Introduction
RNA splicing is a fundamental process, which
contributes to the structural and functional complexity of proteins
and influences their regulatory role and tissue specificity
(1,2).
Splicing enhancers in exons are considered to be responsible for
the inclusion of exonic sequences in the gene transcript. There is
growing evidence that polymorphisms in high impact exonic splicing
enhancers (ESEs) strongly influence the activity of
disease-associated genes and modify their association with
different pathological conditions (3).
Bioinformatic resources are available for evaluating the efficiency
of ESEs (4).
It has been previously demonstrated that a major
role of intragenic DNA methylation is associated with the
regulation of alternative splicing (5,6). It should
therefore be expected that polymorphisms, which modify a G or a C
in a CpG dinucleotide, affect the epigenetic profile in exonic
sequences that are most frequently found to be methylated (7). In sites of tentative DNA methylation,
particularly when located in ESEs, this would lead to
allele-specific methylation differences (8). In view of the recently demonstrated
impact of DNA methylation on the splicing process (6), it is also expected that exonic CpG
polymorphisms may further affect splicing. The presence of ESEs and
their relative potential are predictable by bioinformatic analysis.
Recent experimental evidence verified the consistency of the
computational results with experimentally observed exon inclusion
using a minigene (9). It is also
evident that CpG-single nucleotide polymorphisms (SNPs) in
prominent ESEs of disease-associated genes are of particular
importance (10,11). Based on this evidence, various studies
have focused on genetic variations (SNPs) at CpGs, which may be
responsible for predisposition to various pathological conditions,
including type 2 diabetes (T2D) (11,12).
T2D is a metabolic disorder characterized by high
glucose blood levels associated with insulin resistance and
relatively low levels of insulin. Together with obesity, blood
hypertension and hyperlipidemia, T2D is one of the most frequent
conditions associated with metabolic syndrome, which is currently
considered a major cause for cardiovascular disease. Genetic
association studies for the identification of SNPs associated with
these diseases are performed by genome-wide association study
(13). However, the distinct
epigenetic/splicing-associated role of these SNPs has not, to the
best of our knowledge, been addressed, despite previous evidence
that the expression of different splicing isoforms is a major
factor for disease association even in the heterozygous state
(14,15).
In view of the above, a bioinformatic analysis of
synonymous SNPs in all T2D-associated genes (11) was performed in the present study to
identify prominent CpG-SNPs, which introduce major modifications in
the splicing potential of exonic sequences, which may be
responsible for T2D. This analysis identified two principle
CpG-SNPs, rs5404 in solute carrier family 2, member 2 (SLC2A2), and
rs3749166 in calpain (CAPN10), a membrane protease, which is
involved in glucose transporter (GLUT)4 translocation (16). rs3749166 (A>G) is located in exon 11
of the CAPN10 gene and rs5404 (C>T) in exon 5 of the
SLC2A2 gene. The two CpG-SNPs introduce pronounced changes in the
ESE score (splicing potential) of the corresponding exonic
sequences in these genes. The association of CAPN10 SNP with T2D in
particular, has been addressed in previous studies (17–22).
In the present study, the association of these two
epigenetic CpG-SNPs were analyzed, which introduced the greatest
changes of the splicing potential in the corresponding genes, with
T2D and other metabolic syndrome-associated pathological conditions
(arterial hypertension and obesity). In addition, the possibility
that this association might be observed only in the heterozygotic
state of these SNPs was investigated.
Materials and methods
Study population
The investigated population included 99 non-diabetic
control participants (Table IA) and 71
T2D patients (Table IB). Participants
were classified as having T2D based on the American Diabetic
Association criteria (23) as follows:
i) ≥126 mg/dl fasting plasma glucose concentration; ii)
glycosylated plasma hemoglobin (HbA1c) ≥6.5%; iii) insulin use; iv)
use of other diabetes medication. All participants provided their
medical family history, smoking habits and dietary information,
followed by written informed consent. Their names were anonymized
prior to study completion. The methods followed in the present
study were performed according to the Declaration of Helsinki.
 | Table I.Genotypes and epidemiological
parameters (age, gender, BMI, metabolic, family history, smoking
status, dietary conditions and accompanying diseases) of
non-diabetic control subjects (Table A) and T2D patients (Table
B). |
Table I.
Genotypes and epidemiological
parameters (age, gender, BMI, metabolic, family history, smoking
status, dietary conditions and accompanying diseases) of
non-diabetic control subjects (Table A) and T2D patients (Table
B).
| A, Non-diabetic
control subjects |
|---|
|
|---|
| S/No. | n | Age (years) | Gender | BMI |
| rs3749166 | rs5404 | FG (mg/dl) | HbA1c (%) | Chol (mg/dl) | LDL (mg/dl) | HDL (mg/dl) | TGL (mg/dl) | FH | Smoking status |
| No diet |
| Diseases |
|---|
| 1 |
1 | 65 | M | 23 |
| G/G | C/C | 110 | 5.9 | 106 | 80 | 36 | 95 |
| + |
| + |
|
|
| 2 |
3 | 47 | F | 25 |
| A/A | C/C | 85 | 5.3 | 178 | 139 | 46 | 150 |
| + |
|
|
| HL |
| 3 |
4 | 58 | F | 21 |
| A/G | C/C | 84 | 5.4 | 223 | 182 | 59 | 146 |
|
|
|
|
|
|
| 4 |
6 | 71 | F | 22 |
