Introduction
Mitochondrial uncoupling protein 1 (UCP1), which is
expressed in brown adipose tissue (BAT), is a critical regulator of
adaptive non-shivering thermogenesis via the biological function of
proton leak (1–4). UCP1, a transmembrane protein, is located
at the inner mitochondrial inner membrane, and uncouples the
oxidative phosphorylation to produce heat through decreasing the
proton gradients (1,2). Free fatty acids are the established
activators that reduce the transmembrane potential via UCP1 and
increase UCP1-associated oxygen consumptions (5). Chouchani et al (6) identified the sulfenylation of UCP1
regulated by mitochondrial reactive oxygen species in 2016.
Uncoupling proteins (UCPs) expressed in adipose cells, skeletal
muscle and macrophages participate in fatty acid metabolism
(7,8).
In addition, UCPs have been associated with metabolic diseases,
including obesity and diabetes (9,10).
Subcellular compartments from yeast to mammalian
cells contain numerous proteins and lipids. Phosphatidylcholine and
phosphatidylethanolamine are major mitochondrial phospholipids
(~80%). High cardiolipin content (10–15%) is found in mitochondrial
membranes and its concentration is implicated in human health and
disease (11,12). Protein-lipid interactions are crucial
for protein stability (12).
Cardiolipin directly interacts with ADP/ATP translocase (ANT) at
the mitochondrial inner membrane (13), and in vitro reconstituted
experiments have demonstrated the binding of six molecules of
cardiolipin per ANT dimer (14). The
exact associations of UCP1 with mitochondrial inner membrane lipids
still remain unknown. In the present study, bioinformatics was used
to identify the lipid-binding α-helix regions of UCP1. The results
may provide novel insight into UCP1-lipid binding.
Materials and methods
Prediction of lipid-binding α-helices
of UCP1
Mouse UCP1 protein sequence (P12242) was obtained
from the UNIPROT database (http://www.uniprot.org/) (15). The HeliQuest web server (http://heliquest.ipmc.cnrs.fr/) was used to
predict the lipid-binding α-helices of UCP1 through calculations of
mean hydrophobicity, hydrophobic moment and net charge (16). These scores were further analyzed in
order to calculate the discrimination factor, which was considered
to filter the possible/confident lipid-binding α-helices according
to HeliQuest instructions. The Basic Local Alignment Search Tool
(BLAST; http://blast.ncbi.nlm.nih.gov/) was used to determine
sequence identity.
Helical wheel plots
Helical wheel plots are commonly used to represent
amphiphilic helices. In the present analysis, the helical wheel
diagrams were drawn using the Wenxiang server (http://www.jci-bioinfo.cn/wenxiang2) (17).
Template selection
Template selection for the target protein (UCP1) was
performed in SWISS-MODEL (http://swissmodel.expasy.org/workspace/) and MODELLER
v9.16 (https://salilab.org/modeller/)
(18,19). InterPro Domain Scan (HMMPfam, HMMTigr,
ProfileScan, SuperFamily and BlastProDom), Gapped Blast Query and
HHSearch template library search in SWISS-MODEL were used to search
for templates. Structural resolutions were also considered.
Homology model building
Primary sequences of templates and target protein
were obtained from the UNIPROT database and template PDB files
using MODELLER script. Template-target sequence alignment was also
completed by MODELLER script with the consideration of SWISS-MODEL
results. A three-dimensional (3D) homology model of UCP1 was built
using MODELLER v9.16.
Energy minimization and model
evaluation
The UCP1 homology model generated by MODELLER v9.16
was ranked and scored using the discrete optimized protein energy
(DOPE) score. The best homolog model of UCP1 was evaluated with
PROCHECK (https://www.ebi.ac.uk/thornton-srv/software/PROCHECK/)
(20). The final structure was
visualized in PyMOL 1.7.0.0 (https://pymol.org) (21).
UCP1-lipid docking model
Similar to the homology model building, the docking
model of UCP1 and lipids was generated in MODELLER v9.16. The same
templates were analyzed to predict the docking model. The best
docking model was selected according to the DOPE score. The main
docking molecules contained B-nonylglucoside and cardiolipin.
