Premix (Genet Bio
Inc., Daejeon, Korea), and 7 µl ddH2O were added.
 | Table I.Primer sequences used for the
detection of IFITM3 gene polymorphisms. |
Table I.
Primer sequences used for the
detection of IFITM3 gene polymorphisms.
| Polymorphisms | Sequence (5′→3′) | Restriction
enzyme | Product size
(bp) | Anneling temperature
(°C) |
|---|
| rs7478728 C>T | F:
TTAGCCCTCAGCCCCTCTTTCGTC | Alw26I | T allele: 217,
29 | 53 |
|
| R:
CTGTTGACAGGAGAGAAGAAGGTT |
| C allele: 246 |
|
| rs3888188 T>G | F:
CACAGTGAGGGTTATGGGAGAC | Hpy188I | G allele: 340,
246 | 54 |
|
| R:
ACTGTTGACAGGAGAGAAGAAGGTT |
| T allele: 586 |
|
Amplification was carried out with an initial
denaturation step of 5 min at 95°C followed by 30 cycles of 30 sec
at 95°C, annealing at 53°C for rs7478728 and 54°C for rs3888188 for
30 sec and extension at 72°C for 30 sec. Final extension was
performed at 72°C for 5 min.
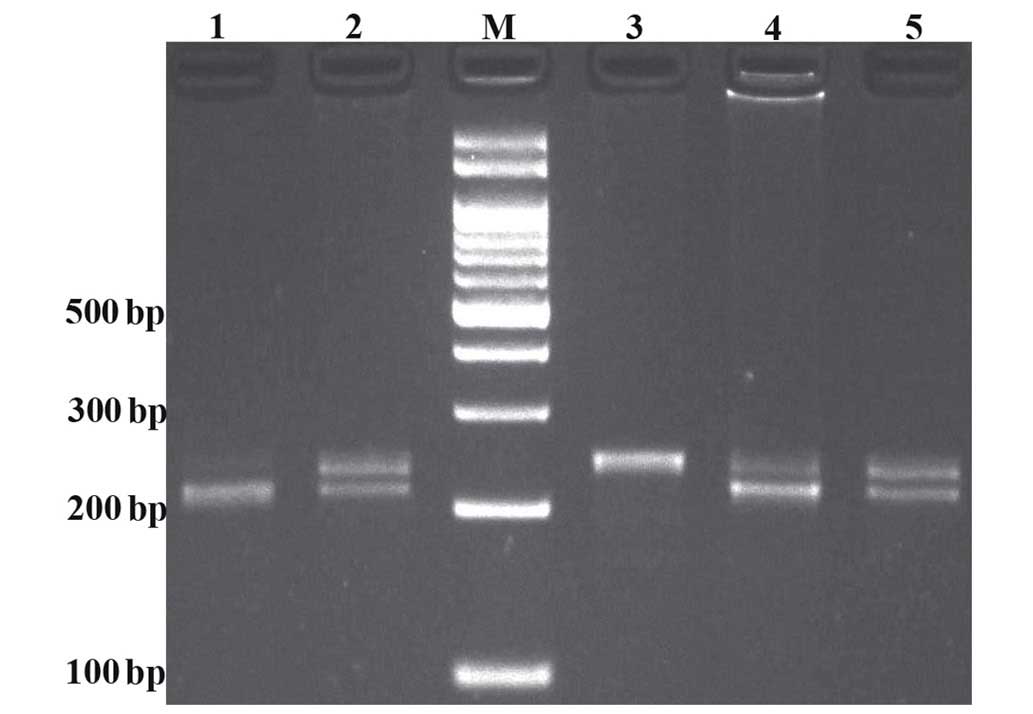
For rs7478728, 10 µl of PCR products was digested
with Alw26I restriction enzyme (Fermentas, Glen Burnie, MD,
USA) and then separated by electrophoresis in 2% agarose gels. The
C allele was undigested (246-bp), while the T allele was digested
and produced 217- and 29-bp fragments (Fig. 1).