| A/G | C/T | 110 | 6.4 | 220 | 182 | 63 | 131 |
|
|
|
|
| HL |
| 5 |
7 | 80 | F | 28 |
| A/A | C/C | 106 | 6.0 | 163 | 141 | 42 | 69 |
|
|
|
|
| HT |
| 6 |
9 | 77 | F | 26 |
| A/A | C/C | 96 | 5.6 | 353 | 305 | 51 | 190 |
|
|
|
|
| HL |
| 7 | 10 | 60 | F | 29 |
| A/G | C/T | 118 | 6.2 | 153 | 111 | 40 | 174 | + |
|
|
|
| HT/HL |
| 8 | 12 | 58 | M | 34 |
| A/G | C/C | 94 | 5.6 | 177 | 143 | 33 | 139 |
| + |
|
|
| HT |
| 9 | 13 | 54 | M | 25 |
| A/G | C/T | 104 | 5.7 | 267 | 219 | 67 | 177 |
|
|
|
|
|
|
| 10 | 14 | 50 | F | 35 |
| A/A | C/C | 98 | 5.6 | 298 | 256 | 53 | 160 |
|
|
|
|
| HL |
| 11 | 16 | 66 | F | 24 |
| G/G | C/C | 110 | 5.9 | 185 | 153 | 60 | 104 |
|
|
|
|
| HT/HL |
| 12 | 19 | 50 | F | 35 |
| G/G | C/T | 82 | 5.6 | 225 | 194 | 31 | 126 |
| + |
| + |
|
|
| 13 | 20 | 68 | F | 32 |
| A/G | C/C | 88 | 5.5 | 215 | 184 | 53 | 103 |
|
|
|
|
| HT |
| 14 | 21 | 75 | M | 28 |
| A/A | C/C | 87 | 5.6 | 112 | 92 | 28 | 76 |
| + |
|
|
| HT |
| 15 | 23 | 50 | M | 22 |
| A/G | C/T | 87 | 5.2 | 239 | 206 | 61 | 106 | + |
|
|
|
|
|
| 16 | 25 | 76 | F | 29 |
| A/G | C/T | 98 | 5.6 | 236 | 186 | 49 | 204 |
|
|
|
|
| HT/HL |
| 17 | 26 | 65 | F | 26 |
| A/G | C/T | 107 | 5.8 | 187 | 152 | 57 | 118 | + |
|
|
|
| HT/HL |
| 18 | 27 | 54 | F | 29 |
| G/G | C/C | 107 | 5.9 | 250 | 218 | 56 | 106 |
| + |
|
|
| HL |
| 19 | 28 | 57 | F | 46 |
| A/G | C/T | 115 | 6.0 | 239 | 200 | 51 | 143 |
|
|
| + |
| HT/HL |
| 20 | 29 | 80 | F | 24 |
| A/G | C/C | 83 | 5.3 | 188 | 161 | 63 | 73 |
|
|
|
|
| HT/HL |
| 21 | 30 | 80 | F | 27 |
| A/A | C/C | 85 | 5.4 | 212 | 180 | 58 | 100 |
|
|
|
|
| HT/HL |
| 22 | 31 | 77 | M | 31 |
| A/G | C/C | 89 | 5.6 | 178 | 154 | 38 | 82 |
|
|
|
|
| HT |
| 23 | 32 | 66 | M | 26 |
| A/G | C/T | 92 | 5.6 | 229 | 188 | 50 | 159 |
|
|
|
|
|
|
| 24 | 37 | 54 | F | 28 |
| A/A | C/C | 110 | 5.8 | 230 | 198 | 40 | 124 | + |
|
|
|
| HT/HL |
| 25 | 38 | 69 | M | 29 |
| A/A | C/T | 101 | 5.9 | 202 | 153 | 40 | 209 |
| + |
|
|
| HT/HL |
| 26 | 39 | 52 | F | 35 |
| A/A | C/T | 95 | 5.6 | 171 | 119 | 40 | 220 |
|
|
|
|
| HL |
| 27 | 40 | 74 | F | 23 |
| G/G | C/C | 100 | 5.7 | 172 | 143 | 69 | 79 |
|
|
|
|
| HT/HL |
| 28 | 43 | 45 | F | 38 |
| A/G | C/T | 100 | 5.7 | 216 | 190 | 38 | 96 |
|
|
|
|
|
|
| 29 | 44 | 56 | M | 38 |
| A/A | C/C | 78 | 5.6 | 179 | 144 | 50 | 128 |
|
|
|
|
|
|
| 30 | 48 | 71 | F | 27 |
| A/G | C/T | 101 | 5.8 | 171 | 122 | 33 | 215 |
|
|
|
|
| HT/HL |
| 31 | 50 | 79 | M | 23 |
| A/G | C/C | 91 | 5.6 | 167 | 134 | 46 | 123 |
|
|
|
|
| HT/HL |
| 32 | 51 | 45 | F | 34 |
| A/G | C/T | 88 | 5.6 | 242 | 199 | 49 | 167 | + |
|
|
|
| HL |
| 33 | 52 | 52 | F | 31 |
| A/A | C/C | 94 | 5.6 | 211 | 162 | 41 | 208 |
|
|
|
|
| HL |
| 34 | 54 | 58 | M | 36 |
| A/G | C/T | 111 | 5.8 | 154 | 113 | 40 | 167 |
|
|
|
|
| HT |
| 35 | 56 | 61 | F | 31 |
| A/G | C/C | 101 | 6.2 | 257 | 216 | 52 | 155 |
|
|
|
|
|
|
| 36 | 58 | 76 | F | 28 |
| A/G | C/C | 107 | 5.8 | 259 | 228 | 73 | 83 |
|
|
|
|
| HT |
| 37 | 59 | 69 | F | 31.5 |
| A/G | C/C | 103 | 5.7 | 202 | 172 | 48 | 103 |
|
|
|
|
| HT |
| 38 | 60 | 73 | F | 25 |
| A/G | C/C | 90 | 5.4 | 222 | 190 | 53 | 109 |
|
|
|
|
| HT |
| 39 | 61 | 45 | F | 34 |
| G/G | C/C | 89 | 5.2 | 173 | 144 | 51 | 97 |
|
|
|
|
|
|
| 40 | 63 | 66 | M | 30 |
| A/A | C/T | 117 | 6.4 | 170 | 138 | 41 | 121 |
| + |
|
|
| HT/HL |
| 41 | 64 | 60 | F | 30 |
| A/G | C/C | 93 | 5.6 | 170 | 131 | 40 | 155 |
|
|
|
|
| HT/HL |
| 42 | 66 | 65 | F | 28 |
| A/G | C/C | 98 | 5.6 | 193 | 150 | 41 | 171 |
|
|
|
|
|
|
| 43 | 67 | 45 | F | 17 |
| G/G | C/T | 98 | 5.0 | 176 | 147 | 53 | 93 |
|
|
|
|
|
|
| 44 | 68 | 63 | F | 28 |
| A/G | C/C | 90 | 5.1 | 214 | 185 | 57 | 92 |
|
|
|
|
| HL |
| 45 | 70 | 50 | F | 33 |
| A/G | C/T | 105 | 6.4 | 217 | 180 | 48 | 137 |
|
|
| + |
| HL |
| 46 | 71 | 62 | F | 28 |
| A/G | C/C | 109 | 6.0 | 263 | 220 | 62 | 152 |
|
|
|
|
| HL |
| 47 | 75 | 60 | F | 29 |
| G/G | C/C | 88 | 5.6 | 225 | 192 | 48 | 114 |
|
|
|
|
| HT |
| 48 | 76 | 52 | M | 25 |
| G/G | C/C | 91 | 5.5 | 197 | 160 | 54 | 134 |
|
|
|
|
|
|
| 49 | 77 | 50 | F | 25 |
| A/A | C/T | 99 | 5.6 | 210 | 168 | 42 | 168 |
|
|
|
|
| HL |
| 50 | 78 | 58 | F | 25 |
| A/G | C/C | 88 | 5.3 | 249 | 214 | 81 | 93 |
|
|
|
|
| HL |
| 51 | 81 | 70 | F | 29 |
| G/G | C/C | 117 | 5.8 | 212 | 172 | 49 | 150 | + |
|
|
|
| HL |
| 52 | 84 | 55 | F | 32 |
| G/G | C/C | 95 | 5.4 | 218 | 187 | 49 | 103 |
|
|
|
|
| HT/HL |
| 53 | 85 | 53 | F | 25 |
| A/A | C/C | 91 | 5.5 | 223 | 183 | 67 | 132 |
|
|
|
|
| HT |
| 54 | 87 | 65 | F | 34 |
| G/G | C/C | 106 | 5.8 | 240 | 202 | 51 | 139 |
|
|
|
|
| HT/HL |
| 55 | 88 | 49 | F | 33 |
| A/G | C/T | 98 | 5.6 | 213 | 170 | 33 | 185 |
| + |
| + |
| HT/HL |
| 56 | 89 | 73 | F | 28 |
| A/A | C/C | 108 | 6.3 | 216 | 188 | 52 | 89 |
|
|
|
|
|
|
| 57 | 90 | 57 | M | 32 |
| A/A | C/C | 91 | 5.5 | 205 | 157 | 30 | 208 |
| + |
|
|
| HT/HL |
| 58 | 92 | 73 | F | 21 |
| A/G | C/T | 109 | 5.9 | 261 | 215 | 55 | 175 |
|
|
|
|
| HT/HL |
| 59 | 94 | 66 | F | 30 |
| A/G | C/C | 115 | 6.3 | 163 | 132 | 52 | 105 |
|
|
|
|
| HT/HL |
| 60 | 99 | 68 | M | 30 |
| A/A | C/C | 85 | 5.5 | 213 | 163 | 45 | 202 |
|
|
|
|
| HL |
| 61 | 100 | 58 | F | 30 |
| A/A | C/C | 95 | 5.6 | 232 | 203 | 44 | 100 |
|
|
|
|
|
|
| 62 | 101 | 53 | F | 27 |
| A/G | C/T | 106 | 5.7 | 218 | 177 | 64 | 140 |
|
|
|
|
|
|
| 63 | 102 | 59 | F | 23 |
| A/G | C/C | 95 | 5.3 | 169 | 142 | 45 | 89 |
|
|
|
|
| HT |
| 64 | 104 | 78 | M | 24 |
| A/A | C/C | 100 | 5.7 | 211 | 186 | 55 | 70 |
|
|
|
|
| HT |
| 65 | 106 | 64 | F | 33 |
| A/G | C/T | 108 | 6.4 | 208 | 164 | 43 | 177 |
|
|
|
|
| HT |
| 66 | 107 | 47 | F | 27 |
| A/A | C/T | 105 | 5.7 | 239 | 196 | 57 | 155 |
|
|
|
|
| HL |
| 67 | 108 | 43 | F | 28 |