Protein-lipid interaction
analysis
The detailed interactions of UCP1 and lipids were
displayed in PyMOL 1.7.0.0 and analyzed with LigPlot+
1.4 (https://www.ebi.ac.uk/thornton-srv/software/LigPlus/)
(22). LigPlot+ draws a
two-dimensional diagram of ligand-protein interactions from 3D
coordinates, which shows the hydrogen-bond interaction patterns and
hydrophobic contacts between the ligands and proteins.
Results and Discussion
Mitochondrial UCP1 is specifically expressed in
mammalian BAT. Thus far, to our knowledge, its high-resolution
structure has not been determined. Reportedly, UCP1 as a
mitochondrial transmembrane protein interacts with mitochondrial
lipids, which considerably affects its biological function
(23). In the current study, the
putative lipid-binding α-helices of UCP1 were determined by
bioinformatics. The HeliQuest algorithm has successfully predicted
the lipid-binding sites of membrane proteins previously (16). The overall HeliQuest-calculated scores
(mean hydrophobicity, hydrophobic moment and net charges) are
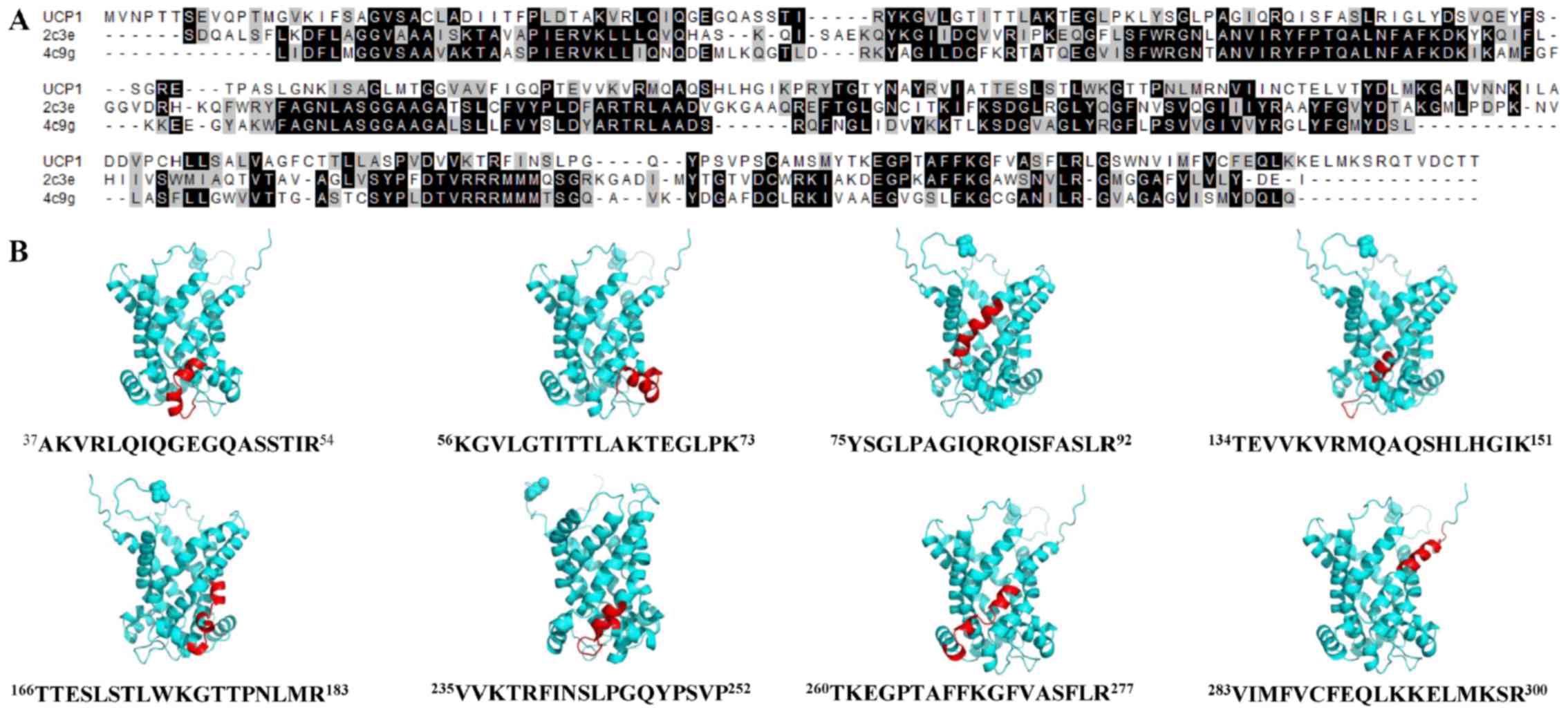
presented in Fig. 1A. Following
further discrimination factor-based analyses, a total of 8
lipid-binding α-helix regions of UCP1 were identified
(37AKVRLQIQGEGQASS TIR54,
56KGVLGTITTLAKTEGLPK73,
75YSGLPAGIQRQISF ASLR92,
134TEVVKVRMQAQSHLHGIK151,
166TTESLSTLWK GTTPNLMR183,
235VVKTRFINSLPGQYPSVP252,
260TKEGPT AFFKGFVASFLR277 and
283VIMFVCFEQLKKELMKSR300). These predicted
results may be considered acceptable among the range of strategies
available to calculate the discrimination factor. The helical wheel
plots of lipid-binding α-helix regions of UCP1 are shown in
Fig. 1B. Helical wheel plots with an
18 amino acid window are considered to depict the best α-helices
(16). As shown in Fig. 1B, the identified lipid-binding α-helix
regions displayed the amphiphilic properties. The amino acid
sequences of the lipid-binding α-helix regions of UCP1 exhibited no
high sequence identities with other known lipid-binding domains
