Introduction
Endometriosis is characterized by the presence of
endometrial tissue (glandular and stromal) abnormally outside the
uterine cavity (1). According to
certain statistics, there is an average of 10.4 years elapse from
the first onset of symptoms to diagnosis, and 74% of patients
receive at least one false diagnosis, which results in economic
costs comparable with certain serious chronic diseases (2). Despite its significant impact on the
quality of life and financial burden on patients, the etiology and
pathophysiology of endometriosis remain unclear. Various theories
have been proposed to explain the pathogenesis; however, none have
interpreted it comprehensively (3).
Sampson's retrograde menstruation theory is the most widely
accepted, which proposes that fragments of the eutopic endometrium
are implanted into the peritoneum or pelvic organs during
menstruation through reflux via the fallopian tubes (4). Subsequently, extensive studies have been
conducted to identify the differences between the eutopic and
normal endometrium; indeed, various studies have demonstrated a
distinct expression pattern in the eutopic endometrium compared
with the normal endometrium, but the evidence is insufficient
(5). In addition, laparoscopy remains
the gold standard for the diagnosis of endometriosis and, to date,
a histological examination is necessary for confirmation. However,
this diagnostic method is a surgically invasive inspection
requiring general anesthesia, which carries surgery-associated
risks, such as hemorrhage, infection and adhesion formation, and
requires an experienced surgeon (6).
As a result of these drawbacks, non-invasive biomarkers using
serum, urine and endometrial tissue as research samples have
emerged; however, these current diagnostic tests for endometriosis
lack sensitivity and specificity, and are difficult to repeat
(7). Therefore, a thorough and
comprehensive description of molecular differences between the
eutopic endometrium in a patient with endometriosis and a control
subject with a normal endometrium is essential to understand the
pathogenesis of this disorder, and identify sensitivity and
specificity biomarkers.
In the current study, it was hypothesized that the
eutopic endometrium in endometriosis patients contains aberrant
expression genes and exhibits dysregulated pathways that predispose
itself to implant, invade and migrate outside the uterus. Messenger
RNA (mRNA) sequencing was performed to detect the transcriptome
expression profiling of eutopic endometrium in women with
endometriosis compared with normal endometrium from healthy control
subjects. Through global mRNA expression profiling, the aim was to
identify candidate pathogenic genes and pathways that are
implicated in the pathogenesis of endometriosis, as well as
potential biomarkers of this common, clinically significant, but
complex disorder.
Materials and methods
Ethics statement
The study protocol was approved by the Local Ethics
Committee of Chinese People's Liberation Army (PLA) General
Hospital (Beijing, China), and each patient was involved in the
study after providing written informed consent.
Patient samples
Twenty-three eutopic endometria from patients with
endometriosis were obtained from the Chinese PLA General Hospital
between February and September 2016. Among them, eight eutopic
endometria were randomly selected and prepared for mRNA sequencing
(mRNA-seq), and the remaining 15 samples were used for validation.
All patients were confirmed to have endometriosis by histological
examination and diagnosed as being of moderate to severe stage
(stage III–IV) according to the revised American Fertility Society
(rAFS) classification (8) during
laparoscopic surgery. None of the patients received hormone therapy
prior to sampling. Twenty women without endometriosis, who
underwent laparoscopic surgery for examination or hydrotubation,
were included in the control group. Five endometria were randomly
selected for sequencing analysis and 15 endometria were used for
validation. Regions potentially exhibiting endometriotic lesions
were confirmed as being negative by biopsy.
The eutopic and normal endometria were obtained via
curettage prior to the laparoscopic procedure. Only patients in the
secretory phase of the menstrual cycle, which was confirmed by the
method of Noyes et al (9) were
included in the study. There were no significant differences
between the ages and body mass index values of the patient and
control groups.
Tissue processing, RNA extraction and
quality control
All tissue samples were divided into two parts:
One-half was fixed and prepared for pathological examination to
identify the endometrial phase of the menstrual cycle and the other
half was placed in RNAlater solution (Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany) at 4°C for 24 h, and subsequently transferred
to −80°C until use. Total RNA was extracted using the single-step
acid guanidinium thiocyanate-phenol-chloroform method (10). The quality and purity of RNA were
examined using a Nanodrop 8000 spectrophotometer (Thermo Fisher
Scientific, Inc., Waltham, MA, USA) and Bioanalyzer 2100 (Agilent
Technologies, Inc., Santa Clara, CA, USA). Samples with RNA
integrity number ≥8 were included.
mRNA sequencing and data analysis
Three micrograms of RNA per sample was prepared for
library construction. The Ribo-Zero Gold kit (Epicentre; Illumina,
Inc., San Diego, CA, USA) and NEB Next Ultra RNA Library Prep kit
(New England BioLabs, Inc., Ipswich, MA, USA) were used for rRNA
removal and library construction according to the manufacturer's
instructions. For high-throughput sequencing, paired-end 150-bp
sequencing of the cDNAs was performed using the Illumina HiSeq4000
system (Illumina, Inc.), which was conducted by Annoroad Genomics
(Beijing, China). Raw data were processed with Perl scripts to
ensure the quality of data used for further analysis. Bowtie2
(v2.2.3; https://sourceforge.net/projects/bowtie-bio/files/bowtie2/)
was used for building the genome index, and clean data was mapped
to the human genome build (hg19) using Tophat (v2.0.12; https://tophat.cbcb.umd.edu/). Read counts of each
gene were counted by HTSeq (v0.6.0; http://www-huber.embl.de/users/anders/HTSeq/doc/overview.html),
and reads per kb of a gene per million reads (RPKM) were
subsequently calculated to estimate the expression level of genes
in each sample. DEGseq (v1.18.0; http://www.bioconductor.org/packages/release/bioc/html/DEGseq.html)
was used for analyzing differentially expressed genes (DEGs) with
the following parameters: False discovery rate (FDR) ≤0.05 and
fold-change (FC) ≥2 or ≤0.5.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Five mRNAs, including matrix metallopeptidase 11
(MMP-11; ENSG00000099953), dual specificity phosphatase 1 (DUSP1;
ENSG00000120129), Fos proto-oncogene, AP-1 transcription factor
subunit (FOS; ENSG00000170345), serpin family E member 1 (SERPINE1;
ENSG00000106366), and adenosine deaminase 2 (ADA2; ENSG00000093072)
were selected for validation analysis, and GAPDH served as an mRNA
endogenous control. The primers are presented in Table I. cDNA synthesis was conducted using a
RevertAid™ First Strand cDNA Synthesis kit (Thermo Fisher
Scientific, Inc.). The relative mRNA expression was determined by
RT-qPCR according to the THUNDERBIRD™ SYBR qPCR Mix (Toyobo, Co.,
Ltd., Osaka, Japan). qRT-PCR was performed on an ABI PRISM 7500
(Applied Biosystems; Thermo Fisher Scientific, Inc.). The relative
gene expression was calculated using ABI PRISM 7500 version 2.0.6
software (Applied Biosystems; Thermo Fisher Scientific, Inc.)
according to the 2−ΔΔCq method (11).
 | Table I.Reverse transcription-quantitative
polymerase chain reaction primers. |
Table I.