| A/A | C/T | 89 | 5.4 | 159 | 130 | 44 | 103 |
| + |
|
|
|
|
| 68 | 109 | 71 | M | 27 |
| A/G | C/C | 101 | 5.7 | 171 | 122 | 33 | 215 |
|
|
|
|
| HT |
| 69 | 110 | 57 | F | 32 |
| A/G | C/T | 104 | 5.8 | 220 | 155 | 40 | 286 |
|
|
|
|
| HL |
| 70 | 111 | 60 | F | 26 |
| G/G | C/C | 99 | 5.3 | 167 | 144 | 49 | 65 |
|
|
|
|
|
|
| 71 | 112 | 65 | F | 26 |
| A/G | C/C | 93 | 5.4 | 201 | 165 | 76 | 104 |
|
|
|
|
| HT/HL |
| 72 | 113 | 47 | F | 29 |
| G/G | C/C | 102 | 5.7 | 216 | 180 | 61 | 122 |
|
|
|
|
|
|
| 73 | 114 | 73 | F | 24 |
| A/G | C/C | 120 | 6.2 | 216 | 188 | 52 | 89 |
|
|
|
|
|
|
| 74 | 118 | 46 | F | 29 |
| A/A | C/T | 88 | 5.5 | 148 | 88 | 30 | 267 |
|
|
|
|
| HT/HL |
| 75 | 120 | 58 | F | 26 |
| A/G | C/C | 104 | 5.7 | 263 | 138 | 61 | 61 |
|
|
|
|
| HL |
| 76 | 121 | 56 | F | 32 |
| G/G | C/C | 87 | 5.6 | 187 | 149 | 70 | 120 |
|
|
|
|
|
|
| 77 | 153 | 60 | F | 33 |
| G/G | C/C | 87 | 5.3 | 235 | 200 | 64 | 112 |
| + |
| + |
| HT/HL |
| 78 | 155 | 66 | M | 26 |
| A/G | C/C | 124 | 5.7 | 256 | 200 | 45 | 232 |
| + |
| + |
| HT/HL |
| 79 | 156 | 50 | F | 26 |
| A/G | C/C | 93 | 5.2 | 256 | 176 | 60 | 337 |
| + |
|
|
| HL |
| 80 | 158 | 54 | F | 24 |
| A/A | C/T | 104 | 5.7 | 257 | 218 | 62 | 131 |
|
|
|
|
| HT |
| 81 | 159 | 63 | F | 25 |
| A/A | C/C | 115 | 5.8 | 181 | 148 | 64 | 99 |
|
|
|
|
| HL |
| 82 | 161 | 49 | F | 22 |
| A/G | C/T | 93 | 5.3 | 213 | 178 | 100 | 75 |
| + |
| + |
|
|
| 83 | 165 | 50 | F | 29 |
| A/G | C/T | 85 | 5.1 | 158 | 84 | 31 | 339 |
|
|
|
|
| HL |
| 84 | 166 | 50 | M | 29 |
| A/G | C/T | 107 | 5.7 | 258 | 215 | 32 | 174 |
| + |
|
|
| HL |
| 85 | 171 | 52 | M | 28 |
| A/A | C/T | 101 | 5.7 | 213 | 175 | 36 | 151 |
| + |
|
|
|
|
| 86 | 173 | 51 | M | 32 |
| A/G | C/T | 78 | 5.3 | 191 | 153 | 38 | 151 |
| + |
| + |
|
|
| 87 | 174 | 50 | M | 28 |
| A/A | C/T | 96 | 5.0 | 171 | 139 | 38 | 120 |
|
|
|
|
|
|
| 88 | 176 | 80 | F | 32 |
| A/G | C/C | 121 | 5.9 | 158 | 112 | 43 | 183 |
|
|
|
|
| HT/HL |
| 89 | 177 | 71 | M | 28 |
| A/G | C/C | 105 | 5.8 | 220 | 175 | 77 | 144 |
|
|
|
|
| HT/HL |
| 90 | 178 | 56 | F | 30 |
| A/G | C/T | 108 | 5.8 | 182 | 152 | 53 | 93 |
|
|
|
|
| HT |
| 91 | 179 | 67 | M | 29 |
| A/G | C/C | 105 | 5.7 | 179 | 149 | 34 | 112 |
|
|
|
|
|
|
| 92 | 180 | 65 | F | 26 |
| A/G | C/C | 91 | 5.7 | 198 | 168 | 73 | 73 |
|
|
|
|
| HT/HL |
| 93 | 189 | 50 | F | 31 |
| A/G | C/T | 82 | 5.2 | 197 | 168 | 58 | 87 |
|
|
| + |
|
|
| 94 | 190 | 50 | M | 31 |
| A/A | C/T | 95 | 5.4 | 286 | 228 | 51 | 187 |
|
|
|
|
| HL |
| 95 | 192 | 58 | F | 58 |
| A/A | C/T | 93 | 5.6 | 244 | 210 | 68 | 100 |
|
|
| + |
| HT/HL |
| 96 | 194 | 52 | F | 25 |
| A/G | C/T | 85 | 5.2 | 219 | 194 | 44 | 81 |
|
|
| + |
| HL |
| 97 | 195 | 61 | F | 27 |
| A/G | C/C | 82 | 5.6 | 242 | 202 | 69 | 93 |
|
|
|
|
| HT/HL |
| 98 | 196 | 77 | F | 39 |
| A/A | C/C | 108 | 6.2 | 218 | 185 | 71 | 90 |
|
|
| + |
| HT/HL |
| 99 | 197 | 61 | F | 35 |
| A/G | C/C | 94 | 5.6 | 182 | 49 | 145 | 122 |
|
|
| + |
| HT/HL |
|
| B, T2D
patients |
|
| S/No. | n | Age (years) | Gender | BMI | Age of
diagnosis | rs3749166 | rs5404 | FG (mg/dl) | HbA1c | Chol (mg/dl) | LDL (mg/dl) | HDL (mg/dl) | TGL (mg/dl) | FH | Smoking status | Medication | No diet | DC | Diseases |
|
| 1 |
2 | 62 | M | 30 | 50 | A/G | C/C | 128 | 9.4 | 154 | 125 | 39 | 109 |
|
|
|
| DNA | HT/HL |
| 2 |
5 | 60 | F | 41 | 35 | A/G | C/C | 183 | 8.9 | 156 | 128 | 39 | 102 |
|
|
| + | DNA | HT/HL |
| 3 |
8 | 80 | F | 32 | 80 | A/G | C/C | 143 | 11 | 177 | 143 | 39 | 134 |
|
| T | + |
| HT/HL |
| 4 | 11 | 54 | F | 27 | 40 | A/G | C/C | 142 | 9.5 | 190 | 159 | 55 | 104 |
|
| T/IN |
|
|
|
| 5 | 15 | 80 | F | 23 | 75 | A/A | C/C | 128 | 9.8 | 135 | 108 | 40 | 98 | + |
| IN |
| DNA | HT |
| 6 | 17 | 71 | F | 44 |
| A/A | C/C | 144 | 7.4 | 150 | 117 | 37 | 129 | + |
|
| + | DNA | HT/HL |
| 7 | 18 | 65 | F | 24 |
| A/G | C/C | 206 | 9.0 | 204 | 168 | 47 | 136 |
|
| T |
|
| HL |
| 8 | 22 | 60 | M | 28 |
| A/G | C/C | 93 | 6.1 | 281 | 188 | 39 | 429 |
|
| T |
|
| HL |
| 9 | 24 | 66 | F | 30 |
| A/G | C/T | 110 | 7.1 | 191 | 164 | 50 | 86 |
|
| T |
| DNA | HT |
| 10 | 33 | 57 | F | 24 |
| A/A | C/C | 177 | 7.8 | 219 | 189 | 55 | 97 |
|
| T/IN |
|
| HT/HL |
| 11 | 34 | 78 | F | 41 | 65 | G/G | C/T | 155 | 7.5 | 157 | 124 | 40 | 126 | + | + | T |
|
| HT |
| 12 | 35 | 63 | M | 36 | 50 | A/A | C/C | 254 | 9.9 | 252 | 198 | 32 | 1077 | + |
| T | + |
| HT/HL |
| 13 | 36 | 71 | F | 33 | 54 | A/G | C/C | 139 | 7.8 | 216 | 175 | 54 | 153 |
|
| T/IN | + | DNA | HL |
| 14 | 41 | 64 | M | 30 | 50 | A/G | C/T | 286 | 8.8 | 219 | 173 | 31 | 202 | + |
| T/IN | + | DNA | HT/HL |
| 15 | 42 | 80 | M | 18 | 60 | A/G | C/T | 346 | 10.5 | 192 | 162 | 51 | 100 | + |
| T |
| DNA |
|
| 16 | 45 | 72 | F | 30 | 65 | A/G | C/C | 83 | 6.4 | 126 | 98 | 29 | 115 |
|
| T |
|
|
|
| 17 | 46 | 62 | F | 31 | 62 | A/A | C/C | 168 | 7.4 | 223 | 184 | 52 | 148 | + |
| Diet |
|
| HT/HL |
| 18 | 47 | 73 | F | 33 | 60 | A/A | C/T | 185 | 8.0 | 189 | 156 | 48 | 117 |
|
| T |
| DNA | HT |
| 19 | 49 | 71 | M | 28 | 65 | A/A | C/C | 111 | 7.6 | 219 | 180 | 45 | 153 |
|
| T |
|
| HT/HL |
| 20 | 53 | 58 | F | 30 | 57 | A/G | C/C | 295 | 8.7 | 201 | 154 | 43 | 195 |
|
| Diet |
|
| HL |
| 21 | 55 | 67 | M | 25 | 55 | A/A | C/C | 295 | 10.6 | 205 | 162 | 36 | 182 |
| + |
|
| DNA | HT/HL |
| 22 | 57 | 66 | F | 29 |
| A/G | C/C | 109 | 7.4 | 200 | 153 | 36 | 200 |
|
| T |
|
| HL |
| 23 | 62 | 79 | F | 30 |
| A/G | C/T | 151 | 7.8 | 169 | 141 | 45 | 99 |
|
| T/IN |
| DNA |
| 24 | 65 | 59 | M | 36 |
| A/A | C/C | 130 | 6.8 | 125 | 84 | 29 | 178 |
|
| T |
|
|
|
| 25 | 69 | 63 | F | 29 | 40 | A/G | C/C | 89 | 7.2 | 178 | 133 | 37 | 190 |
|
| T |
|
|
|
| 26 | 72 | 62 | F | 26 |