(C1, C2, PH, FYVE, PX, ENTH, ANTH, BAR, FERM, PDZ or tubby
domains), indicating that these lipid-binding regions were the
novel lipid-binding sites. Of these lipid-binding regions of UCP1,
the fourth region (134TEVVKVRMQAQSHLHGIK151)
was a highly confident lipid-binding peptide based on the
subsequent screening scores of HeliQuest. The peptide with 100%
sequence identity was indicated to be exclusively found in mice and
rats by BLAST, and may be a crucial interacting sequence for
UCP1-lipid associations and UCP1 stability. Furthermore, there were
some sequence similarities among these novel lipid-binding α-helix
regions despite the lack of peptides with 100% sequence identity.
For example, the above-mentioned fourth lipid-binding protein
region had a 47% sequence similarity to 14NDRTLRRM
RKVVNIINAME33 (Escherichia coli SecA) as
predicted by HeliQuest (24).
To assess the 3D structure of UCP1, acceptable
templates were searched for with high sequence identity. Their
sequence alignment is presented in Fig.
2A. Functional BAT and UCP1 have been identified and
characterized in rodents and human (25,26). In
silico analysis of mouse UCP1 is performed since rodents
generally work as important validated targets prior to clinical
trials, and BAT with UCP1 expression is easily identifiable in
small mammals including mice (26,27). The
crystal structures of Bos taurus and Saccharomyces
cerevisiae ANT carriers (PDB IDs: 2c3e and 4c9g) (28,29) were
used to build a final homology model of UCP1. An available NMR
structure of UCP2 with high sequence identity to UCP1 was not
selected due to its low resolution (30). The PROCHECK program was used to
evaluate the reliability and reasonability of the model, with
>95% residues in most-favored/additionally allowed/generously
allowed regions. Surprisingly, the 8 lipid-binding α-helix regions
were mostly present outside the six main helix bundles of UCP1
(Fig. 2B), suggesting that the
interactions of lipid-binding α-helices belonged to nonannular
protein-lipid interactions (31).
