Introduction
Deleted in leukemia 2 gene (DLEU2) is a long
noncoding RNA (lncRNA), which has been shown to serve a role as a
tumor suppressor gene in several types of blood cancer (1,2). Chronic
lymphocytic leukemia is characterized by DLEU2 deletion as well as
its tumor suppressor microRNAs (miRNAs/miRs) miR-15a/16-1 region in
~55% of cases (3,4). As a tumor suppressor gene, the
DLEU2/miR-15a/miR-16-1 locus has been extensively characterized
(5-7).
Since the DLEU2/miR15a/16-1 locus is present in several somatic
tissues, it also has shown to serve a role in regulating cell
proliferation in various cell types including, vascular endothelial
cells (8), various lymphomas and
leukemias (9), as well as
hepatocytes (10). Physiologically,
miR-15a and miR-16-1 are upregulated as a miRNA cluster from the
intronic 13q14.3 region of the DLEU2 lncRNA (10). DLEU2 itself has also been implicated
in the sponging of several miRNAs, including miR-30a (11) and miR-455, which can regulate several
downstream functions (12). However,
to the best of our knowledge, there are no studies that have
assessed the effects of lncRNA DLEU2 alternative splicing in the
liver.
Large-scale sequencing projects over the last decade
have demonstrated the extent of alternative splicing of mammalian
transcripts. Studies have shown that >95% of human genes
generate transcripts that are alternatively spliced (13-15).
However, not all alternatively spliced transcripts produce
functional proteins (16-18).
In humans, numerous studies have demonstrated an association
between dysregulation of RNA splicing and the
development/progression of several diseases (19,20).
Certain lncRNAs have been shown to serve a crucial role in the
regulation of alternative splicing in response to several stimuli
or during disease (21-23).
Interestingly, Dleu2 lncRNA in mice is differentially regulated,
with 15 different transcripts available over five different
promoter sites (Fig. S1; from
ENSEMBL database: uswest.ensembl.org/Mus_musculus/Gene/Summary?db=core;g=ENSMUSG00000097589;r=14:61602839-61682373)
(24). MiR-15a/15b and miR-16 are
encoded by introns downstream of exons present on some, but not
all, of the Dleu2 transcripts (denoted by a yellow star in Fig. S1). It has previously been shown that
the DLEU2/miR-15a/miR-16-1 locus contributed to cell apoptosis and
progression in liver fibrosis (10),
modulated miR-30a sponging in clear cell renal cell carcinoma
(11), and miR-15a/16 suppression
was implicated in the development of pleural mesothelioma (25).
Dependent on the size of the introns spliced out,
and the exons that are retained, Dleu2 alternative splicing can
have different functions on lncRNA/RNA/miRNA regulation inside the
cell. In an in silico study by Ma et al (26), RNA sequencing (RNASeq) platforms
showed that exon 9 of DLEU2 was a better marker than total DLEU2
expression for predicting unfavorable overall survival rates in
patients with esophageal adenocarcinoma. Though methylation
analysis of the DNA region encoding Dleu2 lncRNA with regard to
several cancer types, including esophageal adenocarcinoma (26), and pediatric acute myeloid leukemia
(27) have been performed, no
specific studies assessing the alternative splicing populations of
Dleu2 in the liver or any other tissues were found. In the present
study, it was shown that Dleu2 alternative splicing transcripts
were affected by silencing or overexpression, and this may improve
our understanding of how Dleu2 dysregulation and modulation affect
progression of various diseases.
Materials and methods
Experimental sample used for
comparison of Dleu2 splicing primer sets
In vitro experiments were performed on the
mouse liver cell line AML-12 using small interfering (si)RNA
knockdown for downregulation of Dleu2 expression, and upregulation
was achieved by treating cells with trans Resveratrol
antioxidant.
