Introduction
Nasopharyngeal carcinoma (NPC) is one of the most
malignant head and neck carcinomas with unique epidemiological
features (1). Each year, >50,000
mortalities of patients with cancer are due to NPC in China
(2). Epidemiologically, the regions
of South China and South East Asia sustain the incidence peaks
(2). Advanced NPC yields high
lethality due to late stage diagnosis and metastasis, which remains
to be the leading cause of therapeutic failure in the clinic
(3). Due to the unique location and a
lack of specific symptoms, NPC is rarely detected during regular
medical examinations (1–3). In this regard, early and accurate
diagnosis continues to be a key approach to obtain an optimal
prognosis for patients with NPC.
The discovery of microRNA (miRNA) has offered novel
perspectives for cancer research. miRNA are classically defined as
a type of RNA transcript (20–22 nucleotides in length), which
negatively regulate the expression of protein-coding genes at the
transcriptional or translational level (4,5). Basal
expression of miRNA in cells or tissues is essential for various
somatic processes, including cell proliferation, differentiation,
the cell cycle and apoptosis (4–6). Numerous
studies have documented that deregulated miRNA expression is
strongly linked to the occurrence of various carcinomas (7,8). It has
become increasingly apparent that miRNA are of crucial importance
for the carcinogenesis and progression of NPC, and therefore they
have been highlighted as powerful screening predictors in
confirming or monitoring NPC (9–15).
Unfortunately, the clinical utility of miRNA signatures remain
unpopular in the clinic. Additionally, it appears that different
studies have presented inconsistent results with regard to miRNA
profiling in identifying NPC. For example, a previous study has
demonstrated that miRNA testing is adequately sensitive and
specific for NPC confirmation, with a diagnostic sensitivity and
specificity of up to 96% (13).
However, some research has reported that miRNA testing achieved an
efficacy <60% (10,14,15).
Therefore, the present study conducted a comprehensive
meta-analysis and aimed to assess the overall diagnostic
performance of miRNA as biomarkers for NPC identification.
Materials and methods
Literature search strategy
The entire contents of the present meta-analysis
followed the guidelines of the Preferred Reporting Items for
Systematic Reviews and Meta-analysis statement (16). Literatures were collected based on the
online PubMed (ncbi.nlm.nih.gov/pubmed) and EMBASE (embase.com/#search) databases up to June 30th 2016,
utilizing the following search words: ‘Nasopharyngeal carcinoma,’
‘microRNA,’ ‘miRNA’ and ‘diagnosis/sensitivity/specificity,’ with
limitations to ‘human’ and ‘English.’ The inclusion criteria were
as follows: i) Studies that evaluated the diagnostic performance of
miRNA for NPC; ii) the final diagnosis of NPC was confirmed by
tissue-proven histopathology; iii) studies provided complete data
to construct 2×2 contingency tables; and iv) studies explicitly
addressed the control sources and size. Studies with the following
criteria were excluded: i) Studies without complete data to
construct 2×2 contingency tables; ii) studies failed to explicitly
state the control group(s); and iii) non-English written papers,
review articles, basic study, letters, commentaries and
meta-analyses.
Data extraction and quality
grading
Eligible data were retrieved by two authors, and the
contents included the name of the first author, year of
publication, origin, patient size, control sources and size, miRNA
expression profiles, and sensitivity and specificity data. The
included studies were further evaluated for the quality grading
based on the diagnostic accuracy (Quality Assessment of Diagnostic
Accuracy Studies; QUADAS) tool issued in 2003 (17). According to the QUADAS scoring
criteria where 14 questions were included, a ‘Yes’ answer
corresponded to a score of ‘1,’ whereas a ‘No’ or ‘Unclear’ answer
received a score of ‘0.’
Statistical analysis
Statistical analyses were performed based on the
platforms of Stata 12.0 (StataCorp LP, College Station, TX, USA)
and Meta-disc 1.4 (Unit of Clinical Biostatistics Team of the Ramón
y Cajal Hospital, Barcelona, Spain) software. Heterogeneity from
the threshold effect was evaluated by the Spearman's correlation
coefficient, and that from the non-threshold effect was assessed by
Cochran's Q test and inconsistency index (I2) test
(P<0.05 or I2 >50%) (18). A random effects model was applied for
the meta-analysis in the case of the existence of heterogeneity
(19). The generated diagnostic
parameters included pooled sensitivity, specificity, positive
likelihood ratio (PLR), negative likelihood ratio (NLR), diagnostic
odds ratio (DOR) and summary receiver operator characteristic
(SROC) curve. The clinical utility (post-test probabilities) were
assessed by Fagan's plot assays. Publication bias among studies was
determined by Deeks' funnel plot asymmetry test with a significant
level of P<0.05.