Reverse transcription-quantitative
polymerase chain reaction primers.
| Primer | Length, bp | Sequence |
|---|
| Matrix
metallopeptidase 11 | 93 | Forward:
GCTGCCTTCCAGGATGCTGAT |
|
|
| Reverse:
GCCTTCCAGAGCCTTCACCTT |
| Dual specificity
phosphatase 1 | 85 | Forward:
GCCACCATCTGCCTTGCTTAC |
|
|
| Reverse
TGCTTCGCCTCTGCTTCACA |
| Fos proto-oncogene,
AP-1 transcription factor subunit | 245 | Forward:
CGAGATTGCCAACCTGCTGAAG |
|
|
| Reverse:
CCATGCTGCTGATGCTCTTGAC |
| Serpin family E
member 1 | 262 | Forward:
TTCAGGCTGACTTCACGAGT |
|
|
| Reverse:
CCAGATGAAGGCGTCTTTCC |
| Adenosine deaminase
2 | 237 | Forward:
GGCTGTCATCGCAGAATCCATC |
|
|
| Reverse:
AGCATCAGAGCATCCAGAATGTTC |
Functional analysis
To exploit the functional roles of DEGs, DAVID
(https://david.ncifcrf.gov/home.jsp)
was used, which integrated the Gene Ontology (GO) and Kyoto
Encyclopedia of Genes and Genomes (KEGG) databases to analyze
biological function. Finally, the enrichment values of the GO
terms, obtained using the hypergeometric test, were considered
significantly enriched when the q-value (adjusted as a P-value) was
0.05.
Results
mRNA filtering and mapping
mRNA sequencing generated 1,396,127,582 reads, with
an average of 107,204,492 reads per sample in the eutopic
endometrium group and 107,698,330 reads per sample in the normal
endometrium group. A fastQC quality test demonstrated that
1,374,958,606 (98.48%) reads had a Q-score ≥30, and thus were
considered for further analyses. Of these reads, 93.71% were mapped
to the hg19 and 98.61% were uniquely aligned. The detailed
filtering and mapping data are presented in Table II.
 | Table II.Summary of the mRNA sequencing data
following filtering and mapping. |
Table II.
Summary of the mRNA sequencing data
following filtering and mapping.
| Sample | Total raw reads | Total Q30 (%) | Total clean
reads | Mapped reads | Unique map
reads | MultiMap reads |
|---|
| EU1 | 108,063,442 | 94.38 | 106,300,690 |
99,490,109 |
98,103,400 | 1,386,709 |
| EU5 | 115,796,574 | 94.93 | 114,245,924 | 108,073,952 | 106,633,173 | 1,440,779 |
| EU6 |
88,538,384 | 94.76 |
87,285,608 |
81,742,080 |
80,563,047 | 1,179,033 |
| EU7 | 125,048,606 | 94.97 | 123,155,952 | 116,419,348 | 114,821,450 | 1,597,898 |
| EU11 | 112,650,384 | 94.35 | 110,637,684 | 102,884,399 | 101,501,805 | 1,382,594 |
| EU18 |
89,731,074 | 94.42 |
88,482,524 |
83,389,862 |
82,191,074 | 1,198,788 |
| EU19 | 105,616,842 | 94.57 | 104,296,544 |
97,924,454 |
96,502,461 | 1,421,993 |
| EU21 | 112,190,626 | 94.68 | 110,717,296 | 103,453,282 | 102,134,657 | 1,318,625 |
| N2 | 119,170,846 | 94.89 | 117,104,548 | 109,135,053 | 107,668,064 | 1,466,989 |
| N8 | 100,224,148 | 95.00 |
98,937,330 |
92,856,399 |
91,660,813 | 1,195,586 |
| N12 |
95,701,846 | 94.50 |
94,451,810 |
88,290,492 |
87,035,145 | 1,255,347 |
| N16 | 106,411,788 | 94.92 | 104,457,624 |
97,033,960 |
95,631,762 | 1,402,198 |
| N19 | 116,983,022 | 93.76 | 114,885,072 | 107,817,828 | 106,214,767 | 1,603,061 |