Cell lines and reagents
AML-12 (ATCC® CRL-2254), the α-mouse
liver cell line 12, was cultured in complete media (DMEM/F12
supplemented with 10% FBS, 1% penicillin/streptomycin, 1X
Insulin/Transferrin/Selenium solution and 40 ng/ml Dexamethasone)
as recommended by the supplier (all form ATCC). SiRNA transfections
were performed using OptiMEM media, Lipofectamine®
RNAiMAX, Silencer Select Negative Control #1 (cat. no. 4390843 5
nm) and mouse Silencer Select n410472 (cat. no. #4390771; sense,
UGCUCUUAAUAAGCAUUAAtt; antisense, UUAAUGCUUAUUAAGAGCAgc; 5 nm) (all
from Thermo Fisher Scientific, Inc. Trans Resveratrol (cat.
no. R5010-100MG) was obtained from Sigma-Aldrich; Merck KGaA.
siRNA transfections
AML-12 cells were seeded at a density of
5x105 cells/well in a 12 well plate and allowed to grow
to 70-80% confluency overnight at 37°C in complete media. After 24
h, the media was removed and cells were washed 3x with room
temperature (RT) 1X PBS, after which pre-warmed (37˚C) OptiMEM was
added. Dleu2 siRNA or Negative Control #1 was added to each well to
a final concentration of 25 nM per ml according to manufacturer's
protocol (Thermo Fisher Scientific, Inc.). After incubation at 37˚C
in 5% CO2, RNA was extracted from cells after 24 h of
transfection.
Trans Resveratrol exposure
In an unrelated project, the effects of several
antioxidants on Dleu2 were investigated. Interestingly, it was
identified that Resveratrol, a potent anti-oxidant, led to an
increase in Dleu2 expression. This upregulation of DLEU2 by
Resveratrol was also observed in the human hepatocellular carcinoma
cell line HepG2 (data not shown). Therefore, Resveratrol was used
as a positive technical control in the present study.
AML-12 cells were seeded as described above. A total
of 1 ml complete media containing 0.1% EtOH, or 50 µM trans
Resveratrol in EtOH was added to each well, and incubated for 20 h
at 37˚C.
RNA extraction and reverse
transcription-quantitative (RT)q-PCR
RNA was extracted using the Qiagen RNeasy Mini Prep
kit (Qiagen, GmbH), according to the manufacturer's protocols, and
RT-qPCR was performed as previously described, using the primer
sequences designed for alternative splicing and the other sequences
shown below (28).
Alternative splicing primer design and
primer sets
SiRNAs were developed to specifically target the
mouse Dleu2 exon (Mouse chromosome 14, Exon 4,
61,632,437-61,632,483 bp) directly upstream of the miR-15a intron
site (Thermo Fisher Scientific, Inc.). Alt1 (Dleu2 alias) primers
for mouse (m) mDleu2 were designed using NCBI BLAST, targeting the
largest cDNA sequences in their database (National Institutes of
Health). Alt1 was designed to bind to 4 of the Dleu2 transcripts at
the same time in ENSEMBL; however, this primer set did not target
several of the other alternative Dleu2 splicing sets in the mouse
ENSEMBL database (ensembl.org). Therefore, 15 different
spliced Dleu2 sequences were investigated, with spliced exon-exon
cDNA sequences retrieved from the ENSEMBL website, and specific
primer sets designed to target different Dleu2 transcripts.
These cDNA sequences were sequentially run on NCBI
BLAST to assess the similarity, with only outlier exon regions not
matching any other mDleu2 cDNA sequences selected. However, several
primer sets still had matching regions on the larger -202 and -207
cDNA sequences yet still targeted their main splicing sequence;
these primer sets were kept. These available selected areas were
then screened using Primer3Plus (primer3plus.com) to find regions capable of producing
standard PCR products (20-25 bp primer length, Tm 60˚C,
60% GC content, 50-200 bp product size). These targeted primer sets
were then screened against the BLAST alignment tool in ENSEMBL to
check for other possible DNA matches in the mouse genome. Primer
sets with low E values (below 0.05) that were specific for the
mDleu2 gene were kept while others were discarded. Primers that
matched all these criteria (Table
SI) were developed and purchased from Sigma-Aldrich; Merck KGaA
for use in the qPCR. Ineffective primer pairs that resulted in
multiple melt curve peaks during qPCR amplification were discarded
post-testing. Primers targeting specific Dleu2 transcripts are
shown outlined in various colors on the ENSEMBL in Fig. 1B.