Previous observations have demonstrated that mutations of certain
residues significantly influence the biological function of UCP1
(9). Of these sites, P79,
R84, R92, E135, M141,
H146, H148, E168, R183,
262EGPTAFFKG270 and R277 are
located at the lipid-binding α-helix regions (9), and these sites associated with the
lipid-binding α-helix regions may involve the biological function
of UCP1. It is hypothesized that mutations alter protein functions
via the conformational disturbances theoretically caused by the
physicochemical properties of amino acid residues (32–34).
Another possible complementary explanation is that these mutations
of membrane proteins may disturb the protein-lipid interactions,
thus causing the protein instability or conformational
abnormalities.
Cardiolipin acts as an insulator and stabilizes
mitochondrial membrane proteins, including mitochondrial
respirasome (35). As shown in
Fig. 3, the putative lipid-binding
α-helix regions of UCP1 in the current UCP1-lipid docking model
appeared to mainly associate with three cardiolipins; an
interaction eluded to previously (23). LigPlot+ analysis
demonstrated a variety of hydrophobic and hydrophilic interactions
between UCP1 and cardiolipins (Fig.
3B); indeed cardiolipin might be a key regulator of optimal
biological activity of many membrane proteins (36). An example is cardiolipin-binding
cytochrome c oxidase. Of these cardiolipin-related interactions
determined by LigPlot+, the main interacting
contributors of UCP1 appeared to be
75YSGLPAGIQRQISFASLR92, 166TTE
SLSTLWKGTTPNLMR183, 235VVKTRFINSLPGQYP
SVP252 and
260TKEGPTAFFKGFVASFLR277. The lysine residues
of cardiolipin-binding cytochrome c seem to be crucial for their
recognition and biological activities (37). In the current docking model, two
lysines (K175 and K269) were revealed to bind
to cardiolipins. The interactions may be implicated in the same
function related to the cytochrome c-cardiolipin complex. However,
the exploration of these interactions between UCP1 and cardiolipins
still requires high-resolution or standard biophysical techniques.
Previous studies have demonstrated that cardiolipin may be a
potential target for therapies against Parkinson's disease; the
current results indicate that it may be a promising therapeutic
target in UCP1-associated diseases.
 | Figure 3.Interactions between UCP1 and lipids.
The detailed interactions of UCP1 and three cardiolipins analyzed
by LigPlot+ are shown, and the relevant
peptides/molecules are highlighted:
37AKVRLQIQGEGQASSTIR54 (red),
56KGVLGTITTLAKTEGLPK73 (blue),
75YSGLPAGIQRQISFASLR92 (orange),
34TEVVKVRMQAQSHLHGIK151 (light blue),
166TTESLSTLWKGTTPNLMR183 (pink),
35VVKTRFINSLPGQYPSVP252 (warm pink),
260TKEGPTAFFKGFVASFLR277 (wheat),
83VIMFVCFEQLKKELMKSR300 (lemon),
B-nonylglucoside (green) and cardiolipin (magenta). UCP1,
uncoupling protein 1. |
Protein-lipid interactions contribute to the
structural stability and biological function of transmembrane
proteins. We herein analyzed the lipid-binding α-helix regions of
mitochondrial UCP1, using bioinformatic approaches due to the
limitations of experimental techniques in this field. A total of 8
plausible lipid-binding α-helix regions of UCP1 were predicted
based on the characteristics of protein-lipid interactions. These
interaction regions also mapped onto the key functional regions of
UCP1, suggesting that these lipid-binding regions are involved in
the biological function of UCP1. The UCP1-lipid docking model
indicate that cardiolipin is a crucial interacting molecule and may
maintain optimal biological function. The findings support the
presence of specific lipid-binding α-helices in UCP1 that may
represent the therapeutic targets for UCP1-associated diseases.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant nos. 31470716, 31000323,
31070672 and 31770981) and the Natural Science Foundation of
Jiangsu Province (grant no. BK20131272).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YJ, YN, CL and KZ performed the experiments and
analyzed the data. DL designed and performed the experiments,
analyzed data, and drafted the manuscript. All authors approved the
final version to be published.