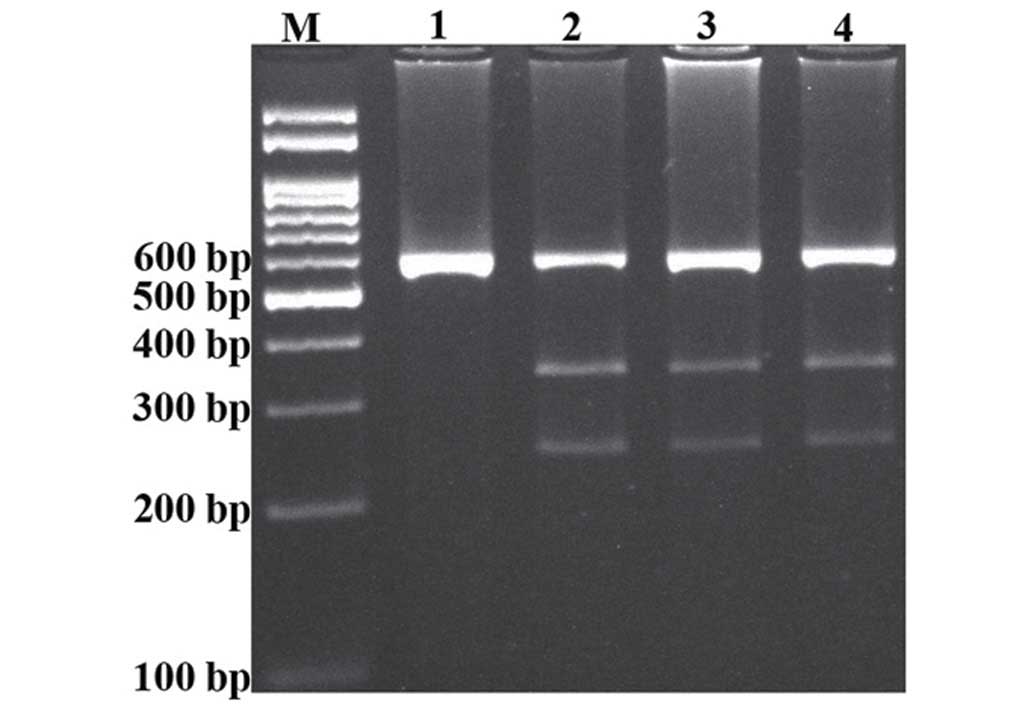
For the rs3888188 variant, the PCR products were
digested with Hpy188I restriction enzyme (Fermentas). The T
allele was undigested (586-bp), while the G allele digested and
produced 340- and 246-bp fragments (Fig.
2).
To confirm the genotyping quality for each
polymorphism, ~20% of random samples were regenotyped and the
findings confirmed the preceding genotyping results.
Statistical analysis
Statistical analysis of the data was performed using
the SPSS 20.0 software (IBM SPSS, Armonk, NY, USA). The analysis
was performed by the χ2 test or independent sample
t-test according to the data. The associations between genotypes
and PTB were calculated by computing the odds ratio (OR) and 95%
confidence interval (CI) from logistic regression analyses. The
Hardy-Weinberg equilibrium (HWE) for cases and controls was
calculated by χ2 test. P<0.05 was considered to
indicate a statistically significant difference.
Results
Patient characteristics
A total of 357 subjects including 188 confirmed PTB
patients (73 males, 115 females; ages 50.0±19.5 years) and 169
unrelated healthy subjects (75 males, 94 females; ages 47.9±15.0
years) were assessed. There was no statistically significant
difference among the groups regarding gender and age
(P>0.05).
Association between the polymorphisms
and PTB risk
Genotypes and allele frequencies of the IFITM3
polymorphisms are shown in Table II.
Regarding rs7478728 polymorphism, the findings indicated that this
variant was not associated with the risk of PTB in codominant
(OR=1.32, 95% CI: 0.80–2.17, P=0.337, CT vs. CC; OR=2.04, 95% CI:
0.63–6.61, P=0.362, TT vs. CC), dominant (OR=1.35, 95% CI:
0.82–2.21, P=0.293, CT+TT vs. CC), and recessive (OR=1.65, 95% CI:
0.54–5.02, P=0.538 TT vs. CC+CT) inheritance model tested. The T
allele was not associated with the risk of PTB (OR=1.16, 95% CI:
0.86–1.56, P=0.377) compared to C allele.
 | Table II.Frequency distribution of
IFITM3 rs7478728 and rs3888188 gene polymorphisms in PTB and
controls. |
Table II.