| A/G | C/C | 228 | 8.6 | 178 | 133 | 50 | 175 |
|
| IN |
|
| HL |
| 27 | 73 | 69 |
| 30 |
| A/A | C/C | 171 | 7.5 | 166 | 132 | 57 | 114 |
|
| T |
|
| HT/HL |
| 28 | 74 | 54 | F | 24 |
| A/G | C/C | 210 | 8.6 | 290 | 216 | 32 | 339 | + |
| T |
|
|
|
| 29 | 79 | 51 | M | 28 |
| A/G | C/C | 127 | 7.6 | 181 | 127 | 26 | 246 |
|
| T/IN |
|
| HT/HL |
| 30 | 80 | 70 | M | 39 |
| A/A | C/T | 134 | 7.7 | 152 | 106 | 33 | 200 |
|
| IN | + | DNA | HT/HL |
| 31 | 82 | 68 | F | 34 | 40 | A/G | C/T | 155 | 8.2 | 243 | 186 | 61 | 224 |
|
| IN |
|
| HL |
| 32 | 83 | 45 | F | 60 | 45 | A/G | C/C | 111 | 7.8 | 240 | 180 | 27 | 275 |
|
| Diet | + |
| HL |
| 33 | 86 | 81 | F | 28 | 68 | A/G | C/C | 132 | 7.4 | 171 | 132 | 41 | 156 |
|
| IN |
| DNA | HT |
| 34 | 91 | 62 | F | 34 | 62 | A/G | C/T | 98 | 7.2 | 312 | 241 | 41 | 313 | + |
| Diet | + |
| HT |
| 35 | 93 | 59 | F | 36 | 58 | A/G | C/C | 91 | 7.2 | 193 | 153 | 40 | 161 |
| + | Diet |
|
| HT |
| 36 | 95 | 52 | F | 40 | 50 | A/G | C/T | 119 | 7.0 | 212 | 172 | 53 | 148 |
|
| T |
|
| HT |
| 37 | 96 | 79 | F | 30 | 60 | G/G | C/C | 145 | 7.6 | 169 | 141 | 49 | 94 | + |
| T |
|
| HT |
| 38 | 97 | 45 | F | 30 | 37 | A/G | C/C | 149 | 7.2 | 144 | 102 | 28 | 182 | + | + | T |
|
| HT |
| 39 | 98 | 45 | F | 22 | 37 | A/G | C/C | 269 | 8.8 | 188 | 162 | 60 | 73 | + | + | IN | + |
|
|
| 40 | 103 | 62 | F | 38 | 52 | A/G | C/T | 152 | 7.8 | 145 | 80 | 45 | 284 |
|
| T | + | DNA | HT |
| 41 | 105 | 67 | M | 32 | 57 | A/G | C/C | 233 | 8.8 | 235 | 182 | 37 | 232 |
| + | T/IN |
| 42 | 115 | 65 | M | 19 |
| A/G | C/C | 422 | 8.9 | 182 | 149 | 41 | 125 | + |
| T |
| DNA |
|
| 43 | 116 | 79 | F | 32 | 75 | A/G | C/T | 151 | 7.8 | 151 | 120 | 36 | 124 |
|
| T |
| DNA | HT |
| 44 | 117 | 74 | F | 38 | 67 | A/A | C/T | 164 | 7.8 | 130 | 93 | 35 | 152 |
|
| T/IN |
|
| HT/HL |
| 45 | 119 | 45 | F | 56 |
| A/G | C/C | 167 | 7.6 | 191 | 134 | 36 | 250 | + | + | Diet |
|
| HL |
| 46 | 151 | 74 | F | 31 |
| A/G | C/C | 102 | 7.2 | 239 | 206 | 62 | 106 | + |
| Diet | + |
| HT/HL |
| 47 | 152 | 66 | F | 30 | 62 | A/G | C/C | 126 | 6.8 | 152 | 120 | 34 | 127 | + | + | T | + |
| HT |
| 48 | 154 | 78 | M | 32 | 58 | A/G | C/C | 179 | 9.1 | 123 | 80 | 33 | 183 | + |
| T/IN | + | DNA | HT/HL |
| 49 | 157 | 73 | M | 33 | 58 | A/G | C/C | 138 | 7.7 | 108 | 85 | 38 | 77 |
|
| T | + | DNA | HT |
| 50 | 160 | 59 | F | 48 | 59 | A/G | C/C | 148 | 7.5 | 190 | 156 | 43 | 130 | + |
| T | + |
| HT/HL |
| 51 | 162 | 68 | F | 33 | 45 | A/G | C/C | 165 | 8.7 | 168 | 122 | 37 | 197 | + |
| IN |
| DNA | HT |
| 52 | 163 | 63 | M | 28 | 58 | A/G | C/C | 257 | 10.3 | 164 | 130 | 38 | 134 |
|
| T |
| DNA | HT/HL |
| 53 | 164 | 74 | F | 30 | 64 | G/G | C/C | 124 | 5.8 | 181 | 130 | 62 | 93 |
|
| Diet |
|
| HT/HL |
| 54 | 167 | 72 | F | 26 | 46 | A/G | C/C | 134 | 7.0 | 162 | 137 | 49 | 77 |
|
| T |
|
| HT/HL |
| 55 | 169 | 67 | F | 28 | 49 | A/G | C/C | 125 | 7.4 | 167 | 130 | 47 | 141 |
|
| T |
| DNA | HT/HL |
| 56 | 170 | 73 | F | 34 | 48 | G/G | C/C | 235 | 10.1 | 177 | 134 | 31 | 187 |
|
| T/IN |
| DNA |
|
| 57 | 172 | 73 | F | 35 | 60 | A/G | C/C | 192 | 12.5 | 153 | 98 | 28 | 248 | + |
| T/IN | + | DNA | HT/HL |
| 58 | 175 | 79 | F | 33 | 69 | A/G | C/C | 217 | 8.8 | 179 | 95 | 30 | 391 |
|
| T | + |
| HT/HL |
| 59 | 181 | 78 | F | 33 | 62 | A/G | C/C | 341 | 8.5 | 212 | 160 | 55 | 207 |
|
| IN |
|
|
|
| 60 | 182 | 80 | M | 29 | 60 | A/G | C/T | 130 | 8.5 | 171 | 126 | 37 | 192 |
|
| T/IN |
| DNA | HT/HL |
| 61 | 183 | 80 | F | 29 | 75 | A/G | C/C | 171 | 7.5 | 152 | 117 | 35 | 143 |
|
| T |
| DNA | HT/HL |
| 62 | 184 | 66 | F | 62 | 51 | A/G | C/T | 68 | 7.8 | 190 | 160 | 47 | 107 |
|
| IN |
|
| HL |
| 63 | 185 | 72 | M | 47 | 55 | A/G | C/C | 269 | 8.8 | 243 | 150 | 37 | 429 |
|
| T/IN |
| DNA | HT/HL |
| 64 | 186 | 78 | M | 30 | 75 | A/A | C/C | 127 | 7.4 | 170 | 145 | 43 | 82 |
|
| T |
|
| HT |
| 65 | 187 | 57 | F | 26 | 53 | A/G | C/C | 240 | 9.5 | 256 | 196 | 33 | 271 | + | + | T | + |
| HL |
| 66 | 188 | 77 | M | 30 | 60 | A/A | C/C | 154 | 8.3 | 229 | 193 | 40 | 141 | + |
| T |
| DNA | HT |
| 67 | 191 | 53 | F | 32 | 51 | A/G | C/C | 114 | 6.9 | 216 | 191 | 54 | 74 | + |
| T |
|
| HT |
| 68 | 193 | 67 | M | 26 |
| A/A | C/C | 155 | 7.2 | 211 | 181 | 58 | 95 |
| + |
| + |
| HT/HL |
| 69 | 198 | 67 | F | 30 | 32 | A/G | C/T | 167 | 6.8 | 228 | 193 | 57 | 122 | + |
| T | + | DNA | HT/HL |
| 70 | 199 | 64 | F | 31 | 55 | A/G | C/C | 174 | 8.2 | 219 | 180 | 47 | 148 | + |
| IN |
| DNA | HL |
| 71 | 200 | 80 | M | 29 | 68 | A/G | C/C | 163 | 7.5 | 155 | 125 | 41 | 113 |
|
| T |
| DNA | HT |
The present study was approved by the Bioethics
Committee of Aristotle University Medical School (Thessaloniki,
Greece; protocol no. 2629; 19 April 2011), the Scientific Council
of Thessaloniki Panagia General Hospital (Thessaloniki, Greece;
protocol no. A9825; 9 June 2011) and the Research Committee of
Aristotle University, Operational Program ‘Education and Lifelong
Learning’ of the National Strategic Reference Framework (NSRF) -
Research Funding Program: Heracleitus II (project no. 87113).
Anthropometric and biochemical
analysis
Anthropometric measurements, including weight and
height were obtained according to standardized protocols. The
epidemiological profile consisted of age, gender, metabolic family
history, smoking status, dietary conditions, and accompanying
diseases (arterial hypertension and hyperlipidemia). Participants
were classified as having an accompanying disease (arterial
hypertension and hyperlipidemia) when the use of antihypertensive
or antihyperlipidemic medication was reported respectively,
independently of their biochemical lipid profile determination.