Results
Literature search and enrollment
The process of study selection is demonstrated in
Fig. 1. According to the predefined
criteria, 275 relevant articles were obtained from online PubMed
and EMBASE databases following elimination of the duplicated
records. The retrieved papers received a detailed review of the
study title and abstract, and 256 records were accordingly
excluded. The possible eligible studies that received full text
evaluation were restricted to 19, and 13 of them failed to meet the
aim of the present study and were finally excluded. Eventually, six
cohorts (including 21 individual studies) comprising 528 NPC cases
and 252 matched controls were available for the meta-analysis
(10–15).
Study characteristics and quality
assessments
The main features for each included study are
summarized in Table I. All six
studies were conducted in China, and the evaluation methods for
miRNA levels were all based on reverse transcription-quantitative
polymerase chain reaction. The test matrices comprised serum,
plasma and tissue, and the reference genes contained U6, miR-454,
miR-39 and miR-238.
 | Table I.Main features of included studies for
miRNA signatures in the identification of nasopharyngeal
carcinoma. |
Table I.
Main features of included studies for
miRNA signatures in the identification of nasopharyngeal
carcinoma.
| Author, year | Study population | Patient/control
size | Specimen type | Test method | miRNA profile | Reference gene | Cut-off value | QUADAS score | (Refs.) |
|---|
| Liu et al,
2013 | Chinese | 217/73 | Plasma | RT-qPCR | miR-16, miR-21,
miR-24, miR-155, miR-378 | U6 | Unclear | 11 | (10) |
| Tang et al,