Identification of DEGs and RT-qPCR
findings
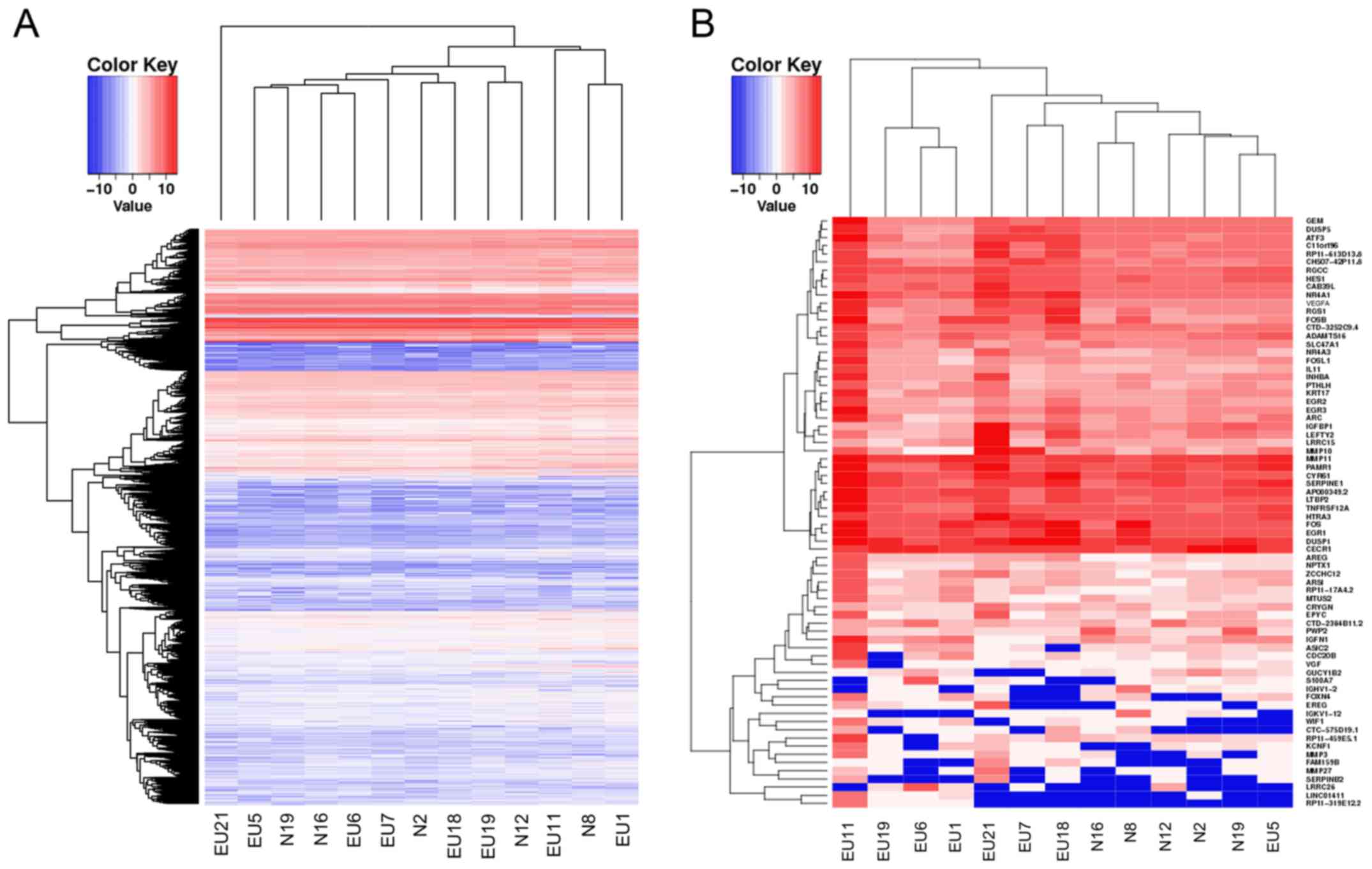
Among the 13 samples, there were 40,576 mRNAs with
an RPKM value of 1 in at least one sample. The transcriptome
expression profiling of eutopic endometria and normal endometria
exhibited very similar expression levels in the evaluated mRNAs,
which directly indicated the homology of the two sequenced groups
(Fig. 1A). On the basis of the
above-mentioned criteria of DEGs, 72 DEGs were identified with 66
upregulated genes and 6 downregulated genes (Table III and Fig.
1B).
 | Table III.Differentially expressed genes (n=72)
in eutopic endometrium samples from women with endometriosis versus
healthy control subjects. |
Table III.
Differentially expressed genes (n=72)
in eutopic endometrium samples from women with endometriosis versus
healthy control subjects.
| Gene symbol | Ensemble ID | Change | Fold-change | False discovery
rate |
|---|
| RP11-319E12.2 |
ENSG00000251459 | Upregulated | 120.99 | 2.32E-07 |
| IGFBP1 |
ENSG00000146678 | Upregulated | 115.01 | 1.79E-03 |
| SERPINB2 |
ENSG00000197632 | Upregulated |
72.95 | 1.51E-04 |
| FOSB |
ENSG00000125740 | Upregulated |
66.12 | 2.01E-19 |
| EREG |
ENSG00000124882 | Upregulated |
52.02 | 2.05E-09 |
| MMP27 |
ENSG00000137675 | Upregulated |
40.93 | 1.69E-04 |
| MMP10 |
ENSG00000166670 | Upregulated |
31.41 | 4.01E-04 |
| LEFTY2 |
ENSG00000143768 | Upregulated |
30.59 | 1.37E-08 |
| WIF1 |
ENSG00000156076 | Upregulated |
30.56 | 1.38E-04 |
| CDC20B |
ENSG00000164287 | Upregulated |
25.21 | 6.06E-09 |
| LRRC15 |
ENSG00000172061 | Upregulated |
25.20 | 3.17E-24 |
| LINC01411 |
ENSG00000249306 | Upregulated |
24.95 | 7.67E-03 |
| RPL10P9 |
ENSG00000233913 | Upregulated |
21.83 | 3.10E-02 |
| IGKV1-12 |
ENSG00000243290 | Upregulated |
21.19 | 2.04E-05 |
| FAM159B |
ENSG00000145642 | Upregulated |
20.34 | 2.34E-02 |
| INHBA |
ENSG00000122641 | Upregulated |
15.33 | 1.19E-15 |
| FOXN4 |
ENSG00000139445 | Upregulated |
14.90 | 1.71E-02 |
| RGS1 |
ENSG00000090104 | Upregulated |
14.80 | 1.19E-15 |
| MMP3 |
ENSG00000149968 | Upregulated |
14.71 | 4.40E-02 |
| FOS |
ENSG00000170345 | Upregulated |
14.54 | 2.55E-24 |
| IGHV1-2 |
ENSG00000211934 | Upregulated |
14.08 | 3.82E-04 |
| NR4A1 |
ENSG00000123358 | Upregulated |
13.80 | 8.80E-17 |
| EGR3 |
ENSG00000179388 | Upregulated |
13.24 | 1.19E-15 |
| VGFA |
ENSG00000112715 | Upregulated |
13.10 | 8.07E-17 |
| RP11-459E5.1 |
ENSG00000253125 | Upregulated |
12.77 | 8.50E-06 |
| ARC |
ENSG00000198576 | Upregulated |
12.72 | 3.82E-04 |
| EPYC |
ENSG00000083782 | Upregulated |
12.28 | 4.50E-02 |
| KCNF1 |
ENSG00000162975 | Upregulated |
10.88 | 1.33E-02 |
| AREG |
ENSG00000109321 | Upregulated |
9.49 | 6.67E-03 |
| FOSL1 |
ENSG00000175592 | Upregulated |
9.42 | 1.87E-08 |
| VGF |
ENSG00000128564 | Upregulated |
9.14 | 3.29E-02 |
| NPTX1 |
ENSG00000171246 | Upregulated |
9.11 | 2.50E-02 |
| ATF3 |
ENSG00000162772 | Upregulated |
8.99 | 3.83E-08 |
| SERPINE1 |
ENSG00000106366 | Upregulated |
