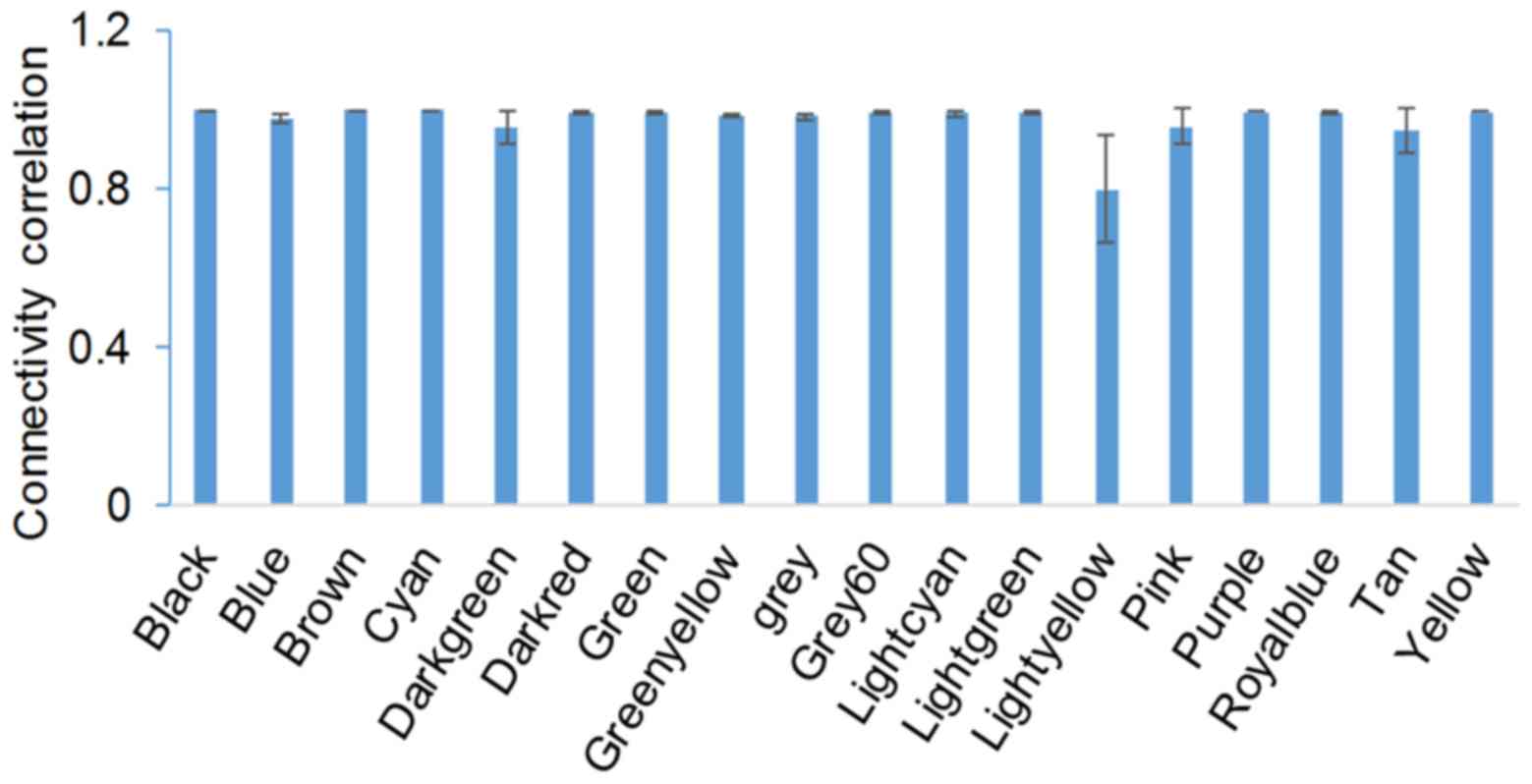
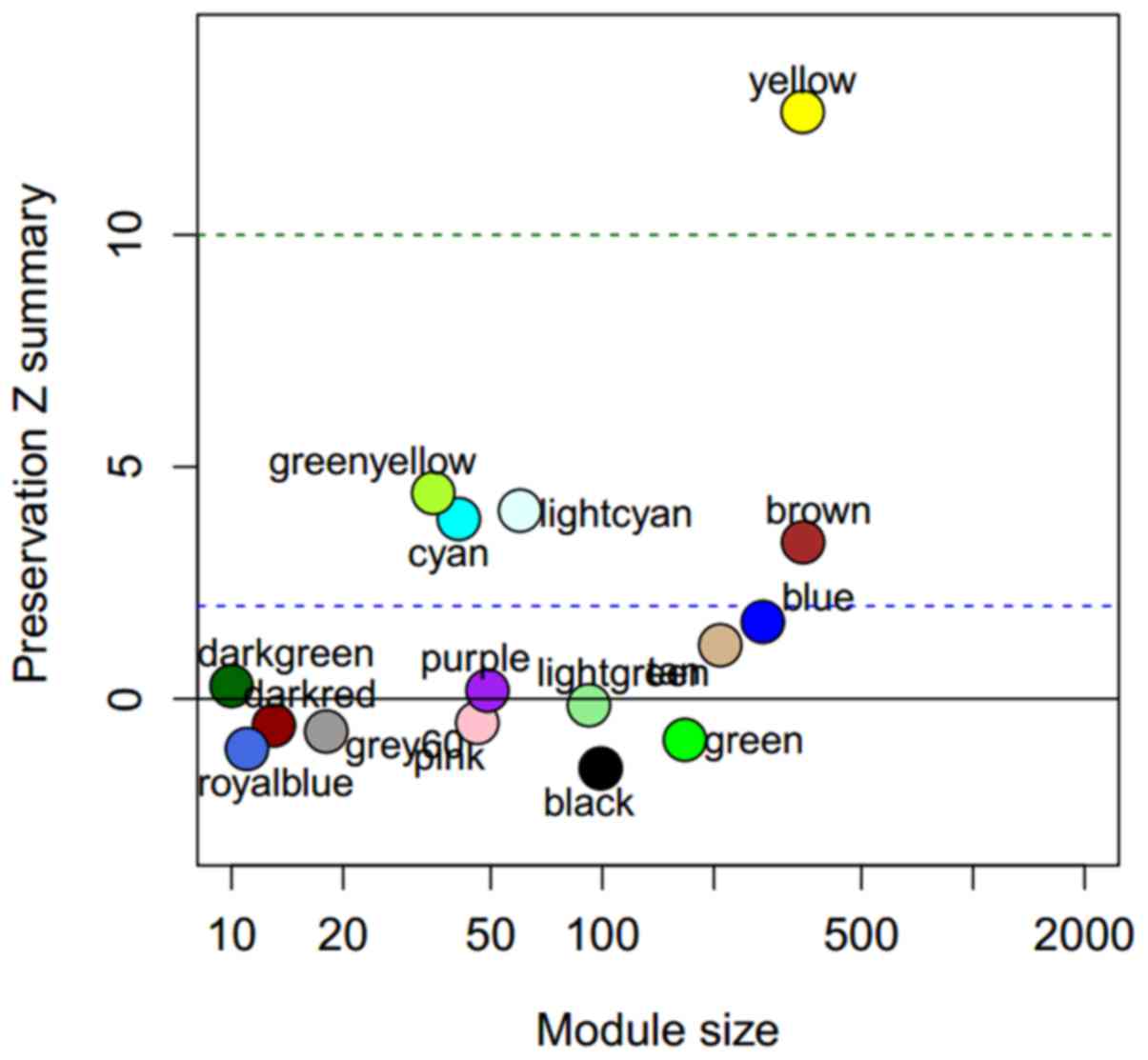
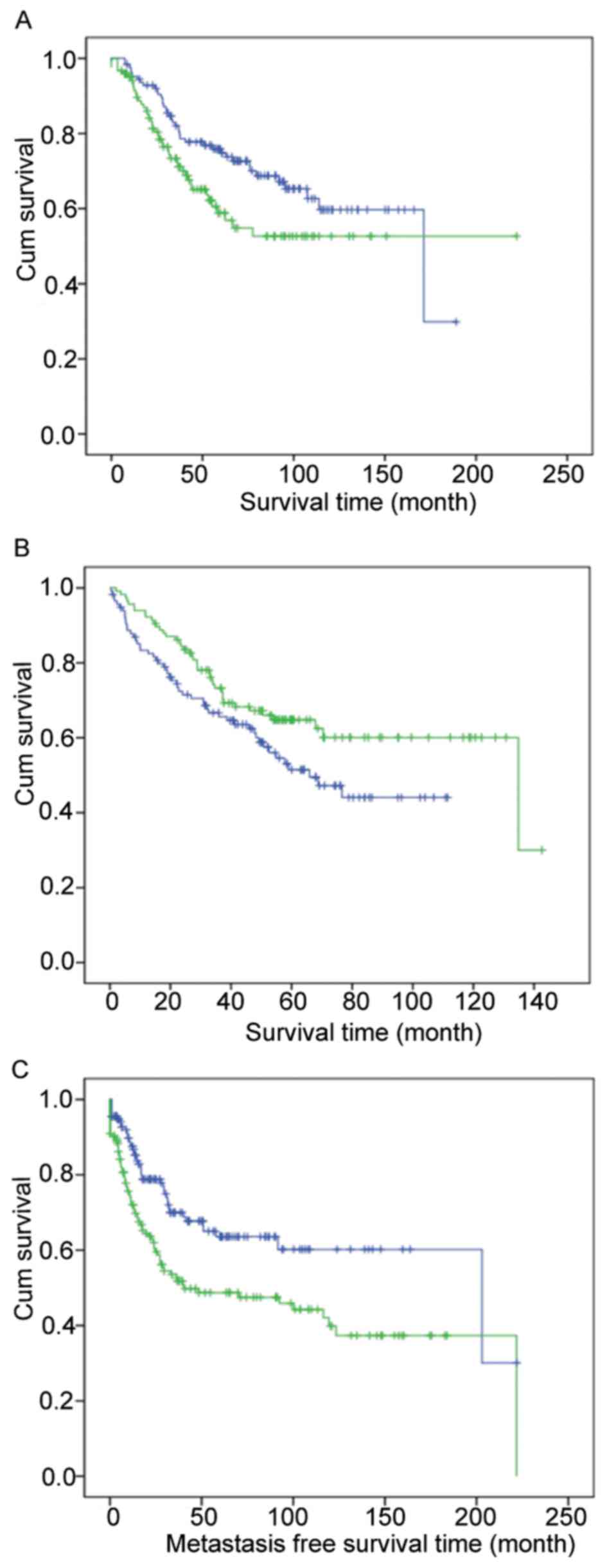
|
1
|
Botstein D, Chervitz SA and Cherry JM:
Yeast as a model organism. Science. 277:1259–1260. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jiang Q, Lin L and Wang T: A new model for
apoptosis research: Yeast. Prog Biochem Biophys. 35:3612008.