2014 | Chinese | 67/25 | Tissue | RT-qPCR | miR-205,
miR-135b | U6 | Unclear | 8 | (11) |
| Wang et al,
2014 | Chinese | 50/50 | Plasma | RT-qPCR | miR-483, miR-103,
miR-29a, let-7c | Unclear | Unclear | 10 | (12) |
| Zeng et al,
2012 | Chinese | 74/57 | Serum | RT-qPCR | miR-17, miR-20a,
miR-29c and miR-223 | miR-39 and
miR-238 | Unclear | 10 | (13) |
| Zhang et al,
2015 | Chinese | 89/28 | Plasma | RT-qPCR | BART-13, BART-7 | U6B | Unclear | 12 | (14) |
| Zheng et al,
2014 | Chinese | 31/19 | Plasma | RT-qPCR | miR-548q,
miR-483-5p | miR-454 | Unclear | 11 | (15) |
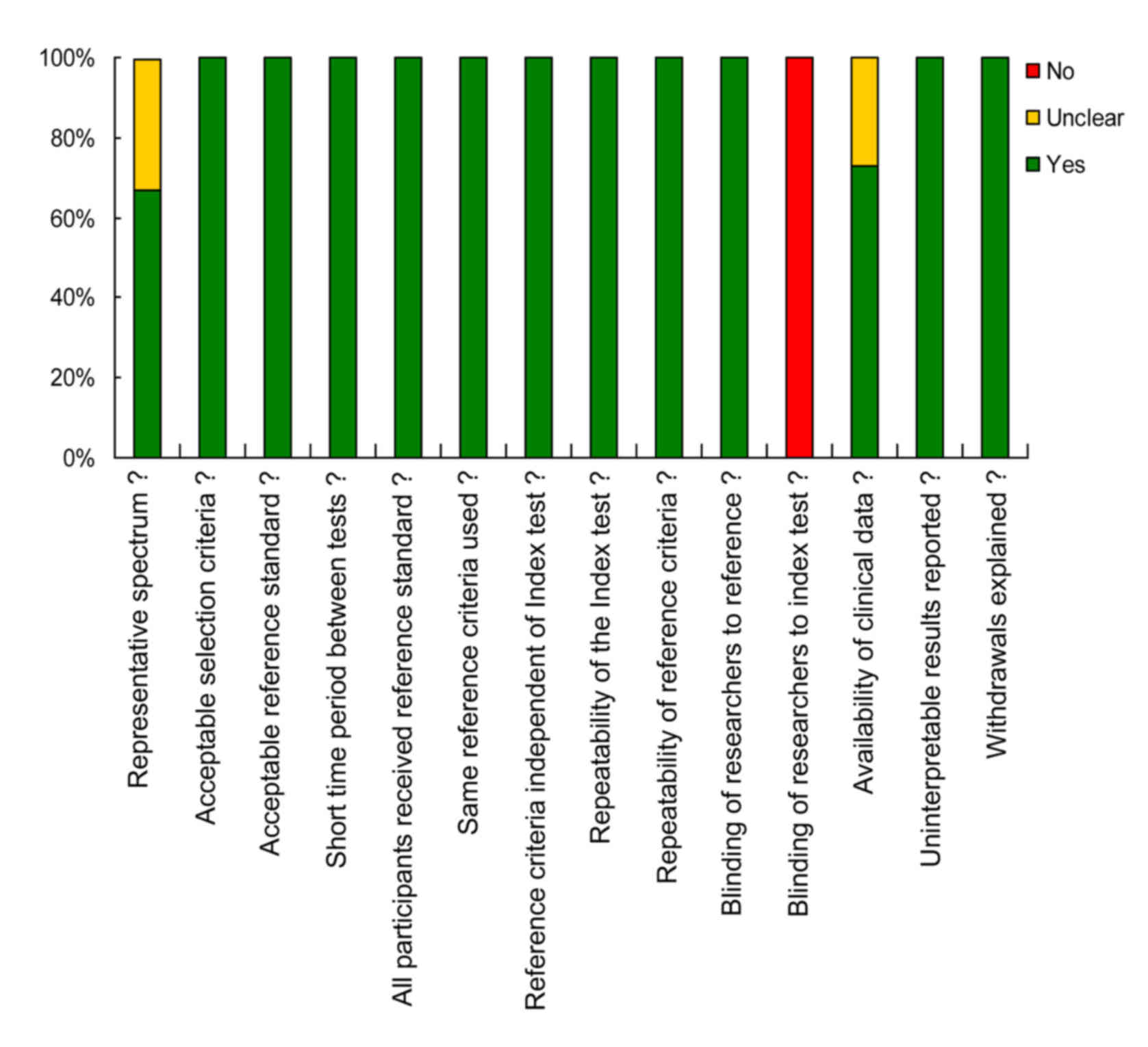
The article quality of each included publication was
evaluated in terms of the 14-item QUADAS checklist (17). All of the studies yielded a QUADAS
score ≥8, suggesting a relatively high quality of the enrolled
studies. The proportions of studies with low, high or unclear
concerns are displayed in Fig. 2.
Heterogeneity
Table II summarizes
the evaluated study heterogeneity from threshold and non-threshold
effects. As indicated in Table II,
the P-value obtained from Spearman's correlation coefficient in the
overall pooled studies was estimated to be 0.012, indicating a
significant heterogeneity generated from threshold effect.
Additionally, the Cochran's Q test for the overall pooled analysis
yielded a P-value of P<0.0001, along with an I2 value
of 83.8%, suggesting that the non-threshold effect is likely to be
a source of heterogeneity as well. As a result, a random analysis
model was selected for the final meta-analysis.
 | Table II.Analyses of study heterogeneity of all
pooled studies. |
Table II.
Analyses of study heterogeneity of all
pooled studies.