8.17 | 2.00E-04 |
| IL11 |
ENSG00000095752 | Upregulated |
7.97 | 7.57E-04 |
| IGFN1 |
ENSG00000163395 | Upregulated |
7.79 | 2.78E-05 |
| ASIC2 |
ENSG00000108684 | Upregulated |
7.40 | 1.31E-02 |
| NR4A3 |
ENSG00000119508 | Upregulated |
7.30 | 4.57E-06 |
| CRYGN |
ENSG00000127377 | Upregulated |
7.25 | 9.11E-03 |
| AP000349.2 |
ENSG00000280178 | Upregulated |
7.07 | 4.06E-13 |
| MTUS2 |
ENSG00000132938 | Upregulated |
6.87 | 5.42E-03 |
| ZCCHC12 |
ENSG00000174460 | Upregulated |
6.82 | 1.05E-03 |
| MMP11 |
ENSG00000099953 | Upregulated |
6.77 | 5.21E-13 |
| ARSI |
ENSG00000183876 | Upregulated |
6.46 | 9.06E-03 |
| LOC101929415 |
ENSG00000254254 | Upregulated |
5.90 | 4.56E-02 |
| RP11-613D13.8 |
ENSG00000244953 | Upregulated |
5.72 | 2.98E-03 |
| KRT17 |
ENSG00000128422 | Upregulated |
5.56 | 9.34E-05 |
| EGR1 |
ENSG00000120738 | Upregulated |
5.47 | 1.43E-09 |
| CYR61 |
ENSG00000142871 | Upregulated |
5.38 | 2.70E-09 |
| GEM |
ENSG00000164949 | Upregulated |
4.72 | 7.61E-06 |
| C11orf96 |
ENSG00000187479 | Upregulated |
4.57 | 2.05E-04 |
| PTHLH |
ENSG00000087494 | Upregulated |
4.51 | 3.73E-02 |
| EGR2 |
ENSG00000122877 | Upregulated |
4.41 | 1.33E-02 |
| DUSP1 |
ENSG00000120129 | Upregulated |
4.35 | 2.05E-04 |
| LOC102724428 |
ENSG00000275993 | Upregulated |
4.35 | 8.21E-03 |
| PAMR1 |
ENSG00000149090 | Upregulated |
4.23 | 1.34E-03 |
| TNFRSF12A |
ENSG00000006327 | Upregulated |
3.83 | 5.70E-03 |
| DUSP5 |
ENSG00000138166 | Upregulated |
3.66 | 1.60E-03 |
| SLC47A1 |
ENSG00000142494 | Upregulated |
3.62 | 8.33E-03 |
| ADAMTS16 |
ENSG00000145536 | Upregulated |
3.39 | 1.45E-02 |
| CAB39L |
ENSG00000102547 | Upregulated |
3.38 | 1.65E-03 |
| HES1 |
ENSG00000114315 | Upregulated |
3.23 | 6.51E-03 |
| HTRA3 |
ENSG00000170801 | Upregulated |
3.17 | 1.60E-03 |
| LTBP2 |
ENSG00000119681 | Upregulated |
2.92 | 6.92E-03 |
| LOC284454 |
ENSG00000267519 | Upregulated |
2.89 | 4.88E-02 |
| RGCC |
ENSG00000102760 | Upregulated |
2.74 | 4.03E-02 |
| LRRC26 |
ENSG00000184709 | Downregulated | 109.57 | 2.98E-03 |
| S100A7 |
ENSG00000143556 | Downregulated |
40.68 | 5.10E-08 |
| PWP2 |
ENSG00000241945 | Downregulated |
18.29 | 8.21E-03 |
| GUCY1B2 |
ENSG00000123201 | Downregulated |
11.64 | 6.54E-03 |
| CTD-2384B11.2 |
ENSG00000225407 | Downregulated |
7.54 | 6.54E-03 |
| ADA2 |
ENSG00000093072 | Downregulated |
3.23 | 1.30E-02 |
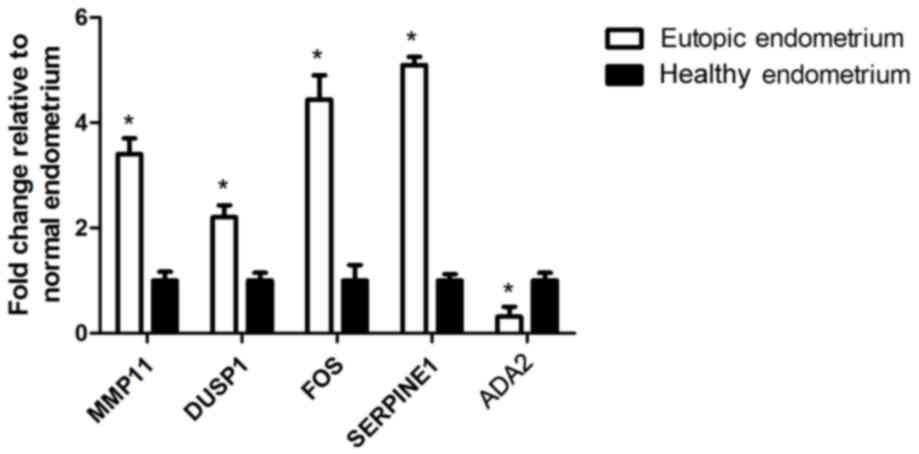
Two DEGs, ADA2 and MMP-11, were significantly
different in the current study, but were not previously selected
for further validation. FOS, which exhibited contradictory results
in previous studies, was selected. In addition, two DEGs, SERPINE1
and DUSP1, which have been associated with endometriosis in cell
lines and animal models, but have not been reported in human
tissue, were simultaneously selected. Although the counts of DEGs
in mRNA sequencing were relatively low in the two groups, the
RT-qPCR analysis indicated easily detectable expression levels.