Information regarding the type of medication (tablets and insulin)
and potential diabetic complications were recorded for the diabetic
patients.
The biochemical analysis included determination of
fasting plasma glucose, HbA1c, total serum cholesterol, low-density
lipoprotein (LDL) cholesterol, high-density lipoprotein (HDL)
cholesterol and serum triglycerides. Peripheral blood samples (2
ml) from all 170 participants for molecular genetic analysis were
collected in tubes containing EDTA and centrifuged at 4,500 × g for
20 min at room temperature. Buffy coat leukocytes were then
isolated and stored at −20°C.
DNA extraction and genotype
analysis
Genomic DNA was extracted from the buffy coat
fraction prepared as described above using PureLink Genomic DNA kit
(Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA),
according to the manufacturer's instructions. DNA integrity was
verified by gel electrophoresis (70 V/cm for 20 min) using 0.8%
agarose gel and ethidium bromide staining. DNA purity was
determined by the optical density
(OD)260/OD280 nm absorption ratio using an
Eppendorf Biophotometer. Genomic sequences containing SNPs
(rs3749166 and rs5404) were amplified by DNA polymerase chain
reaction (PCR) using Platinum Taq DNA polymerase (Invitrogen;
Thermo Fisher Scientific, Inc.). The PCR conditions for rs3749166
amplification were as follows: 94°C for 2 min, 35 cycles of 94°C
for 45 sec, 60°C for 45 sec and 72°C for 1.5 min followed by 72°C
for 10 min. A forward primer (5′-CAGGTCCCAGAGGGTGGAA-3′) and a
reverse primer (5′-CAGGTAGGTGGAGGGCACAA-3′) were used for
amplifying a 153-bp fragment containing SNP rs3749166. A 344-bp
fragment containing SNP rs5404, was amplified by PCR using a
forward primer (5′-TCAGGGAGGGGCTTTCATTC-3′) and a reverse primer
(5′-CAGTCAGGGAGGGACGAGA-3′) under the following conditions: 94°C
for 2 min, 35 cycles of 94°C for 45 sec, 58°C for 45 sec and 72°C
for 1.5 min followed by 72°C for 10 min. Primer design was
facilitated by Primer-BLAST (https://www.ncbi.nlm.nih.gov/tools/primer-blast/),
an online primer designing tool (24).
Twelve microliters of each PCR product were separated (70 V/cm for
20 min) on a 2% agarose gel and visualized using ethidium bromide
staining. In addition, the PCR products were purified using a
PureLink PCR Purification kit (Invitrogen; Thermo Fisher
Scientific, Inc.), according to the manufacturer's instructions.
The sequence of the purified PCR products was verified by
commercial sequence analysis (VBC-Biotech Service GmbH, Vienna,
Austria) using the forward primer for rs3749166 and the reverse
primer for rs5404). Nucleotide sequence analysis was performed
using the Chromas software (version 2.6.2).
Personal Genome Project (PGP)
data
To validate the results of the analysis, the allele
frequencies of the rs5404 and rs3749166 polymorphisms were
evaluated using public genome and exome data, available through the
PGP repository (25). Cases matching
the patient and non-diabetic control profiles of the present study
were selected and 40 additional cases (35 non-diabetic controls and
5 T2D patients) were included, containing allele information of the
rs3749166 polymorphism. In addition, 71 cases (48 non-diabetic
controls and 23 T2D patients) with allele information of the rs5404
polymorphism were evaluated. Among these, 35 non-diabetic controls
and 5 T2D patients contained genetic data for the two
polymorphisms. Of the 71 PGP individuals, 32.4% were female and the
mean age was 58 years (standard deviation, 10.85).
Statistical analysis
Graphpad online tool (https://www.graphpad.com) was used to perform
statistical analyses. Student's t-test was used to compare groups
of continuous variables, and the χ2 and Fisher's test
were used to compare the proportions of genotypes or alleles. A
two-tailed P<0.05 was considered to indicate a statistically
significant difference. The difference of the ESE scores between
the major SA and minor Sa alleles was
calculated as the ΔScore = |SA-Sa|.
Results
Clinical data and statistical analysis
of epidemiological parameters
Statistical analysis of the data from T2D patients
and non-diabetic control subjects (Table
II) demonstrated that among the T2D patients, fasting glucose
levels and ΗbΑ1c were significantly higher (P<0.0001); however,
there was no correlation with accompanying diseases, such as
arterial hypertension or hyperlipidemia. By contrast, LDL and
triglyceride levels were significantly higher among T2D patients
(P<0.0001; P=0.0052) and HDL levels were significantly lower
(P<0.0001). The observed age difference among T2D patients and
non-diabetic control subjects was significant (P<0.0001),
potentially because the majority of individuals with T2D are
diagnosed at an older age (data not shown). All other parameters,
such as smoking status, did not differ among T2D patients and
non-diabetic control subjects in the present study.
 | Table II.Statistical analysis of
epidemiological parameters of individuals included in Table I. |
Table II.
Statistical analysis of
epidemiological parameters of individuals included in Table I.
| Parameter | Non-diabetic
controlsa (n=99) | T2D
patientsb (n=71) | χ2 | P-value |
|---|
| Male | 24 | 21 | 0.605 | 0.4368 |
| Female | 75 | 50 |
|
|
| Age (years) | 60.62±10.11 | 66.94±9.65 |
| <0.0001 |
| Body mass index
(kg/m2) | 28.96±5.34 | 32.30±7.98 |
| 0.0013 |
| Age of T2D
diagnosis |
| 56.37±10.95 |
|
|
| T2D duration
(years) |
| 11.65±8.33 |
|
|
| Fasting glucose
(mg/dl) | 98.31±10.45 | 170.32±6.63 |
| <0.0001 |
| Glycosylated
hemoglobin (%) | 5.65±0.31 | 8.13±1.21 |
| <0.0001 |
| Total cholesterol
(mg/dl) | 208.57±38.52 | 188.96±40.46 |
| 0.0016 |
| Low-density
lipoprotein cholesterol (mg/dl) | 147.14±35.00 | 169.78±36.81 |
| <0.0001 |
| High-density
lipoprotein cholesterol (mg/dl) | 50.93±13.06 | 42.14±9.46 |
| <0.0001 |
| Trigycerides
(mg/dl) | 137.18±54.49 | 179.45±134.09 |
| 0.0052 |
| Family history
(positive) |
6 | 25 | 23.565 | <0.0001 |
| Smoking status | 20 | 10 | 1.065 | 0.3021 |
| Not following
dietary instructions | 13 | 20 | 5.977 | 0.0014 |
| Diet |
|
8 |
|
|
| Medication
(tablets) |
| 35 |
|
|
| Medication
(insulin) |
| 10 |
|
|
| Medication (tablets
and insulin) |
| 13 |
|
|
| No medication or
dietary intervention |
|
5 |
|
|
| Diabetic
complications (neuropathy, angiopathy) |
| 30 |
|
|
| Diabetic
complications and disease duration (>4 years) |
| 24 |
| 0.0014 |
| Diabetic
complications and disease duration (≤4 years) |
|
1 |
|
|
| Accompanying
diseases | 72 | 59 | 2.516 | 0.1127 |
| Accompanying
diseases (arterial hypertension) | 17 | 19 | 2.278 | 0.1313 |
| Accompanying
diseases (hyperlipidemia) | 24 | 12 | 1.335 | 0.2479 |
| Accompanying
diseases (arterial hypertension and hyperlipidemia) | 31 | 28 | 1.204 | 0.2725 |
Genotype frequencies for rs3749166 and
rs5404 SNPs in T2D patients and non-diabetic control subjects
The rs3749166 polymorphism was detected by PCR
amplification (data not shown) and sequencing of a 153-bp PCR
fragment, which included the SNP (Fig.
1A). Similarly, a 344-bp fragment, including the rs5404 SNP was
amplified by PCR (data not shown) and analyzed by sequencing
(Fig. 1B).