Frequency distribution of
IFITM3 rs7478728 and rs3888188 gene polymorphisms in PTB and
controls.
| Polymorphisms | Case n (%) | Control n (%) | OR (95% CI) | P-value |
|---|
| rs7478728 C>T |
|
|
|
|
| Codominant |
|
|
|
|
| CC | 38 (20.2) | 43 (25.4) | 1.00 | – |
| CT | 141 (75.0) | 121 (71.6) | 1.32 (0.80–2.17) | 0.337 |
| TT | 9 (4.8) | 5 (3.0) | 2.04 (0.63–6.61) | 0.362 |
| Dominant |
|
|
|
|
| CC | 38 (20.2) | 43 (25.4) | 1.00 | – |
|
CT+TT | 150 (79.8) | 126 (74.6) | 1.35 (0.82–2.21) | 0.293 |
| Recessive |
|
|
|
|
|
CC+CT | 179 (95..2) | 164 (97.0) | 1.00 |
|
| TT | 9 (4.8) | 5 (3.0) | 1.65 (0.54–5.02) | 0.538 |
| Allele |
|
|
|
|
| C | 217 (57.7) | 207 (61.2) | 1.00 | – |
| T | 159 (42.3) | 131 (38.8) | 1.16
(0.86–1.56) | 0.377 |
| rs3888188
T>G |
|
|
|
|
| TT | 139 (73.9) | 148 (87.6) | 1.00 | – |
| TG | 49 (26.1) | 21 (12.4) | 2.48
(1.42–4.35) | 0.002 |
| GG | 0 (0.0) | 0 (0.0) | – | – |
| Allele |
|
|
|
|
| T | 327 (87.0) | 317 (93.8) | 1.00 | – |
| G | 49 (13.0) | 21 (6.2) | 2.26
(1.33–3.86) | 0.003 |
Regarding the rs3888188 variant, the results
revealed that TG genotype significantly increased the risk of PTB
compared to TT genotype (OR=2.48, 95% CI: 1.42–4.35; P=0.002).
Similarly, the G allele increased the risk of PTB in comparison
with T allele (OR=2.26, 95% CI: 1.33–3.86; P=0.003).
The interaction of the two variants of the
IFITM3 gene was analyzed (Table
III) and the findings suggested that the CT/TG genotype
significantly increased the risk of PTB compared to CC/TT genotype
(P=0.006).
 | Table III.Interaction of IFITM3
rs7478728 and rs3888188 gene polymorphisms on PTB risk. |
Table III.
Interaction of IFITM3
rs7478728 and rs3888188 gene polymorphisms on PTB risk.
| rs7478728
C>T | rs3888188
T>G | Case n (%) | Control n (%) | OR (95% CI) | P-value |
|---|
| CC | TT | 29
(15.4) | 39
(23.1) | 1.00 | – |
| CT | TT | 104 (55.3) | 104 (61.5) | 1.34
(0.77–2.34) | 0.329 |
| CT | TG | 37
(19.7) | 17
(10.1) | 2.93
(1.38–6.19) | 0.006 |
| TT | TT | 6
(3.2) | 5
(3.0) | 1.61
(0.45–5.81) | 0.524 |
| CC | TG | 9
(4.8) | 4
(2.4) | 3.03
(0.85–10.80) | 0.128 |
| TT | TG | 3
(1.6) | 0
(0.0) | – | – |
The genotype of IFITM3 rs7478728 variant in
cases and controls was not in HWE (χ2=54.1, P<0.001
and χ2=43.64, P<0.001, respectively). Regarding the
IFITM3 rs3888188 variant, the genotype in controls
(χ2=0.74, P=0.389) but not in cases (χ2=4.22,
P=0.040) was in HWE.
Discussion
In the present study, we examined the possible
association between IFITM3 rs7478728 and rs3888188
polymorphisms and the risk of PTB in a sample of Iranian
population. Our findings did not support an association between
rs7478728 variant and risk of PTB in the population studied.
However, we found that TG genotype as well as G allele of rs3888188
polymorphism significantly increased the risk of PTB. There is only
one study concerning the possible association between IFITM3
variants and risk of TB (16). Shen
et al (16) have found that the
rs3888188 G allele increased the risk of pediatric TB (OR=1.30, 95%
CI: 1.08–1.56; P=0.039). In addition, they found that the rs7478728
T allele was significantly associated with pediatric TB (OR=1.34,
95% CI: 1.07–1.68; P=0.010), but not after Bonferroni correction
(P=0.082). Authors of that study also evaluated the effect of
rs3888188 (−204 T>G) variant on IFITM3 transcription
in vitro and found that the promoter activity of rs3888188 G
allele was lower than that of the T allele. Similarly,
peripheral-blood mononuclear cells carrying the rs3888188 GG
genotype showed a reduced IFITM3 mRNA level compared to
cells carrying TT or GT genotype. It was concluded that the
rs3888188 variant is a functional promoter polymorphism of
IFITM3 that increased the risk of pediatric TB in the Han
Chinese population (16). IFITM3 has
been recognized as a key component of the IFNγ signaling pathway
and downregulation of IFITM3 via siRNA significantly reduced
the antiviral activities of IFNγ by 40–70% (12,13). It is
thus a potential candidate gene for TB susceptibility.