Data analysis indicated that the results from RT-qPCR were
consistent with the mRNA sequencing data (Fig. 2).
Functional analysis
To gain an overall understanding of the functional
roles in these DEGs, GO term and KEGG pathway analysis were
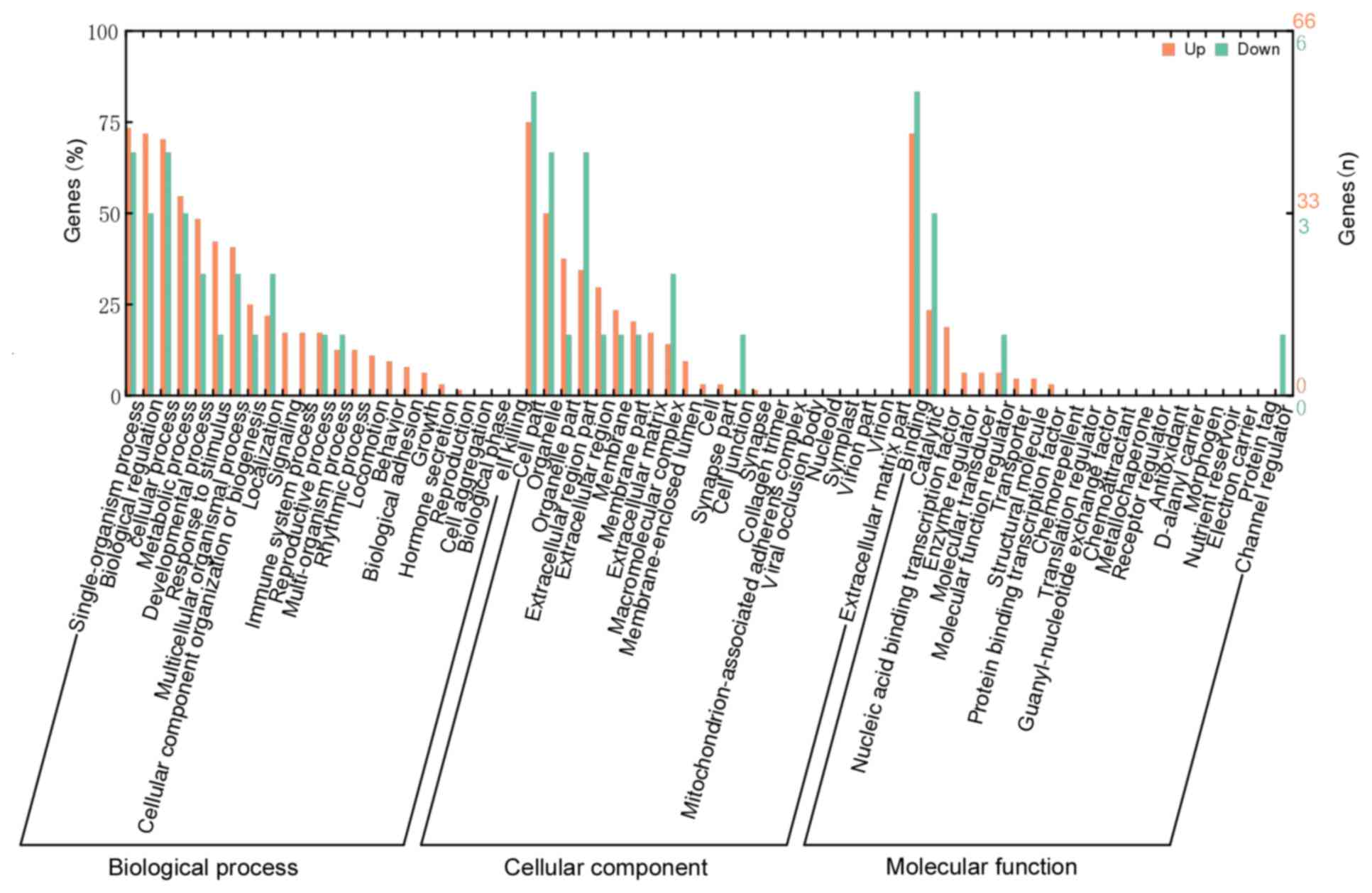
conducted. The results revealed that significantly enriched GO
terms under the cellular component (CC) category were extracellular
matrix (ECM; GO:0031012), proteinaceous ECM (GO:0005578) and
extracellular space (GO:0005615). The molecular function (MF)
category included nine enriched terms, particularly in
metalloendopeptidase activity (GO:0004222), growth factor activity
(GO:0008083), and RNA polymerase II core promoter proximal region
sequence-specific DNA binding transcription factor activity
(GO:0000982). The biological process (BP) category contained 88
enriched terms, the top three of which were response to endogenous
stimulus (GO:0009719), cellular response to endogenous stimulus
(GO:0071495) and response to cyclic adenosine monophosphate
(GO:0051591). In addition, certain GO terms that are commonly
observed in tumor-like diseases were also significantly enriched in
the current results, such as growth (GO:0040007), angiogenesis
(GO:0001525) and cell migration (GO:0016477). These results are
presented in Fig. 3 and Table IV. Due to the limited number of DEGs,
none of the pathways were identified to be significantly enriched
in the KEGG analysis with the above-mentioned thresholds.
 | Table IV.Top 10 enriched GO terms of DEGs in
the endometrium from women with endometriosis compared with healthy
control subjects. |
Table IV.