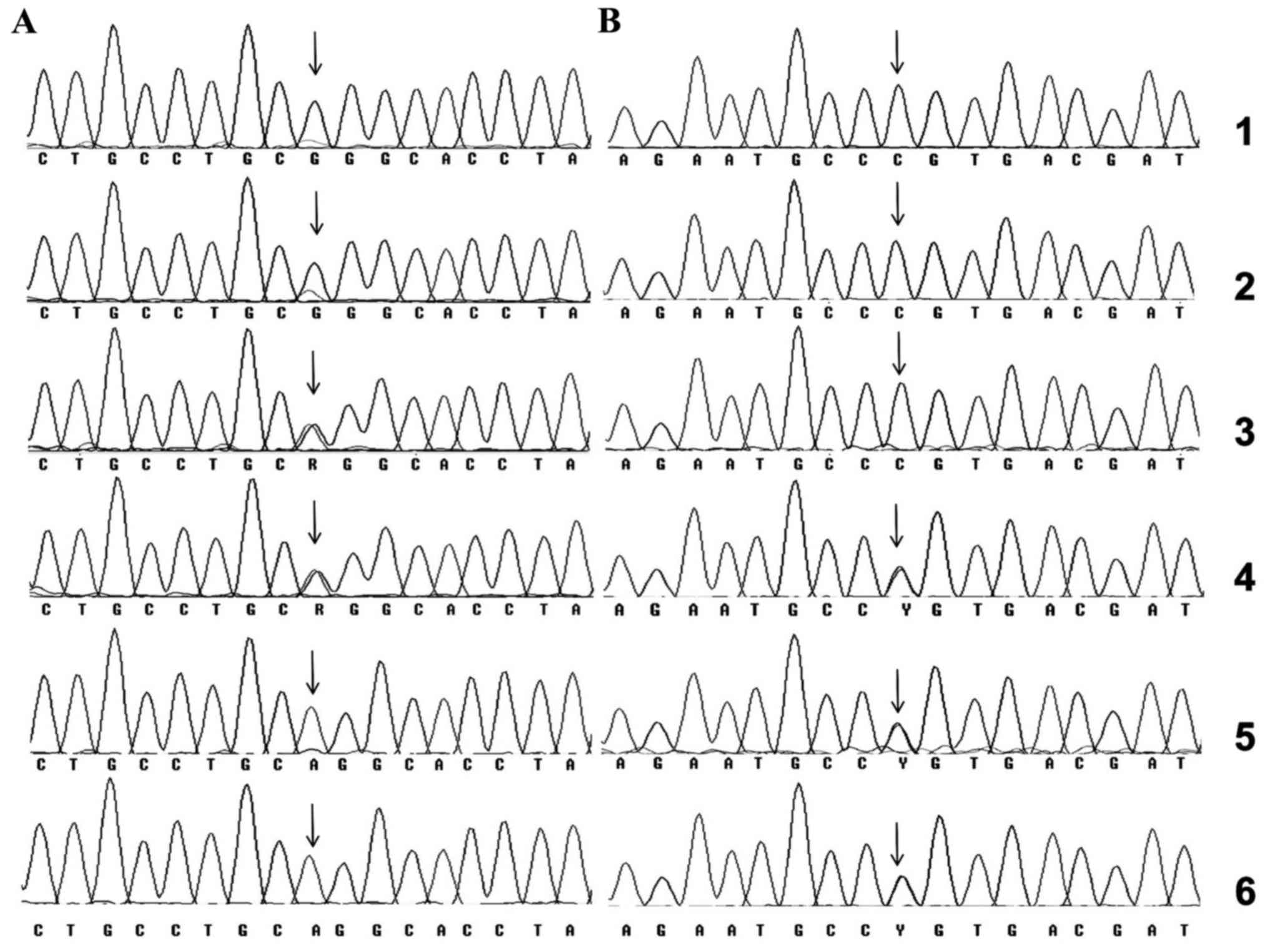 | Figure 1.(A) Nucleotide sequence of the
amplified fragments containing the rs3749166 (A>G) calpain 10
polymorphism for samples 34, 40, 187, 200, 46 and 117 (rows 1–6,
respectively). (B) Nucleotide sequences of the amplified fragments
of rs5404 (C>T) solute carrier family 2 member 2 for the samples
3, 68, 74, 19, 47 and 77 (rows 1–6, respectively). rs3749166 and
rs5404 polymorphic sites are indicated using arrows. |
The rs3749166 and rs5404 frequencies for the Greek
T2D patients and non-diabetic controls are summarized in Table III. Statistical analysis of these
data revealed that only the heterozygous rs3749166 genotype (AG,
partially epigenetic) was associated with T2D, while the epigenetic
genotype (GG) appeared to be protective for the disease (P=0.0262;
Table IIIA). A more significant
positive correlation was obtained when the PGP data were
incorporated into the study (P=0.0039; Table IIIA).
 | Table III.Statistical evaluation of (A)
rs3749166 and (B) rs5404 SNP frequencies and genotypes among total
and Greek T2D patients and non-diabetic controls. (C) Association
of observed rs3749166 and rs5404 genotype combinations with disease
among total, and Greek T2D patients and non-diabetic control
subjects. |
Table III.
Statistical evaluation of (A)
rs3749166 and (B) rs5404 SNP frequencies and genotypes among total
and Greek T2D patients and non-diabetic controls. (C) Association
of observed rs3749166 and rs5404 genotype combinations with disease
among total, and Greek T2D patients and non-diabetic control
subjects.
| A, rs3749166
genotypes |
|---|
|
|---|
| Subject | AA, n (%) | AG, n (%) | GG, n (%) | χ2 | P-value |
|
|---|
| Non-diabetic
control (total) | 40 (29.85) | 71 (52.99) | 23 (17.16) | 11.09 | 0.0039 |
|
| T2D patients
(total) | 15 (19.74) | 57 (75.00) | 4 (5.26) |
|
|
|
| Non-diabetic
control (Greek) | 29 (29.29) | 54 (54.55) | 16 (16.16) | 7.28 | 0.0262 |
|
| T2D patients
(Greek) | 15 (21.13) | 52 (73.24) | 4 (5.63) |
|
|
|
|
| B, rs5404
genotypes |
|
| Subject | TT, n (%) | CT, n (%) | CC, n (%) | χ2 | P-value |
|
|
| Non-diabetic
control (total) | 0 (0) | 51 (34.69) | 96 (65.31) | 4.967 | 0.0258 |
|
| T2D patients
(total) | 0 (0) | 20 (21.28) | 74 (78.72) |
|
|
|
| Non-diabetic
control (Greek) | 0 (0) | 39 (39.39) | 60 (60.61) | 5.369 | 0.0205 |
|
| T2D patients
(Greek) | 0 (0) | 16 (22.54) | 55 (77.46) |
|
|
|
|
| C, rs3749166 and
rs5404 combined genotypes |
|
| Subject | GG/CC, n (%) | GG/CT, n (%) | AG/CC, n (%) | AG/CT, n (%) | AA/CC, n (%) | AA/CT, n (%) |
|
| Non-diabetic
control (total) | 18 (13.43) | 5 (3.73) | 44 (32.84) | 27 (20.15) | 23 (17.16) | 17 (12.69) |
| T2D patients
(total) | 3 (3.95) | 1 (1.32) | 43 (56.58) | 14 (18.42) | 12 (15.79) | 3 (3.95) |
| P-value | 0.031 | 0.4211 | 0.0008a | 0.7614 | 0.7973 | 0.0491 |
| Non-diabetic
control (Greek) | 14 (14.14) | 2 (2.02) | 29 (29.29) | 25 (25.25) | 17 (17.17) | 12 (12.12) |
| T2D patients
(Greek) | 3 (4.22) | 1 (1.40) | 40 (56.33) | 12 (16.90) | 12 (16.90) | 3 (4.22) |
| P-value | 0.039 | 1.000 | 0.004b | 0.169 | 0.963 | 0.100 |
Analysis of the rs5404 polymorphism from the two
sets of data revealed that the homozygous TT genotype was not
observed, although the CT frequency was significant (21.28% in T2D
patients and 34.69% in non-diabetic control subjects) and that the
T genotype may be protective for the disease (Table IIIB; P=0.0205 and P=0.0258).