IFITM proteins are key mediators of the host
antiviral response (11–13,25,26). Everitt et al (25) showed that mice lacking IFITM3
gene display fulminant viral pneumonia following infection with a
low-pathogenicity influenza virus. Similarly, in an in vitro
study, an increase in viral replication was observed in the absence
of IFITM3, and re-introduction of IFITM3 limited the replication of
the influenza A virus (25).
One of the limitations of the present study is the
relatively small sample sizes. There is no clear explanation for
deviation from HWE for the IFITM3 rs7478728 variant in our
population. The probable reason may be due to genetic drift.
In conclusion, our findings suggest that
IFITM3 rs3888188 polymorphism significantly increased the
risk of PTB in a sample of Iranian population. Additional studies
with larger sample sizes and diverse ethnicities are necessary to
confirm these findings.
Acknowledgements
The present study was funded by a dissertation
research grant (M.D. thesis of FA no. 7265) from the Zahedan
University of Medical Sciences. The authors would like to thank the
patients and healthy subjects who willingly participated in the
study.
References
|
1
|
Lin PL and Flynn JL: Understanding latent
tuberculosis: A moving target. J Immunol. 185:15–22. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Oxlade O, Schwartzman K, Behr MA,
Benedetti A, Pai M, Heymann J and Menzies D: Global tuberculosis
trends: A reflection of changes in tuberculosis control or in
population health? Int J Tuberc Lung Dis. 13:1238–1246.
2009.PubMed/NCBI
|
|
3
|
Zumla A, George A, Sharma V and Herbert N;
Baroness Masham of Ilton: WHO's 2013 global report on tuberculosis:
Successes, threats, and opportunities. Lancet. 382:1765–1767. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Stark GR: How cells respond to interferons
revisited: From early history to current complexity. Cytokine
Growth Factor Rev. 18:419–423. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lee J and Kornfeld H: Interferon-γ
regulates the death of M. tuberculosis-infected macrophages. J Cell
Death. 3:1–11. 2010.PubMed/NCBI
|
|
6
|
Ottenhoff TH, Kumararatne D and Casanova
JL: Novel human immunodeficiencies reveal the essential role of
type-I cytokines in immunity to intracellular bacteria. Immunol
Today. 19:491–494. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Naderi M, Hashemi M, Rezaei M and Safdari
A: Association of genetic polymorphisms of IFNGR1 with the risk of
pulmonary tuberculosis in Zahedan, Southeast Iran. Tuberc Res
Treat. 2015:2925052015.PubMed/NCBI
|
|
8
|
Hashemi M, Sharifi-Mood B, Nezamdoost M,
Moazeni-Roodi A, Naderi M, Kouhpayeh H, Taheri M and Ghavami S:
Functional polymorphism of interferon-γ (IFN-γ) gene +874T/A
polymorphism is associated with pulmonary tuberculosis in Zahedan,
Southeast Iran. Prague Med Rep. 112:38–43. 2011.PubMed/NCBI
|
|
9
|
Seyfried NT, Huysentruyt LC, Atwood JA
III, Xia Q, Seyfried TN and Orlando R: Up-regulation of NG2
proteoglycan and interferon-induced transmembrane proteins 1 and 3
in mouse astrocytoma: A membrane proteomics approach. Cancer Lett.
263:243–252. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Fan J, Peng Z, Zhou C, Qiu G, Tang H, Sun
Y, Wang X, Li Q, Le X and Xie K: Gene-expression profiling in
Chinese patients with colon cancer by coupling experimental and
bioinformatic genomewide gene-expression analyses: Identification
and validation of IFITM3 as a biomarker of early colon
carcinogenesis. Cancer. 113:266–275. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Brass AL, Huang IC, Benita Y, John SP,
Krishnan MN, Feeley EM, Ryan BJ, Weyer JL, van der Weyden L, Fikrig
E, et al: The IFITM proteins mediate cellular resistance to
influenza A H1N1 virus, West Nile virus, and dengue virus. Cell.