|
|
3
|
Kachroo AH, Laurent JM, Yellman CM, Meyer
AG, Wilke CO and Marcotte EM: Evolution. Systematic humanization of
yeast genes reveals conserved functions and genetic modularity.
Science. 348:921–925. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Barrett T, Troup DB, Wilhite SE, Ledoux P,
Rudnev D, Evangelista C, Kim IF, Soboleva A, Tomashevsky M and
Edgar R: NCBI GEO: Mining tens of millions of expression profiles -
database and tools update. Nucleic Acids Res. 35(Database):
D760–D765. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhu J, Zhang B, Smith EN, Drees B, Brem
RB, Kruglyak L, Bumgarner RE and Schadt EE: Integrating large-scale
functional genomic data to dissect the complexity of yeast
regulatory networks. Nat Genet. 40:854–861. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Guelzim N, Bottani S, Bourgine P and Képès
F: Topological and causal structure of the yeast transcriptional
regulatory network. Nat Genet. 31:60–63. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang B and Horvath S: A general framework
for weighted gene co-expression network analysis. Stat Appl Genet
Mol Biol. 4:: 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Barrett T, Troup DB, Wilhite SE, Ledoux P,
Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH,
Sherman PM, et al: NCBI GEO: Archive for functional genomics data
sets − 10 years on. Nucleic Acids Res. 39(Database): D1005–D1010.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Miller JA, Horvath S and Geschwind DH:
Divergence of human and mouse brain transcriptome highlights
Alzheimer disease pathways. Proc Natl Acad Sci USA.
107:12698–12703. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Oldham MC, Konopka G, Iwamoto K,
Langfelder P, Kato T, Horvath S and Geschwind DH: Functional
organization of the transcriptome in human brain. Nat Neurosci.
11:1271–1282. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Langfelder P and Horvath S: WGCNA: An R
package for weighted correlation network analysis. BMC
Bioinformatics. 9:5592008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Huang W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Huang W, Sherman BT and Lempicki RA:
Bioinformatics enrichment tools: Paths toward the comprehensive
functional analysis of large gene lists. Nucleic Acids Res.
37:1–13. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: The Gene Ontology Consortium: Gene ontology: Tool for the
unification of biology. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Benjamini Y and Hochberg Y: Controlling
the false discovery rate: A practical and powerful approach to
multiple testing. J R Stat Soc B. 57:289–300. 1995.
|
|
16
|
Langfelder P, Mischel PS and Horvath S:
When is hub gene selection better than standard meta-analysis? PLoS
One. 8:e615052013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gallina I, Colding C, Henriksen P, Beli P,
Nakamura K, Offman J, Mathiasen DP, Silva S, Hoffmann E, Groth A,
et al: Cmr1/WDR76 defines a nuclear genotoxic stress body linking
genome integrity and protein quality control. Nat Commun.
6:65332015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Brar GA, Yassour M, Friedman N, Regev A,
Ingolia NT and Weissman JS: High-resolution view of the yeast
meiotic program revealed by ribosome profiling. Science.
335:552–557. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhan J, Thakare D, Ma C, Lloyd A, Nixon
NM, Arakaki AM, Burnett WJ, Logan KO, Wang D, Wang X, et al: RNA
sequencing of laser-capture microdissected compartments of the
maize kernel identifies regulatory modules associated with
endosperm cell differentiation. Plant Cell. 27:513–531. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liu W and Ye H: Co-expression network
analysis identifies transcriptional modules in the mouse liver. Mol
Genet Genomics. 289:847–853. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liu W, Li L and Li W: Gene co-expression
analysis identifies common modules related to prognosis and drug
resistance in cancer cell lines. Int J Cancer. 135:2795–2803. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Childs KL, Davidson RM and Buell CR: Gene
coexpression network analysis as a source of functional annotation
for rice genes. PLoS One. 6:e221962011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Novick P and Botstein D: Phenotypic
analysis of temperature-sensitive yeast actin mutants. Cell.
40:405–416. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li C, Qian W, Maclean CJ and Zhang J: The
fitness landscape of a tRNA gene. Science. 352:837–840. 2016.
View Article : Google Scholar : PubMed/NCBI
|