|
|
|
|
| Heterogeneity |
|---|
|
|
|
|
|
|
|---|
| Analysis | Spearman's
correlation coefficient | Cochran's Q test
test | I2
(%) | Threshold effect | Non-threshold
effect |
|---|
| Single miRNA | −0.165, P=0.557 | 37.16, P=0.0007 | 62.3 | No | Yes |
| Multiple miRNA | −0.928, P=0.008 | 27.01, P=0.0001 | 81.5 | Yes | Yes |
| Plasma | −0.275, P=0.285 | 80.95,
P<0.0001 | 80.2 | No | Yes |
| Serum/tissue | −0.600, P=0.400 | 17.63, P=0.0005 | 83 | No | Yes |
| Outlier excluded | −0.539, P=0.021 | 75.76,
P<0.0001 | 77.6 | Yes | Yes |
| Overall | −0.535,
P=0.012 | 123.43,
P<0.0001 | 83.8 | Yes | Yes |
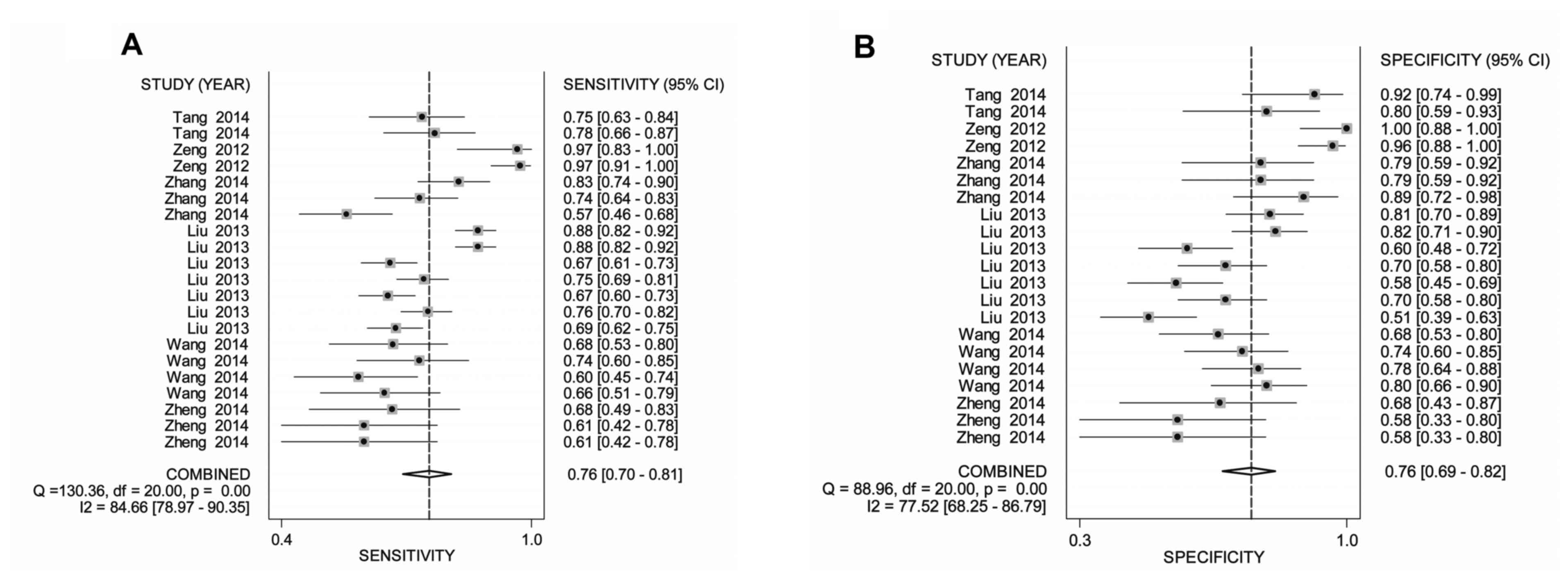
Diagnostic performance and clinical
utility
As indicated in Table
III, the pooled sensitivity, specificity, PLR, NLR, DOR and
area under the curve (AUC) for miRNA profiling were 0.76 [95%
confidence interval (CI), 0.70–0.81], 0.76 (95% CI, 0.69–0.82),
2.81 (95% CI, 2.19–3.61), 0.35 (95% CI, 0.28–0.44), 9.01 (95% CI,
5.62–14.44) and 0.83, respectively. Following adjustment of the
outlier studies, the combined sensitivity, specificity, PLR, NLR,
DOR and AUC for miRNA profiling were estimated to be 0.78 (95% CI,
0.76–0.80), 0.78 (95% CI, 0.75–0.81), 3.19 (95% CI, 2.48–4.11),
0.32 (95% CI, 0.25–0.40), 11.43 (95% CI, 7.02–18.61) and 0.84,
respectively. Forest plots of pooled sensitivity, specificity as
well as the SROC curve are demonstrated in Fig. 3A-C.
 | Table III.Pooled diagnostic indices of miRNA
profiling in confirming nasopharyngeal carcinoma. |
Table III.