139:1243–1254. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Jiang D, Weidner JM, Qing M, Pan XB, Guo
H, Xu C, Zhang X, Birk A, Chang J, Shi PY, et al: Identification of
five interferon-induced cellular proteins that inhibit west nile
virus and dengue virus infections. J Virol. 84:8332–8341. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Weidner JM, Jiang D, Pan XB, Chang J,
Block TM and Guo JT: Interferon-induced cell membrane proteins,
IFITM3 and tetherin, inhibit vesicular stomatitis virus infection
via distinct mechanisms. J Virol. 84:12646–12657. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lange UC, Saitou M, Western PS, Barton SC
and Surani MA: The fragilis interferon-inducible gene family of
transmembrane proteins is associated with germ cell specification
in mice. BMC Dev Biol. 3:12003. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Stein CM, Zalwango S, Malone LL, Won S,
Mayanja-Kizza H, Mugerwa RD, Leontiev DV, Thompson CL, Cartier KC,
Elston RC, et al: Genome scan of M. tuberculosis infection and
disease in Ugandans. PLoS One. 3:e40942008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Shen C, Wu XR, Jiao WW, Sun L, Feng WX,
Xiao J, Miao Q, Liu F, Yin QQ, Zhang CG, et al: A functional
promoter polymorphism of IFITM3 is associated with susceptibility
to pediatric tuberculosis in Han Chinese population. PLoS One.
8:e678162013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hashemi M, Sharifi-Mood B, Rasouli A,
Amininia S, Naderi M and Taheri M: Macrophage migration inhibitory
factor −173 G/C polymorphism is associated with an increased risk
of pulmonary tuberculosis in Zahedan, Southeast Iran. EXCLI J.
14:117–122. 2015.PubMed/NCBI
|
|
18
|
Naderi M, Hashemi M, Taheri M, Pesarakli
H, Eskandari-Nasab E and Bahari G: CD209 promoter −336 A/G
(rs4804803) polymorphism is associated with susceptibility to
pulmonary tuberculosis in Zahedan, southeast Iran. J Microbiol
Immunol Infect. 47:171–175. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Naderi M, Hashemi M, Pourmontaseri Z,
Eskandari-Nasab E, Bahari G and Taheri M: TIRAP rs8177374 gene
polymorphism increased the risk of pulmonary tuberculosis in
Zahedan, southeast Iran. Asian Pac J Trop Med. 7:451–455. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Naderi M, Hashemi M, Hazire-Yazdi L,
Taheri M, Moazeni-Roodi A, Eskandari-Nasab E and Bahari G:
Association between toll-like receptor2 Arg677Trp and 597T/C gene
polymorphisms and pulmonary tuberculosis in Zahedan, Southeast
Iran. Braz J Infect Dis. 15:516–520. 2013. View Article : Google Scholar
|
|
21
|
Hashemi M, Eskandari-Nasab E,
Moazeni-Roodi A, Naderi M, Sharifi-Mood B and Taheri M: Association
of CTSZ rs34069356 and MC3R rs6127698 gene polymorphisms with
pulmonary tuberculosis. Int J Tuberc Lung Dis. 17:1224–1228. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Naderi M, Hashemi M and Amininia S:
Association of TAP1 and TAP2 Gene Polymorphisms with Susceptibility
to Pulmonary Tuberculosis. Iran J Allergy Asthma Immunol. 15:62–68.
2016.PubMed/NCBI
|
|
23
|
Bahari G, Hashemi M, Taheri M, Naderi M,
Eskandari-Nasab E and Atabaki M: Association of IRGM polymorphisms
and susceptibility to pulmonary tuberculosis in Zahedan, Southeast
Iran. Sci World J. 2012:9508012012. View Article : Google Scholar
|
|
24
|
Hashemi M, Bojd H Hanafi, Nasab E
Eskandari, Bahari A, Hashemzehi NA, Shafieipour S, Narouie B,
Taheri M and Ghavami S: Association of Adiponectin rs1501299 and
rs266729 Gene Polymorphisms With Nonalcoholic Fatty Liver Disease.
Hepat Mon. 13:e95272013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Everitt AR, Clare S, Pertel T, John SP,
Wash RS, Smith SE, Chin CR, Feeley EM, Sims JS, Adams DJ, et al:
MOSAIC Investigators: IFITM3 restricts the morbidity and mortality
associated with influenza. Nature. 484:519–523. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Feeley EM, Sims JS, John SP, Chin CR,
Pertel T, Chen LM, Gaiha GD, Ryan BJ, Donis RO, Elledge SJ, et al:
IFITM3 inhibits influenza A virus infection by preventing cytosolic
entry. PLoS Pathog. 7:e10023372011. View Article : Google Scholar : PubMed/NCBI
|