Top 10 enriched GO terms of DEGs in
the endometrium from women with endometriosis compared with healthy
control subjects.
| Category | GO ID | Description | Q value | n | Involved DEGs |
|---|
| BP | GO:0009719 | Response to
endogenous stimulus | 4.47E-07 | 21 | CAB39L, SERPINE1,
AREG, HES1, NR4A3, LTBP2, DUSP1, EGR1, INHBA, EGR2, NR4A1, EREG,
FOSB, VGF, LEFTY2, IGFBP1, MMP3, HTRA3, FOSL1, EGR3, VEGFA |
| BP | GO:0071495 | Cellular response
to endogenous stimulus | 1.54E-06 | 17 | CAB39L, SERPINE1,
HES1, LTBP2, DUSP1, EGR1, INHBA, EGR2, NR4A1, EREG, FOSB, LEFTY2,
IGFBP1, MMP3, HTRA3, EGR3, VEGFA |
| BP | GO:0051591 | Response to cyclic
adenosine monophosphate | 1.54E-06 | 8 | AREG, DUSP1, EGR1,
EGR2, FOSB, VGF, FOSL1, EGR3 |
| CC | GO:0031012 | Extracellular
matrix | 4.5E-06 | 11 | EPYC, MMP11,
SERPINE1, LTBP2, MMP27, CYR61, LEFTY2, ADAMTS16, MMP3, MMP10,
AP000349.2 |
| BP | GO:0001525 | Angiogenesis | 1.2E-05 | 9 | TNFRSF12A, RGCC,
SERPINE1, NR4A1, EREG, CYR61, S100A7, EGR3, VEGFA |
| BP | GO:0046683 | Response to
organophosphorus | 1.49E-05 | 8 | AREG, DUSP1, EGR1,
EGR2, FOSB, VGF, FOSL1, EGR3 |
| BP | GO:0014074 | Response to
purine-containing compound | 1.95E-05 | 8 | AREG, DUSP1, EGR1,
EGR2, FOSB, VGF, FOSL1, EGR3 |
| BP | GO:0048646 | Anatomical
structure formation involved in morphogenesis | 3.16E-05 | 17 | TNFRSF12A, PTHLH,
RGCC, SERPINE1, NR4A3, DUSP1, INHBA, EGR2, NR4A1, EREG, DUSP5,
FOXN4, CYR61, S100A7, ADAMTS16, EGR3, VEGFA |
| BP | GO:0010243 | Response to
organonitrogen compound | 5.42E-05 | 14 | CAB39L, AREG, HES1,
NR4A3, DUSP1, EGR1, EGR2, EREG, FOSB, VGF, IGFBP1, MMP3, FOSL1,
EGR3 |
| BP | GO:0009725 | Response to hormone
stimulus | 8.50E-05 | 14 | CAB39L, AREG, HES1,
NR4A3, DUSP1, EGR1, INHBA, EGR2, EREG, FOSB, VGF, IGFBP1, FOSL1,
EGR3 |
Discussion
To date, the majority of studies that focused on the
eutopic endometrium of women with endometriosis were
hypothesis-based studies, which evaluated a limited number of
previously susceptible genes (7). A
recent systematic review from 1984 to 2010 summarized >200
potential endometrial biomarkers in the endometrium, but did not
identify a standard biomarker in a clinical study (12). In addition, various studies using a
microarray-based method have identified hundreds of potential
pathogenic genes and pathways; however, these studies present few
overlapping results (13–15). This inconsistency may be caused by
various factors, including a small number of samples, poorly
defined controls, different rAFS stages, different phases of the
menstrual cycle, methodology limitations, various types of
endometriosis, and interference by coexistent diseases. Therefore,
future studies should continue to search for the important DEGs and
focus on these confounding factors. To the best of our knowledge,
the current study is the first to present the genome-wide gene
expression profiling of the eutopic endometrium in women with
endometriosis using a transcriptome sequencing technique.
Considering that the genome profiling of normal endometria
demonstrated marked molecular differences between samples obtained
from the proliferative and secretory phases of the menstrual cycle,
only mid- and late-secretory phase endometria were investigated in
the patient and control groups, which represents most closely the
reflux endometrium. To avoid other confounding factors, the
endometriosis patients included were restricted to those in the
moderate to severe stages (stages III–IV) and only those patients
with ovarian endometriosis without combined diseases were enrolled.
Finally, 72 DEGs enriched in 100 functional GO terms were
identified. The top enriched terms in each category were ECM
(GO:0031012) in CC, metalloendopeptidase activity (GO:0004222) in
MF, and cellular response to endogenous stimulus (GO:0009719) in
BP. Notably, various DEGs may be candidates for potential
biomarkers in the eutopic endometrium of women with
endometriosis.
It has been reported that human endometrium
undergoes cyclic tissue remodeling during the menstrual period,
during which several MMPs and ECM-associated proteins are activated
(16). These proteins are suggested to
facilitate the degradation and invasion of ECM and facilitate with
the attachment of reflux endometrial tissue to the peritoneum and
ovarian surface. In the current study, four MMP members, MMP-3,
MMP-10, MMP-11, and MMP-27, were identified as upregulated in
eutopic endometrium (17).
Gilabert-Estellés et al (18)
and Ramón et al (19)
demonstrated that eutopic endometria from women with endometriosis
have increased expression levels of MMP-3, which is consistent with
the present result. Uzan et al (20) examined the immunohistochemical
expression of MMP-11, although no difference was identified between
the patient and control groups. Cominelli et al (21) suggested that MMP-27 is maximally
expressed during the menstrual phase in the normal endometrium and
no difference in ectopic versus eutopic endometria was observed;
however, the authors did not compare between eutopic and normal
endometria. Prior studies reported that the overexpression of
SERPINE1 (also termed PAI-1) may result in the impairment of the
fibrinolytic system, rendering the woman prone to endometriosis
(22). Braza-Boïls et al
(23) demonstrated that the protein
expression levels of SERPINE1 were significantly higher in
endometriotic lesions than in control endometrial tissue samples,
but identified no difference between eutopic and control
endometria. Unlike other members mentioned above, MMP-10,
epiphycan, latent transforming growth factor β binding protein 2,
cysteine rich angiogenic inducer 61, left-right determination
factor 2, ADAM metallopeptidase with thrombospondin type 1 motif
16, and AP000349.2 have received little attention in endometriosis
research and therefore require further confirmation.
The FOS gene family comprises four members, namely
FOS, FosB proto-oncogene, AP-1 transcription factor subunit (FOSB),
FOS like 1, AP-1 transcription factor subunit (FOSL1) and FOSL2.
Their encoded proteins dimerize with the Jun family members to form
the group of AP-1 proteins, and are involved in various
physiological and pathological processes, such as cell
proliferation, apoptosis, and differentiation and transformation
(24). In the present study,
abnormally high expression levels of FOS, FOSB, and FOSL1 were
observed in eutopic endometria from patients with endometriosis,
which were predominantly enriched in response to endogenous
stimulus terms, nucleic acid-binding transcription factor activity
terms, and response to hormone stimulus terms. FOS, as an early
response gene, is critical in estrogen-mediated proliferation of
endometrial cells (25). Pan et
al (26) reported that FOS protein
expression levels in eutopic and ectopic endometria samples from
females with endometriosis were significantly higher than those in
the endometria samples from healthy control subjects; however, the
findings of Morsch et al (27)
were not similar. Therefore, the FOS gene was selected to validate
RT-qPCR in the present study, and the result was consistent with
the results of Pan et al (26).