Pooled diagnostic indices of miRNA
profiling in confirming nasopharyngeal carcinoma.
|
| Diagnostic
indices |
|---|
|
|
|
|---|
| Analysis | Pooled sensitivity
(95% CI) | Pooled specificity
(95% CI) | Pooled positive
likelihood ratio (95% CI) | Pooled negative
likelihood ratio (95% CI) | Pooled diagnostic
odds ratio (95% CI) | Area under the
curve |
|---|
| miRNA profile |
|
|
|
|
|
|
| Single
miRNA | 0.7 | 0.69 | 2.25 | 0.44 | 5.34 | 0.75 |
|
| (0.68–0.72) | (0.65–0.72) | (1.83–2.76) | (0.38–0.51) | (3.78–7.52) |
|
|
Multiple miRNA | 0.88 | 0.85 | 5.23 | 0.16 | 41.29 | 0.95 |
|
| (0.85–0.90) | (0.81–0.89) | (2.91–9.39) | (0.09–0.27) | (13.83–123.22) |
|
| Sample type |
|
|
|
|
|
|
|
Plasma | 0.73 | 0.71 | 2.5 | 0.39 | 6.46 | 0.78 |
|
| (0.68–0.77) | (0.66–0.76) | (2.02–3.08) | (0.32–0.49) | (4.31–9.68) |
|
|
Serum/tissue | 0.91 | 0.94 | 15.8 | 0.1 | 157.53 | 0.97 |
|
| (0.73–0.97) | (0.82–0.98) | (4.33–57.70) | (0.03–0.33) |
(15.34–1617.17) |
|
| Outlier
eliminated | 0.78 | 0.78 | 3.19 | 0.32 | 11.43 | 0.84 |
|
| (0.76–0.80) | (0.75–0.81) | (2.48–4.11) | (0.25–0.40) | (7.02–18.61) |
|
| Overall | 0.76 | 0.76 | 2.81 | 0.35 | 9.01 | 0.83 |
|
| (0.70–0.81) | (0.69–0.82) | (2.19–3.61) | (0.28–0.44) | (5.62–14.44) |
|
For the clinical utility assessed by Fagan's plot
assay, apparent improvements of post-test probabilities were
displayed in the pooled analysis, with a post-test probability of a
positive result of 45% and negative result of 7% (Fig. 3D).
Subgroup analysis
Subgroup analysis was stratified by miRNA profiling
(single or in parallel) and the test matrix. As exemplified in
Table III, paralleled testing of
miRNA achieved better diagnostic efficacy than single miRNA
analysis: The pooled sensitivity was 0.88 (95% CI, 0.85–0.90) vs.
0.70 (95% CI, 0.68–0.72), specificity was 0.85 (95% CI, 0.81–0.89)
vs. 0.69 (95% CI, 0.65–0.72) and AUC was 0.95 vs. 0.75. When the
studies were stratified by the test matrix, the data manifested
that serum/tissue-based tests conferred higher accuracy than
plasma-based miRNA analysis in confirming NPC (sensitivity, 0.91
vs. 0.73; specificity, 0.94 vs. 0.71; AUC, 0.97 vs. 0.78).
Influence assay and
meta-regression
Influence analysis and meta-regression tests were
performed to deeply trace the heterogeneity sources. As
demonstrated in Fig. 4, three
individual studies were evaluated to undergo a deviation status.
Following adjustment of the data by eliminating the outliers, the
P-value of Spearman's correlation coefficient was altered from
0.012 to 0.021, and the I2 value declined from 83.8 to
77.6%, hinting that the deviation data is likely a source of
heterogeneity (Table II).
The meta-regression test was conducted based on five
pre-specified covariates: Test matrix (plasma, serum or tissue),
reference gene (U6 or other), test pattern (panel or single), NPC
cases (<100 or ≥100), control size (<50 or ≥50) and article
quality (QUADAS≥10 or <10) (20).
The results revealed that the test pattern (P=0.0001) and reference
gene (P=0.0026) were the key factors that contributed to the
heterogeneity sources (Table
IV).
 | Table IV.Inverse variance weights for the
potential source of heterogeneity assessed by meta-regression
tests. |
Table IV.
Inverse variance weights for the
potential source of heterogeneity assessed by meta-regression
tests.