Ethics approval and consent to
participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
ANT
|
ADP/ATP translocase
|
|
BAT
|
brown adipose tissue
|
|
DOPE
|
discrete optimized protein energy
|
|
UCP1
|
uncoupling protein 1
|
|
UCPs
|
uncoupling proteins
|
|
3D
|
three-dimensional
|
References
|
1
|
Bertholet AM and Kirichok Y: UCP1: A
transporter for H+ and fatty acid anions. Biochimie.
134:28–34. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bonet ML, Mercader J and Palou A: A
nutritional perspective on UCP1-dependent thermogenesis. Biochimie.
134:99–117. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Crichton PG, Lee Y and Kunji ERS: The
molecular features of uncoupling protein 1 support a conventional
mitochondrial carrier-like mechanism. Biochimie. 134:35–50. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Marlatt KL and Ravussin E: Brown adipose
tissue: an update on recent findings. Curr Obes Rep. 6:389–396.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Fedorenko A, Lishko PV and Kirichok Y:
Mechanism of fatty-acid-dependent UCP1 uncoupling in brown fat
mitochondria. Cell. 151:400–413. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chouchani ET, Kazak L, Jedrychowski MP, Lu
GZ, Erickson BK, Szpyt J, Pierce KA, Laznik-Bogoslavski D,
Vetrivelan R, Clish CB, et al: Mitochondrial ROS regulate
thermogenic energy expenditure and sulfenylation of UCP1. Nature.
532:112–116. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Rousset S, Alves-Guerra MC, Mozo J, Miroux
B, Cassard-Doulcier AM, Bouillaud F and Ricquier D: The biology of
mitochondrial uncoupling proteins. Diabetes. 53:S130–S135. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kopecky J, Rossmeisl M, Flachs P, Bardova
K and Brauner P: Mitochondrial uncoupling and lipid metabolism in
adipocytes. Biochem Soc T. 29:791–797. 2001. View Article : Google Scholar
|
|
9
|
Klingenspor M, Fromme T, Hughes DA Jr,
Manzke L, Polymeropoulos E, Riemann T, Trzcionka M, Hirschberg V
and Jastroch M: An ancient look at UCP1. Biochim Biophys
Acta-Bioenergetics. 1777:S24. 2008. View Article : Google Scholar
|
|
10
|
Sreedhar A and Zhao YF: Uncoupling protein
2 and metabolic diseases. Mitochondrion. 34:135–140. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zinser E and Daum G: Isolation and
biochemical characterization of organelles from the Yeast,
Saccharomyces cerevisiae. Yeast. 11:493–536. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Horvath SE and Daum G: Lipids of
mitochondria. Prog Lipid Res. 52:590–614. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lawson JE, Gawaz M, Klingenberg M and
Douglas MG: Structure-function studies of adenine nucleotide
transport in mitochondria. I. Construction and genetic analysis of
yeast mutants encoding the ADP/ATP carrier protein of mitochondria.
J Biol Chem. 265:14195–14201. 1990.PubMed/NCBI
|
|
14
|
Claypool SM: Cardiolipin, a critical
determinant of mitochondrial carrier protein assembly and function.
Biochim Biophys Acta. 1788:2059–2068. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
The UniProt C: UniProt: the universal
protein knowledgebase. Nucleic Acids Res. 45:D158–D169. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gautier R, Douguet D, Antonny B and Drin
G: HELIQUEST: a web server to screen sequences with specific
α-helical properties. Bioinformatics. 24:2101–2102. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chou KC, Lin WZ and Xiao X: Wenxiang: a
web-server for drawing wenxiang diagrams. Nat Sci. 3:862–865.
2011.
|
|
18
|
Biasini M, Bienert S, Waterhouse A, Arnold
K, Studer G, Schmidt T, Kiefer F, Gallo Cassarino T, Bertoni M,
Bordoli L, et al: SWISS-MODEL: modelling protein tertiary and
quaternary structure using evolutionary information. Nucleic Acids
Res. 42:W252–W258. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Webb B and Sali A: Protein structure
modeling with MODELLER. Methods Mol Biol. 1654:39–54. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Laskowski RA, Rullmannn JA, MacArthur MW,
Kaptein R and Thornton JM: AQUA and PROCHECK-NMR: programs for
checking the quality of protein structures solved by NMR. J Biomol
NMR. 8:477–486. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yuan S, Chan HCS, Filipek S and Vogel H:
PyMOL and Inkscape bridge the data and the data visualization.