To the best of our knowledge, the association between FOSB and
FOSL1, and endometriosis have not yet been reported. As with FOS,
the early growth response (EGR) family of transcription regulatory
factors was predicted to be key in cellular growth and
differentiation (28). Three members,
EGR1, EGR2 and EGR3 were identified to be highly expressed by
sequencing, which were also significantly enriched in response to
endogenous stimulus terms, response to gonadotropin stimulus terms,
and growth terms. Birt et al (29) reported the overexpression of EGR1 in
endometriotic animal models and inferred that EGR1 may affect
downstream protease pathways impeding ovulation in endometriosis.
The roles of EGR1and EGR3 in angiogenesis has also recently been
recognized and were considered to regulate certain important
angiogenic factors, such as vascular endothelial growth factor A
(VEGFA), fibroblast growth factor 2, and C-X-C motif chemokine
ligand 1. Angiogenesis is considered to be pivotal to the implant
and growth of endometriotic lesions in the pelvic microenvironment
(30). The endometrium, which contains
robust stem cell populations and striking regenerative ability, is
a rich source of angiogenic factors (31). In the present study, enriched
angiogenesis and blood vessel development terms were also observed.
Aberrant upregulation of VEGFA in eutopic endometria demonstrated
concordance with two earlier results: Taylor et al (32) emphasized the importance of VEGFA in the
endometrium of women with endometriosis, as it may be activated by
inflammatory-, oxidative-, hormonal- and endoplasmic
reticulum-stress signals. Bourlev et al (33) reported a high expression level of VEGFA
in the eutopic endometrium of women with endometriosis, as well as
high concentrations of VEGFA in the peritoneal fluid. In addition,
dysregulation of various angiogenic factors, including regulator of
cell cycle, TNF receptor superfamily member 12A, nuclear receptor
subfamily 4 group A member 1, epiregulin, cysteine rich angiogenic
inducer 61, and S100 calcium binding protein A7, was observed in
the present study in women with endometriosis when compared with
those without endometriosis. Although these preliminary data
require further characterization, the current findings provide
novel information for future experimental studies.
In addition to gaining an improved understanding of
pathogenesis, the present study attempted to identify various
potential biomarkers. Although certain studies have questioned the
unpleasant sensation of endometrial biopsy, the majority of
participants are willing to undergo the procedure (34). In the current study, elevated
expression levels of ADA2, MMP-11, FOS, SERPINE1, and DUSP1 in
women with endometriosis were revealed by mRNA sequencing and
RT-qPCR and, thus, these genes were considered as candidate
biomarkers. Due to the limited sample size, the sensitivity,
specificity, and receiver operating characteristic curve analysis
were not calculated for a diagnostic test of endometriosis, which
would be vital for a large sample-size study in future.
Despite the novel results, there were limitations of
the current study. The primary limitation of the study is the
relatively small sample size. In addition, it is difficult to
evaluate whether the control endometria are from completely healthy
women, as ~6% of endometriotic lesions are only visible under a
microscope, and these women macroscopically presented a normal
appearance (35). In addition, these
preliminary results require validation by downstream
experiments.
In conclusion, to the best of our knowledge, the
current study presents the first genome-wide gene expression
profile of eutopic endometria from women with endometriosis using a
high-throughput sequencing technique. Seventy-two DEGs in eutopic
endometria from women with endometriosis were compared with normal
endometria from control subjects. GO analysis further revealed the
important roles of these DEGs in the pathogenesis of endometriosis.
Five genes, including MMP-11, DUSP1, FOS, SERPINE1, and ADA2 were
further confirmed by RT-qPCR, and the results were consistent with
the mRNA sequencing, indicating that these genes may present as
novel biomarkers in the endometrium of women with endometriosis.
The current study provides a comprehensive, but preliminary insight
into elucidating the underlying mechanisms of this complex
disorder, which merits further in-depth studies for
confirmation.
Acknowledgements
The current study was funded by the National Natural
Science Foundation of China (grant no. 81571411).
References
|
1
|
Kennedy S, Bergqvist A, Chapron C,
D'Hooghe T, Dunselman G, Greb R, Hummelshoj L, Prentice A and
Saridogan E: ESHRE Special Interest Group for Endometriosis and
Endometrium Guideline Development Group: ESHRE guideline for the
diagnosis and treatment of endometriosis. Hum Reprod. 20:2698–2704.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hudelist G, Fritzer N, Thomas A, Niehues
C, Oppelt P, Haas D, Tammaa A and Salzer H: Diagnostic delay for
endometriosis in Austria and Germany: Causes and possible
consequences. Hum Reprod. 27:3412–3416. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sourial S, Tempest N and Hapangama DK:
Theories on the pathogenesis of endometriosis. Int J Reprod Med.
2014:1795152014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sampson JA: Metastatic or Embolic
Endometriosis, due to the Menstrual Dissemination of Endometrial
Tissue into the Venous Circulation. Am J Pathol. 3:93–110.43.
1927.PubMed/NCBI
|
|
5
|
Liu H and Lang JH: Is abnormal eutopic
endometrium the cause of endometriosis? The role of eutopic
endometrium in pathogenesis of endometriosis. Med Sci Monit.
17:RA92–RA99. 2011.PubMed/NCBI
|
|
6
|
Lattarulo S, Pezzolla A, Fabiano G and
Palasciano N: Intestinal endometriosis: Role of laparoscopy in
diagnosis and treatment. Int Surg. 94:310–314. 2009.PubMed/NCBI
|
|
7
|
Fassbender A, Vodolazkaia A, Saunders P,
Lebovic D, Waelkens E, De Moor B and D'Hooghe T: Biomarkers of
endometriosis. Fertil Steril. 99:1135–1145. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
American Society for Reproductive, .