| Study
characteristic | P-value | Relative diagnostic
odds ratio (95% confidence interval) |
|---|
| Test pattern (panel
vs. single) | 0.0001 | 7.53
(3.27–17.32) |
| Test matrix (plasma
vs. serum vs. tissue) | 0.0763 | 0.28
(0.07–1.16) |
| Reference gene (U6
vs. other) | 0.0026 | 0.61
(0.45–0.82) |
| Nasopharyngeal
carcinoma cases (<100 vs. ≥100) | 0.1735 | 0.51
(0.19–1.39) |
| Control size
(<50 vs. ≥50) | 0.1101 | 3.40
(0.73–15.88) |
| Article quality
(QUADAS≥10 vs. QUADAS<10) | 0.0703 | 0.71
(0.49–1.03) |
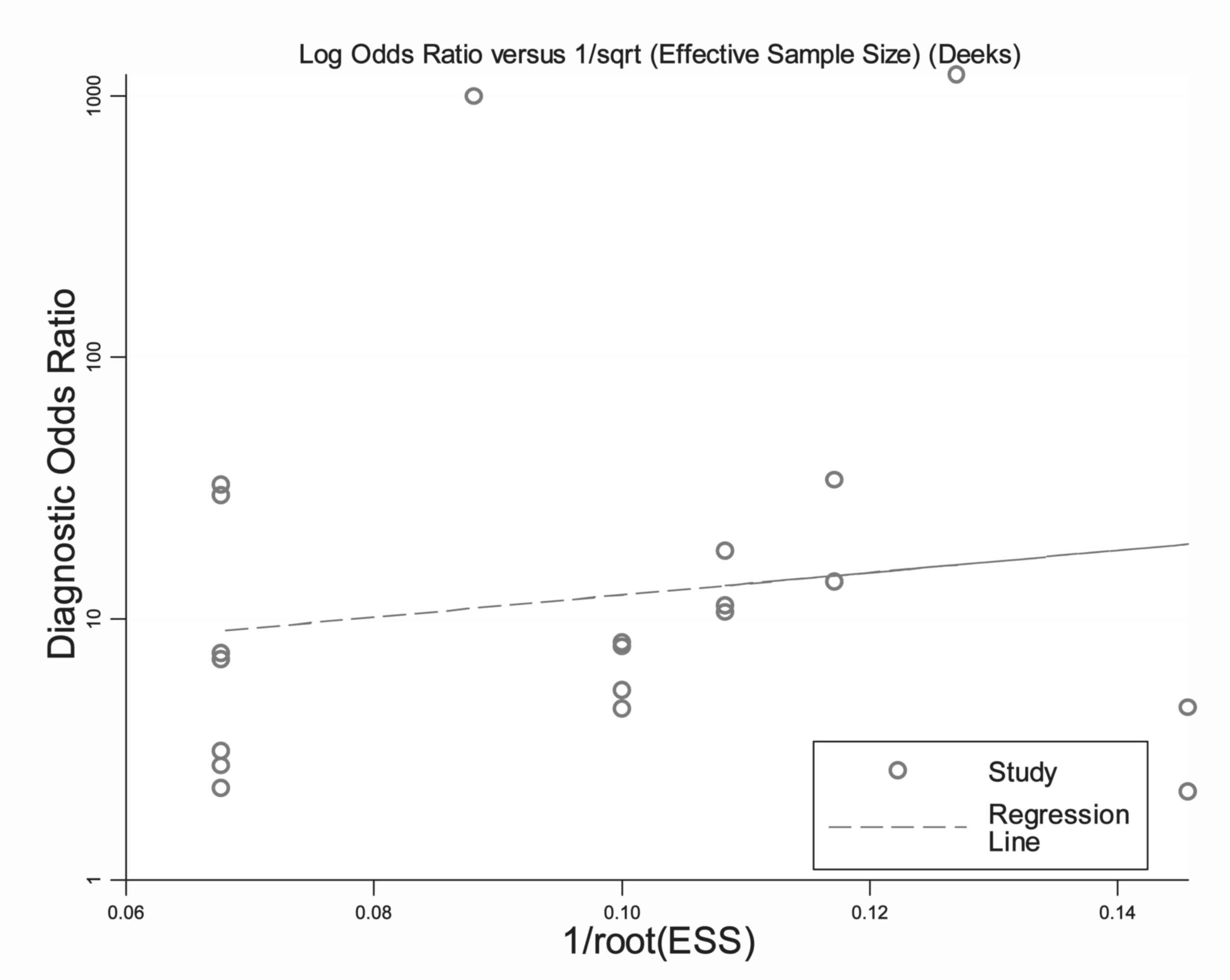
Publication bias
The funnel plots for publication bias demonstrated
no asymmetry for the overall pooled analysis, along with a P-value
of 0.702, indicating that there was no bias from the publications
(Fig. 5).
Discussion
Advanced NPC sustains high lethality and delayed
diagnosis remains to be the leading cause for therapeutic failure
(1,2).