Expression of genes associated with
proliferation
RT-qPCR was also used to measure expression of genes
associated with proliferation, including proliferating cell nuclear
antigen, transforming growth factor-β and epidermal growth factor
receptor (EGFR) in Dleu2-silenced and control AML-12 cells. The
sequences of the primers used are listed in Table SI.
Statistical analysis
All data are presented as the mean ± the standard
error of the mean. Comparisons between groups were performed using
a Student's t-test (two groups). P<0.05 was considered to
indicate a statistically significant difference. Statistical
analysis was performed using GraphPad Prism version 6.0 (GraphPad
Software, Inc.).
Results
Alternative splicing modulates Dleu2
for targeted primers
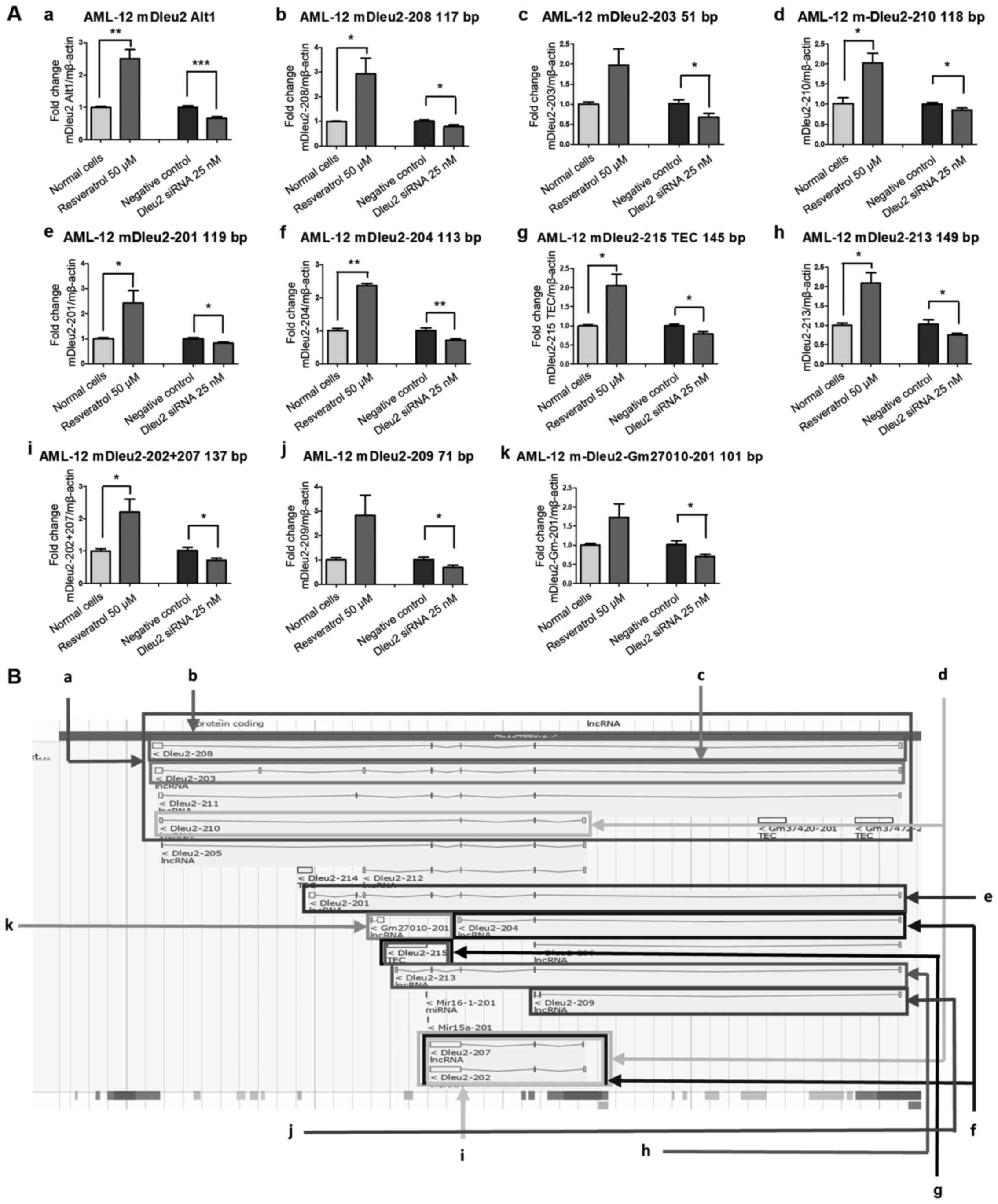
The Alt1 primer set targets mDleu2-208, -203, -210
and -211, four of the mDleu2 transcripts produced in the Dleu2
transcript map with overlapping regions (Fig. 1Ba). As such, this primer set provides
a general outlook on how mDleu2 is expressed overall in the mouse
under downregulating/upregulating experimental conditions.