Revised American Society for Reproductive Medicine classification
of endometriosis: 1996. Fertil Steril. 67:817–821. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Noyes RW, Hertig AT and Rock J: Dating the
endometrial biopsy. Am J Obstet Gynecol. 122:262–263. 1975.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chomczynski P and Sacchi N: The
single-step method of RNA isolation by acid guanidinium
thiocyanate-phenol-chloroform extraction: Twenty-something years
on. Nat Protoc. 1:581–585. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
May KE, Villar J, Kirtley S, Kennedy SH
and Becker CM: Endometrial alterations in endometriosis: A
systematic review of putative biomarkers. Hum Reprod Update.
17:637–653. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Burney RO, Talbi S, Hamilton AE, Vo KC,
Nyegaard M, Nezhat CR, Lessey BA and Giudice LC: Gene expression
analysis of endometrium reveals progesterone resistance and
candidate susceptibility genes in women with endometriosis.
Endocrinology. 148:3814–3826. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sherwin JR, Sharkey AM, Mihalyi A, Simsa
P, Catalano RD and D'Hooghe TM: Global gene analysis of late
secretory phase, eutopic endometrium does not provide the basis for
a minimally invasive test of endometriosis. Hum Reprod.
23:1063–1068. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tamaresis JS, Irwin JC, Goldfien GA,
Rabban JT, Burney RO, Nezhat C, DePaolo LV and Giudice LC:
Molecular classification of endometriosis and disease stage using
high-dimensional genomic data. Endocrinology. 155:4986–4999. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Muramatsu T and Miyauchi T: Basigin
(CD147): A multifunctional transmembrane protein involved in
reproduction, neural function, inflammation and tumor invasion.
Histol Histopathol. 18:981–987. 2003.PubMed/NCBI
|
|
17
|
Pitsos M and Kanakas N: The role of matrix
metalloproteinases in the pathogenesis of endometriosis. Reprod
Sci. 16:717–726. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gilabert-Estellés J, Ramón LA, España F,
Gilabert J, Vila V, Réganon E, Castelló R, Chirivella M and
Estellés A: Expression of angiogenic factors in endometriosis:
Relationship to fibrinolytic and metalloproteinase systems. Hum
Reprod. 22:2120–2127. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ramón L, Gilabert-Estellés J, Castelló R,
Gilabert J, España F, Romeu A, Chirivella M, Aznar J and Estellés
A: mRNA analysis of several components of the plasminogen activator
and matrix metalloproteinase systems in endometriosis using a
real-time quantitative RT-PCR assay. Hum Reprod. 20:272–278. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Uzan C, Cortez A, Dufournet C, Fauvet R,
Siffroi JP and Daraï E: Eutopic endometrium and peritoneal, ovarian
and bowel endometriotic tissues express a different profile of
matrix metalloproteinases-2, −3 and −11, and of tissue inhibitor
metalloproteinases-1 and −2. Virchows Arch. 445:603–609. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cominelli A, Chevronnay HP Gaide, Lemoine
P, Courtoy PJ, Marbaix E and Henriet P: Matrix metalloproteinase-27
is expressed in CD163+/CD206+ M2 macrophages in the cycling human
endometrium and in superficial endometriotic lesions. Mol Hum
Reprod. 20:767–775. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhao L, Gu C and Meng Y: Meta-analysis of
the association between endometriosis and polymorphisms in ACE and
PAI-1. Int J Clin Exp Med. 9:10602–10614. 2016.
|
|
23
|
Braza-Boïls A, Marí-Alexandre J, Gilabert
J, Sánchez-Izquierdo D, España F, Estellés A and Gilabert-Estellés
J: MicroRNA expression profile in endometriosis: Its relation to
angiogenesis and fibrinolytic factors. Hum Reprod. 29:978–988.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Milde-Langosch K: The Fos family of
transcription factors and their role in tumourigenesis. Eur J
Cancer. 41:2449–2461. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Nemos C, Delage-Mourroux R, Jouvenot M and
Adami P: Onset of direct 17-beta estradiol effects on proliferation
and c-fos expression during oncogenesis of endometrial glandular
epithelial cells. Exp Cell Res. 296:109–122. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Pan H, Sheng JZ, Tang L, Zhu R, Zhou TH
and Huang HF: Increased expression of c-fos protein associated with
increased matrix metalloproteinase-9 protein expression in the
endometrium of endometriotic patients. Fertil Steril. 90:1000–1007.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Morsch DM, Carneiro MM, Lecke SB, Araújo
FC, Camargos AF, Reis FM and Spritzer PM: c-fos gene and protein
expression in pelvic endometriosis: A local marker of estrogen
action. J Mol Histol. 40:53–58. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
O'Donovan KJ, Tourtellotte WG, Millbrandt
J and Baraban JM: The EGR family of transcription-regulatory
factors: Progress at the interface of molecular and systems
neuroscience. Trends Neurosci. 22:167–173. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Birt JA, Nabli H, Stilley JA, Windham EA,
Frazier SR and Sharpe-Timms KL: Elevated peritoneal fluid TNF-α
incites ovarian early growth response factor 1 expression and
downstream protease mediators: A correlation with ovulatory
dysfunction in endometriosis. Reprod Sci. 20:514–523. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Djokovic D and Calhaz-Jorge C:
Angiogenesis as a therapeutic target in endometriosis. Acta Med
Port. 27:489–497. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Groothuis PG, Nap AW, Winterhager E and
Grümmer R: Vascular development in endometriosis. Angiogenesis.
8:147–156. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Taylor RN, Yu J, Torres PB, Schickedanz
AC, Park JK, Mueller MD and Sidell N: Mechanistic and therapeutic
implications of angiogenesis in endometriosis. Reprod Sci.
16:140–146. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Bourlev V, Volkov N, Pavlovitch S, Lets N,
Larsson A and Olovsson M: The relationship between microvessel
density, proliferative activity and expression of vascular
endothelial growth factor-A and its receptors in eutopic
endometrium and endometriotic lesions. Reproduction. 132:501–509.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
de Iaco P, Marabini A, Stefanetti M, Del
Vecchio C and Bovicelli L: Acceptability and pain of outpatient
hysteroscopy. J Am Assoc Gynecol Laparosc. 7:71–75. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Nisolle M, Paindaveine B, Bourdon A,
Berlière M, Casanas-Roux F and Donnez J: Histologic study of
peritoneal endometriosis in infertile women. Fertil Steril.
53:984–988. 1990. View Article : Google Scholar : PubMed/NCBI
|