The treatment outcomes, as well as NPC management, may be improved
by the development of non-invasive biomarker assays that help to
reinforce the overall diagnostic efficacy (3). Large quantities of research regarding
the diagnostic value of miRNA profiling for NPC are available
(9–15); however, there are currently no
consistent results among these studies and the clinical utility of
miRNA profiling for NPC management is hotly debated. In this
regard, the present meta-analysis was conducted and a comprehensive
evaluation of the predictive efficacy of miRNA signatures for NPC
identification was performed.
Results from the present meta-analysis demonstrated
that miRNA profiling retained a pooled sensitivity of 0.76,
specificity of 0.76 and AUC of 0.83 for its capacity to
discriminate the patients with NPC from cancer-free individuals.
Another important indicator, termed the DOR (18), was estimated to be 9.01 in the present
analysis, which indicated a relatively high discriminatory
performance of miRNA testing in the management of NPC.
Additionally, the pooled PLR of 2.81 suggested that miRNA profiling
yielded a ratio of nearly 3 between the true positive rate and
false positive rate. Correspondingly, the pooled NLR indicated that
the probability of NPC cases that tested negative for miRNA vs. the
probability of cases that tested positive for these miRNA achieved
a ratio of 0.35. For the clinical utility, miRNA signatures raised
the post-test probability of a positive result to 46% and lowered
the post-test probability of a negative result to 7%. Overall, the
present data demonstrated that miRNA signatures may be popularized
as auxiliary biomarkers for NPC identification.
Stratified analysis was also conducted based on
miRNA profiling (single or parallel) and test matrices. The data
demonstrated that multiple testing of miRNA achieved higher
diagnostic efficacy than single miRNA analysis. A study by Liu et
al (10) evidenced that a combination
of five miRNA (miR-16, −21, −24, −155 and −378) reached a
diagnostic sensitivity of 0.88 and specificity of 0.82 for NPC,
which is better than the single tests. On the other hand, the
analysis of miRNA signature test matrix revealed that
serum/tissue-based testing achieved a higher diagnostic accuracy
than that of plasma-based analysis. A study by Wang et al (21) indicated that the coagulation process
is likely to affect the spectrum of extracellular molecules in the
blood, hinting that different matrices, including serum or plasma,
may sustain altered diagnostic efficacy. Nevertheless, the analysis
stratified by the matrix yielded a small study size and displayed
high heterogeneity among studies. Hence, more investigations are
warranted to reinforce this preliminary evidence.
The present pooled analysis demonstrated significant
heterogeneity among studies. Heterogeneity mainly derives from
threshold and non-threshold effects (18–20). The
threshold effect is predominantly generated by the different
cut-off value settings or thresholds used in different studies,
whereas the non-threshold effect could be caused by different
ethnicities, testing methods, sample types, as well as the severity
of disease conditions (18). In the
overall pooled analysis, the Spearman's correlation coefficient,
Cochran's Q and I2 tests all presented significant
results, indicating that heterogeneity came from both threshold and
non-threshold effects. As a result, sensitivity analysis and
meta-regression tests were conducted in the present study to deeply
trace the heterogeneity sources. The results revealed that
different miRNA test patterns, as well as the non-unified reference
gene, appeared to be a contributor of study heterogeneity, whereas
the control types, study size and article quality demonstrated a
low likelihood of influencing the heterogeneity sources.
In conclusion, the present analyses evaluated the
diagnostic value of miRNA profiling for NPC identification, in
particular elucidating that parallel testing and non-plasma based
miRNA signatures yield improved efficacy. Nevertheless, due to the
small number of studies, obvious heterogeneity, as well as
complicated control sources in the present study, the overall
pooled accuracy is compromised. More investigations are warranted
to further testify the present preliminary evidence.
Acknowledgements
The present study was supported by the National
Clinical Key Specialty Construction Program of China (2013) and
Provincial Natural Science Fund of Fujian (grant no.
2016J01511).
References
|
1
|
Petersson F: Nasopharyngeal carcinoma: A
review. Semin Diagn Pathol. 32:54–73. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wei KR, Zheng RS, Zhang SW, Liang ZH, Ou
ZX and Chen WQ: Nasopharyngeal carcinoma incidence and mortality in
China in 2010. Chin J Cancer. 33:381–387. 2014.PubMed/NCBI
|
|
3
|
Cao SM, Simons MJ and Qian CN: The
prevalence and prevention of nasopharyngeal carcinoma in China.
Chin J Cancer. 30:114–119. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Pritchard CC, Cheng HH and Tewari M:
MicroRNA profiling: Approaches and considerations. Nat Rev Genet.