Resveratrol treatment for 20 h resulted in significant upregulation
(P<0.01) in the mDleu2 Alt1 subset, and transfection of mDleu2
siRNA significantly downregulated its expression (P<0.001;
Fig. 1Aa). The mDleu2-208 primer set
only targets mDleu2-208 in the Dleu2 transcript map (Fig. 1Ab and Bb). After 20 h Resveratrol treatment
significant upregulation (P<0.05) in the mDleu2-208 subset was
observed, whereas siRNA-mediated knockdown resulted in significant
downregulation (P<0.05), although the effect was less potent
than that on the Alt1 set. The mDleu2-203 primer set only targets
mDleu2-203 in the Dleu2 transcript map (Fig. 1Ac and Bc). The Resveratrol treatment had no
significant effect on the mDleu2-203 subset, whereas mDleu2 siRNA
resulted in knockdown (P<0.05), although the effect was less
potent than that on the Alt1 set. The mDleu2-210 primer set targets
mDleu2-210, and overlaps -202 and -207 in the Dleu2 transcript map
(Fig. 1Ad and 1Bd). Resveratrol treatment resulted in
significant upregulation (P<0.05) in the mDleu2-210 subset, and
mDleu2 siRNA knockdown resulted in significant downregulation
(P<0.05), although it was less significantly associated than
that of the Alt1 set.
The mDleu2-201 primer set only targets mDleu2-201 in
the Dleu2 transcript map (Fig. 1Ae
and Be). Resveratrol treatment
resulted in significant upregulation (P<0.05) in the mDleu2-201
subset, and mDleu2 siRNA knockdown resulted in significant
downregulation (P<0.05). The mDleu2-204 primer set targets
mDleu2-204, and overlaps part of -202 and -207 in the Dleu2
transcript map (Fig. 1Af and
Bf). Resveratrol exposure resulted
in significant upregulation (P<0.01) of the mDleu2-204 subset,
and mDleu2 siRNA knockdown significantly downregulated expression
of this subset (P<0.01).
The mDleu2-215 TEC primer set, mDleu2-213 primer
set, and mDleu2-202+207 primer set targets mDleu2-215 TEC,
mDleu2-213 and overlaps -215 TEC, and mDleu2-202 and -207 in the
Dleu2 transcript map, respectively (Fig.
1Ag and Bg; Fig. 1Ah and Bh; Fig. 1Ai
and Bi). Resveratrol treatment
resulted in significant upregulation (P<0.05) of the mDleu2-215
TEC subset, mDleu2-213 subset, and mDleu2-202+207 subset; and
mDleu2 siRNA knockdown resulted in significant downregulation of
all three sets (P<0.05).
The mDleu2-209 primer set targets mDleu2-209 in the
Dleu2 transcript map (Fig. 1Aj and
Bj). Resveratrol exposure did not
significantly affect the mDleu2-209 subset; however, the mDleu2
siRNA did significantly downregulate its expression (P<0.05).
The mDleu2-Gm27010-201 primer set only targets mDleu2-Gm27010-201
in the Dleu2 transcript map (Fig.