Finally, the association of these splicing-affecting
genotype combinations with TD2 was analyzed. The results are
presented in Table IIIC and reveal
that only the AG/CC genotype is strongly associated with T2D in all
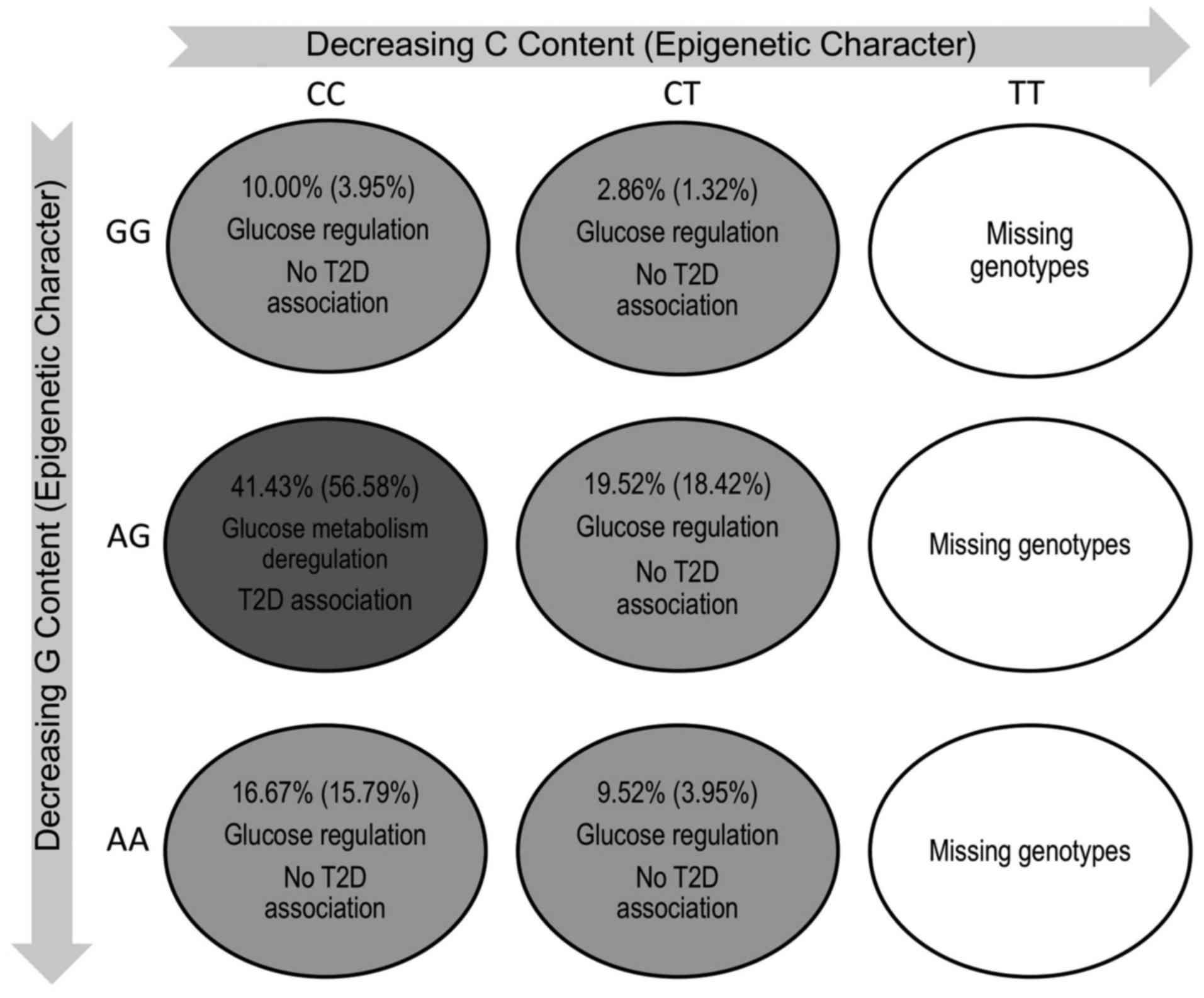
cases examined [Greek: P=0.004 and odds ratio (OR), 3.11; PGP:
P=0.0008 and OR, 2.67]. Furthermore, the GG/CC and AG/CT genotypes
may be protective for the disease. In addition, the T allele was
infrequent among individuals who were homozygous for rs3749166
(GG/CT epigenetic genotype).
Association of the rs3749166/rs5404
genotype combinations with glucose metabolism
Another common characteristic among carriers of the
AG/CC genotype (disease-associated) is the presence of high HbA1c
levels (≥8.5%) (Table IV; P=0.0002,
OR, 5.10) although neither of the polymorphisms was found to be
independently associated with the T2D criteria (elevated fasting
glucose levels and HbA1c).
 | Table IV.AG/CC genotype combination among all
individuals and T2D patients, relative to the HbA1c levels (≥8.5%).
T2D patients are shown in parenthesis. |
Table IV.
AG/CC genotype combination among all
individuals and T2D patients, relative to the HbA1c levels (≥8.5%).
T2D patients are shown in parenthesis.
|
| AG/CC | AA/CC | GG/CC | AG/CT | AA/CT | GG/CT |
|---|
| HbA1c ≥8.5% | 19 (19) | 3 (3) | 1 (1) | 3 (3) |
0 |
0 |
| HbA1c <8.5% | 50 (21) | 26 (9) | 16 (2) | 34 (9) | 15 (3) | 3 (1) |
| Total | 69
(40)a | 29 (12) | 17 (3) | 37 (12) | 15 (3) | 3 (1) |
Discussion
Elucidating the impact of epigenetic synonymous
SNPs, particularly those involved in the regulation of
alternatively spliced exons, is critical for understanding the
pathogenesis of complex diseases. The polymorphisms included in the
present study were selected on the basis of their epigenetic
character and because they are the only synonymous SNPs strongly
modifying the splicing-associated exonic enhancers associated with
glucose transport (11). CAPN10 and
GLUT2 participate in complementary transporter systems, which might
be expected to act in a concerted manner. CAPN10 is a
T2D-associated protease, which facilitates insulin-stimulated GLUT4
translocation via its activity on the distal secretory pathway
(16). Although the association of
rs3749166 with T2D has been the subject of various reviews and
meta-analytic studies, it is still questioned if it may influence
the development of T2D independently or in combination with other
CAPN10 gene polymorphisms (17,18).
Furthermore, the second SNP investigated in the present study,
rs5404 in SLC2A2, has been evaluated in association with T2D
(19–22); however, the obtained results were
contradictory. In certain studies (20,21) a
significant risk was observed among homozygotes, which was similar
to the present results, while in another study (19) the minor allele was found to be
associated with increased disease risk and with reduced
postprandial glucose levels. To the best of our knowledge, the
present study is the first to provide a comprehensive concurrent
analysis of two SNPs (rs3749166 and rs5404) under investigation,
which appear to be critical for the splicing of genes involved in
complementary glucose transport systems.
Computational analysis has shown that the two
polymorphisms interfere with splicing regulation (11). However, according to the data reported
by Karambataki et al (11) and
summarized in Table V (4), these SNPs modify the binding potential of
splicing factors in different ways. rs3749166 (A allele) in CAPN10
strongly modifies the binding site of 3SS_U2 splicing enhancer of
alternatively expressed exon 11 and, thus, may lead to the
production of more than one splicing product in AG heterozygotes.
By contrast, in its heterozygotic state, rs5404 (T allele) in
SLC2A2 modifies the response of the ESE elements in this sequence
to serine/arginine-rich (SR) proteins, particularly SRp40 and
SF2/ASF (IgM-BRCA1) (26). As the two
SNPs modify CpG sequences, they also perturb epigenetic regulation
for the homozygotic genotypes AA and TT (but not GG and CC, or AG
and CT genotypes); however, they do not introduce functionally
significant single amino acid modifications (coding
synonymous).
 | Table V.Evaluation of the ESE modifications
introduced by the rs3749166 (A>G) and rs5404 (C>T) SNPs using
ESE finder [Cartegni et al (4)]. |
Table V.
Evaluation of the ESE modifications
introduced by the rs3749166 (A>G) and rs5404 (C>T) SNPs using
ESE finder [Cartegni et al (4)].
| Gene | SNP | Exon type | Splice site/SR
protein binding | Major allele ESE
finder score | Minor allele ESE
finder score | ΔScorea |
|---|
| CAPN10 | rs3749166 | Alternative | Exon 11 splice site
(3SS_U2_human) | 10.350 | −3.220 | 13.570 |
| SLC2A2 | rs5404 | Constitutive | SRp40 | 3.793 | 6.324 | 2.531 |
|
|
|
| SF2/ASF
(IgM-BRCA1) | 2.337 | 3.769 | 1.402 |
|
|
|
| SF2/ASF | 3.162 | 3.778 | 0.616 |
The present results, summarized in Fig. 2, indicate that the most protective
genotype for T2D is the fully epigenetic genotype. In accordance
with the above-mentioned analysis, the heterozygous rs3749166 and
rs5404 genotypes are also differently associated with T2D. Carriers
of the AG/CC genotype (heterozygous for rs3749166) are
significantly more frequent among the T2D patients and exhibit
particularly high levels of HbA1c, probably indicating resistance
to pharmaceutical intervention for T2D. Similar findings regarding
the negative effect associated with the synthesis of two different
isoforms have been recently reported in association with
heterozygous SNPs causing alternative splicing. For example, Tian
et al (27) reported a number
of disorders associated with alternative exon expression and
splicing. In addition, Kurzawski et al (14) reported on the effect of epigenetic SNP
rs5030952 in CAPN10, which exhibits a heterozygotic association
with post-transplant diabetes mellitus.
By contrast, the presence of the apparently
protective rs5404 SNP (CT genotype in SLC2A2) is potentially
associated with the modified response of CT heterozygotes to
different stimuli (based on the data from ESE score analysis at
least one novel ESE is formed, which significantly responds to
SRp40 proteins). This appears to be particularly significant for
carriers of the rs3749166 CAPN10 AG genotype (the combined AG/CT
genotype is not T2D-associated, and its carriers do not exhibit
particularly high levels of HbA1c. The advantage of epigenetic
regulation provided by the C allele and, thus, the ESE response may
be lost among TT homozygotes. This could be a possible explanation
for the absence of TT homozygotes regardless of the relatively high
total frequency of the T allele (21.28%, non-Mendelian
genetics).