13:358–369. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pileczki V, Cojocneanu-Petric R, Maralani
M, Neagoe IB and Sandulescu R: MicroRNAs as regulators of apoptosis
mechanisms in cancer. Clujul medical 1957. 89:50–55.
2016.https://doi.org/10.15386/cjmed-512 View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Frixa T, Donzelli S and Blandino G:
Oncogenic MicroRNAs: Key Players in Malignant Transformation.
Cancers (Basel). 7:2466–2485. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tuna M, Machado AS and Calin GA: Genetic
and epigenetic alterations of microRNAs and implications for human
cancers and other diseases. Genes Chromosomes Cancer. 55:193–214.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lee KT, Tan JK, Lam AK and Gan SY:
MicroRNAs serving as potential biomarkers and therapeutic targets
in nasopharyngeal carcinoma: A critical review. Crit Rev Oncol
Hematol. 103:1–9. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liu X, Luo HN, Tian WD, Lu J, Li G, Wang
L, Zhang B, Liang BJ, Peng XH, Lin SX, et al: Diagnostic and
prognostic value of plasma microRNA deregulation in nasopharyngeal
carcinoma. Cancer Biol Ther. 14:1133–1142. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tang JF, Yu ZH, Liu T, Lin ZY, Wang YH,
Yang LW, He HJ, Cao J, Huang HL and Liu G: Five miRNAs as novel
diagnostic biomarker candidates for primary nasopharyngeal
carcinoma. Asian Pac J Cancer Prev. 15:7575–7581. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang HY, Yan LX, Shao Q, Fu S, Zhang ZC,
Ye W, Zeng YX and Shao JY: Profiling plasma microRNA in
nasopharyngeal carcinoma with deep sequencing. Clin Chem.
60:773–782. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zeng X, Xiang J, Wu M, Xiong W, Tang H,
Deng M, Li X, Liao Q, Su B, Luo Z, et al: Circulating miR-17,
miR-20a, miR-29c, and miR-223 combined as non-invasive biomarkers
in nasopharyngeal carcinoma. PLoS One. 7:e463672012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang G, Zong J, Lin S, Verhoeven RJ, Tong
S, Chen Y, Ji M, Cheng W, Tsao SW, Lung M, et al: Circulating
Epstein-Barr virus microRNAs miR-BART7 and miR-BART13 as biomarkers
for nasopharyngeal carcinoma diagnosis and treatment. Int J Cancer.
136:E301–E312. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zheng XH, Cui C, Ruan HL, Xue WQ, Zhang
SD, Hu YZ, Zhou XX and Jia WH: Plasma microRNA profiling in
nasopharyngeal carcinoma patients reveals miR-548q and miR-483-5p
as potential biomarkers. Chin J Cancer. 33:330–338. 2014.PubMed/NCBI
|
|
16
|
Moher D, Liberati A, Tetzlaff J and Altman
DG; PRISMA Group, : Preferred reporting items for systematic
reviews and meta-analyses: The PRISMA statement. J Clin Epidemiol.
62:1006–1012. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Whiting P, Rutjes AW, Reitsma JB, Bossuyt
PM and Kleijnen J: The development of QUADAS: A tool for the
quality assessment of studies of diagnostic accuracy included in
systematic reviews. BMC Med Res Methodol. 3:252003. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cui ZL, Zheng DZ, Liu YH, Chen LY, Lin DH
and Feng-Hua Lan: Diagnostic Accuracies of the TUNEL SCD, and Comet
Based Sperm DNA Fragmentation Assays for Male Infertility: A
Meta-analysis Study. Clin Lab. 61:525–535. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chen Y, Cui Z, Xiao Z, Hu M, Jiang C, Lin
Y and Chen Y: PAX1 and SOX1 methylation as an initial screening
method for cervical cancer: A meta-analysis of individual studies
in Asians. Ann Transl Med. 4:3652016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cui Z, Chen Y, Xiao Z, Hu M, Lin Y, Chen Y
and Zheng Y: Long noncoding RNAs as auxiliary biomarkers for
gastric cancer screening: A pooled analysis of individual studies.
Oncotarget. 7:25791–25800. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang K, Yuan Y, Cho JH, McClarty S, Baxter
D and Galas DJ: Comparing the MicroRNA spectrum between serum and
plasma. PLoS One. 7:e415612012. View Article : Google Scholar : PubMed/NCBI
|