Structure. 24:2041–2042. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Laskowski RA and Swindells MB:
LigPlot+multiple ligand-protein interaction diagrams for
drug discovery. J Chem Inf Model. 51:2778–2786. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lee Y, Willers C, Kunji ERS and Crichton
PG: Uncoupling protein 1 binds one nucleotide per monomer and is
stabilized by tightly bound cardiolipin. Proc Natl Acad Sci USA.
112:pp. 6973–6978. 2015; View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Keller RC: The prediction of novel
multiple lipid-binding regions in protein translocation motor
proteins: A possible general feature. Cell Mol Biol Lett. 16:40–54.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Cypess AM, Lehman S, Williams G, Tal I,
Rodman D, Goldfine AB, Kuo FC, Palmer EL, Tseng YH, Doria A, et al:
Identification and importance of brown adipose tissue in adult
humans. N Engl J Med. 360:1509–1517. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lau AZ, Chen AP, Gu Y, Ladouceur-Wodzak M,
Nayak KS and Cunningham CH: Noninvasive identification and
assessment of functional brown adipose tissue in rodents using
hyperpolarized 13C imaging. Int J Obes (Lond). 38:126–131. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ong FJ, Ahmed BA, Oreskovich SM, Blondin
DP, Haq T, Konyer NB, Noseworthy MD, Haman F, Carpentier AC,
Morrison KM, et al: Recent advances in the detection of brown
adipose tissue in adult humans: a review. Clin Sci (Lond).
132:1039–1054. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Nury H, Dahout-Gonzalez C, Trezeguet V,
Lauquin G, Brandolin G and Pebay-Peyroula E: Structural basis for
lipid-mediated interactions between mitochondrial ADP/ATP carrier
monomers. FEBS Lett. 579:6031–6036. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ruprecht JJ, Hellawell AM, Harding M,
Crichton PG, McCoy AJ and Kunji ER: Structures of yeast
mitochondrial ADP/ATP carriers support a domain-based
alternating-access transport mechanism. Proc Natl Acad Sci USA.
111:pp. E426–E434. 2014; View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Berardi MJ, Shih WM, Harrison SC and Chou
JJ: Mitochondrial uncoupling protein 2 structure determined by NMR
molecular fragment searching. Nature. 476:109–113. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Contreras FX, Ernst AM, Wieland F and
Brugger B: Specificity of intramembrane protein-lipid interactions.
Cold Spring Harb Perspect Biol. 3:a0047052011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Gromiha MM, Oobatake M, Kono H, Uedaira H
and Sarai A: Relationship between amino acid properties and protein
stability: buried mutations. J Protein Chem. 18:565–578. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Gromiha MM, Oobatake M, Kono H, Uedaira H
and Sarai A: Role of structural and sequence information in the
prediction of protein stability changes: comparison between buried
and partially buried mutations. Protein Eng. 12:549–555. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Gromiha MM, Oobatake M and Sarai A:
Important amino acid properties for enhanced thermostability from
mesophilic to thermophilic proteins. Biophys Chem. 82:51–67. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Guo R, Zong S, Wu M, Gu J and Yang M:
Architecture of human mitochondrial respiratory megacomplex
I2III2IV2. Cell. 170:1247–1257. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Planas-Iglesias J, Dwarakanath H,
Mohammadyani D, Yanamala N, Kagan VE and Klein-Seetharaman J:
Cardiolipin interactions with proteins. Biophys J. 109:1282–1294.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Sinibaldi F, Howes BD, Droghetti E,
Polticelli F, Piro MC, Di Pierro D, Fiorucci L, Coletta M,
Smulevich G and Santucci R: Role of lysines in cytochrome
c-cardiolipin interaction. Biochemistry. 52:4578–4588. 2013.
View Article : Google Scholar : PubMed/NCBI
|