These findings provide the hypothesis that mutations
modifying the response to splicing regulatory mechanisms
(epigenetic and ESE) may be associated with strong negative
functional changes, and exhibit complex, nonlinear disease
associations (28). Provided that
functionally significant epigenetic SNPs are frequent (11), this type of genetic variation is
expected to have a strong impact on disease and evolution.
A major obstacle in investigating complex
pathological conditions, such as metabolic syndrome, is the limited
understanding of the regulatory factors involved in the expression
of interacting components. Recent evidence indicates the key role
of alternative RNA expression in developmental changes (29), and the production of coding and
non-coding RNA sequences. Another factor is the complex epigenetic
modifications, which may also lead to the expression of different
RNA isoforms. The current results indicate likely synergies between
synonymous splicing-regulatory epigenetic SNPs, which modify the
splicing potential of two different glucose transport-associated
genes, and reveal that bioinformatic analysis and careful
investigation of the SNPs under investigation may become a powerful
tool for identifying potentially significant genetic modifications
with respect to splicing.
In conclusion, the results presented above indicate
for the first time, to the best of our knowledge, the correlation
and disease association of two synonymous epigenetic SNPs, which
participate in the regulation of the glucose transport system and
introduce exclusively splicing-associated modifications. Taken
together, these results reveal that T2D is subject to deregulation
by complex splicing mechanisms, which may exhibit heterozygous
disease association or protection, depending on the
splicing-affecting genetic variation. A detailed bioinformatic
analysis of the changes introduced by SNPs would facilitate the
understanding of the impact of functional changes introduced by
genetic variation.
Acknowledgements
The present study was co-financed by the European
Union (the European Social Fund) and Greek national funds through
the Operational Program, ‘Education and Lifelong Learning’ of the
NSRF - Research Funding Program: Heracleitus II. Investing in
knowledge society through the European Social Fund (project no.
87113).
References
|
1
|
Romero PR, Zaidi S, Fang YY, Uversky VN,
Radivojac P, Oldfield CJ, Cortese MS, Sickmeier M, LeGall T,
Obradovic Z, et al: Alternative splicing in concert with protein
intrinsic disorder enables increased functional diversity in
multicellular organisms. Proc Natl Acad Sci USA. 103:8390–8395.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Stamm S, BenAri S, Rafalska I, Tang Y,
Zhang Z, Toiber D, Thanaraj TA and Soreq H: Function of alternative
splicing. Gene. 344:1–20. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Soukarieh O, Gaildrat P, Hamieh M, Drouet
A, BaertDesurmont S, Frébourg T, Tosi M and Martins A: Exonic
splicing mutations are more prevalent than currently estimated and
can be predicted by using in silico tools. PLoS Genet.
12:e10057562016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cartegni L, Wang J, Zhu Z, Zhang MQ and
Krainer AR: ESEfinder: a web resource to identify exonic splicing
enhancers. Nucleic Acids Res. 31:3568–3571. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Anastasiadou C, Malousi A, Maglaveras N
and Kouidou S: Human epigenome data reveal increased CpG
methylation in alternatively spliced sites and putative exonic
splicing enhancers. DNA Cell Biol. 30:267–275. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ong CT and Corces VG: CTCF: an
architectural protein bridging genome topology and function. Nat
Rev Genet. 15:234–246. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Malousi A and Kouidou S: DNA
hypermethylation of alternatively spliced and repeat sequences in
humans. Mol Genet Genomics. 287:631–642. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Shoemaker R, Deng J, Wang W and Zhang K:
Allele-specific methylation is prevalent and is contributed by
CpG-SNPs in the human genome. Genome Res. 20:883–889. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Scalet D, Balestra D, Rohban S, Bovolenta
M, Perrone D, Bernardi F, Campaner S and Pinotti M: Exploring
splicing-switching molecules for seckel syndrome therapy. Biochim
Biophys Acta. 1863:15–20. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Karambataki M, Malousi A, Maglaveras N and
Kouidou S: Synonymous polymorphisms at splicing regulatory sites
are associated with CpGs in neurodegenerative disease-related
genes. Neuromolecular Med. 12:260–269. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Karambataki M, Malousi A and Kouidou S:
Risk-associated coding synonymous SNPs in type 2 diabetes and
neurodegenerative diseases: Genetic silence and the underrated
association with splicing regulation and epigenetics. Mutat Res.
770:85–93. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Harlid S, Ivarsson MI, Butt S, Hussain S,
Grzybowska E, Eyfjörd JE, Lenner P, Försti A, Hemminki K, Manjer J,
et al: A candidate CpG SNP approach identifies a breast cancer
associated ESR1-SNP. Int J Cancer. 129:1689–1698. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Imamura M and Maeda S: Genetics of type 2
diabetes: the GWAS era and future perspectives (Review). Endocr J.
58:723–739. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kurzawski M, Dziewanowski K, Kedzierska K,
Gornik W, Banas A and Drozdzik M: Association of calpain-10 gene
polymorphism and posttransplant diabetes mellitus in kidney
transplant patients medicated with tacrolimus. Pharmacogenomics J.
10:120–125. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Shchetynsky K, Protsyuk D, Ronninger M,
DiazGallo LM, Klareskog L and Padyukov L: Gene-gene interaction and
RNA splicing profiles of MAP2K4 gene in rheumatoid arthritis. Clin
Immunol. 158:19–28. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Brown AE, Yeaman SJ and Walker M: Targeted
suppression of calpain-10 expression impairs insulin-stimulated
glucose uptake in cultured primary human skeletal muscle cells. Mol
Genet Metab. 91:318–324. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Alsaraj F, O'Gorman D, McAteer S,
McDermott J, Hawi Z and Sreenan S: Haplotype association of calpain
10 gene variants with type 2 diabetes mellitus in an Irish sample.
Ir J Med Sci. 179:269–272. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Song Y, You NC, Hsu YH, Sul J, Wang L,
Tinker L, Eaton CB and Liu S: Common genetic variation in
calpain-10 gene (CAPN10) and diabetes risk in a multi-ethnic cohort
of American postmenopausal women. Hum Mol Genet. 16:2960–2971.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Barroso I, Luan J, Middelberg RP, Harding
AH, Franks PW, Jakes RW, Clayton D, Schafer AJ, O'Rahilly S and
Wareham NJ: Candidate gene association study in type 2 diabetes
indicates a role for genes involved in beta-cell function as well
as insulin action. PLoS Biol. 1:E202003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Laukkanen O, Lindström J, Eriksson J,
Valle TT, Hämäläinen H, Ilanne-Parikka P, Keinänen-Kiukaanniemi S,
Tuomilehto J, Uusitupa M and Laakso M: Finnish Diabetes Prevention
Study: Polymorphisms in the SLC2A2 (GLUT2) gene are associated with
the conversion from impaired glucose tolerance to type 2 diabetes:
The Finnish Diabetes Prevention Study. Diabetes. 54:2256–2260.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kilpeläinen TO, Lakka TA, Laaksonen DE,
Mager U, Salopuro T, Kubaszek A, Todorova B, Laukkanen O, Lindström
J, Eriksson JG, et al: Finnish Diabetes Prevention Study Group:
Interaction of single nucleotide polymorphisms in ADRB2, ADRB3,
TNF, IL6, IGF1R, LIPC, LEPR, and GHRL with physical activity on the
risk of type 2 diabetes mellitus and changes in characteristics of
the metabolic syndrome: The Finnish Diabetes Prevention Study.
Metabolism. 57:428–436. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Willer CJ, Bonnycastle LL, Conneely KN,
Duren WL, Jackson AU, Scott LJ, Narisu N, Chines PS, Skol A,
Stringham HM, et al: Screening of 134 single nucleotide
polymorphisms (SNPs) previously associated with type 2 diabetes
replicates association with 12 SNPs in nine genes. Diabetes.
56:256–264. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
American Diabetes Association, . Standards
of medical care in diabetes - 2013. Diabetes Care. 36:(Suppl 1).
S11–S66. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ye J, Coulouris G, Zaretskaya I,
Cutcutache I, Rozen S and Madden TL: Primer-BLAST: A tool to design
target-specific primers for polymerase chain reaction. BMC
Bioinformatics. 13:1342012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Church GM: The personal genome project.
Mol Syst Biol. 1:2005.0030. 2005. View Article : Google Scholar
|
|
26
|
Chen HH, Wang YC and Fann MJ:
Identification and characterization of the CDK12/cyclin L1 complex
involved in alternative splicing regulation. Mol Cell Biol.
26:2736–2745. 2006. View Article : Google Scholar
|
|
27
|
Tian C, Yan R, Wen S, Li X, Li T, Cai Z,
Li X, Du H and Chen H: A splice mutation and mRNA decay of EXT2
provoke hereditary multiple exostoses. PLoS One. 9:e948482014.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Strohman RC: Linear genetics, non-linear
epigenetics: complementary approaches to understanding complex
diseases. Integr Physiol Behav Sci. 30:273–282. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lau E: Non-coding RNA: Zooming in on
lncRNA functions. Nat Rev Genet. 15:574–575. 2014. View Article : Google Scholar : PubMed/NCBI
|