1Ak and Bk). Resveratrol
treatment did not have a significant effect on the mDleu2-201
subset, whereas mDleu2 siRNA did significantly downregulate its
expression (P<0.05).
Expression of genes associated with
proliferation
Expression of proliferating cell nuclear antigen,
transforming growth factor-β and epidermal growth factor receptor
was not altered significantly when Dleu2 expression was knocked
down (Fig. S2).
Discussion
DLEU2 is a lncRNA that was only discovered a decade
ago, and has more recently been implicated in several types of
cancer, including multiple types of blood cancer, as well as
hepatocellular carcinoma. As such, it is possible that changes to
the expression patterns of one or several of its multiple
transcripts, and thus dysregulation of the downstream lncRNAs,
miRNAs and/or RNAs interactions, may lead to abnormalities. Dleu2
has been shown to regulate several physiological pathways,
including proliferation (29-32),
although, it does not alter proliferation of the non-cancerous
AML-12 cells (Fig. S2).
Interestingly, Dleu2 has a unique alternative splicing
transcriptome, with multiple patterns for exon-exon splicing and
intron removal (2,26). How these populations of various
transcriptions arise may be important in interpreting the
pathological outcomes. For example, changes in the expression
levels of various exons within Dleu2 have been shown to be
correlated with unfavorable overall survival rates in patients with
esophageal adenocarcinoma (26).
Therefore, the ability to monitor and investigate changes to these
expression patterns or alternative transcription motifs may be of
use in identifying dysregulated cellular biology and cancer
progression.
Based on the results of the individual alternative
splicing primer sets, a repeating pattern for all lncRNA Dleu2
primers used to measure Dleu2 transcript expression was observed.
The mDleu2-Alt1 primer set targeted the mDleu2-208, -203, -210 and
-211 transcripts, and the expression of these transcripts increased
when treated with Resveratrol, and decreased when expression was
knocked down using siRNAs. Most individual primer sets displayed
this same pattern with significant upregulation of mDleu2 upon
exposure to Resveratrol, and significant downregulation in the
siRNA knockdown experiments, although there were some exceptions;
specifically Resveratrol did not result in significant
downregulation of mDleu2-203, -209 and -Gm27010-201, a downward
trend was observed in these cases as well. Whereas the Resveratrol
results showed some inconsistencies, what was noteworthy about
these results was that in the Dleu2 transcripts which lacked the
Exon 4 sequence, the target of the siRNAs, they were still knocked
down significantly overall. Specifically, mDleu2-210, -204,
-202+207, -209 and -Gm27010-201 transcripts lacked Exon 4 from the
ENSEMBL alternative transcription map (Fig. S1), yet were still significantly
knocked in the cells transfected with the siRNAs. It is possible
that there are interactions present between the various alternative
transcripts that modulate the expression of the entire Dleu2
transcript family, or perhaps the targeted siRNAs also targeted
other areas similar to Exon 4 on the other transcripts. Future
experiments using other targeted siRNAs that specifically target
other exons on Dleu2 may provide additional information as to how
this principle within Dleu2 is regulated.
Overall, the results found evidence of Dleu2
modulation across the spectrum of transcripts, with upregulation
due to Resveratrol exposure and downregulation due to siRNA
treatment. Upregulation of Dleu2 occurred across all transcripts
generally uniformly (though not always significantly), with
increased lncRNA expression from the 5'-promoter sites available
(Fig. S1, bottom panel, red bars).
c-Myb and PPAR are known to be positive regulators of these
promoter binding sites in Dleu2(33). Furthermore, noted shifts in splice
patterns are present in diseased states as compared with the global
splicing map of the human genome, which can modulate the frequency
of Poly-A choices on transcripts (21).
It is possible that the limited changes in Dleu2
expression from 20 h treatment with Resveratrol and 24 h siRNA
silencing treatments using the individual transcript primer sets
may be due to the limited time span of the AML-12 cell culture
experiments, and that longer exposures may have resulted in more
significant results. Some transcripts, particularly mDleu2-203,
-209 and -Gm27010-201, were not as significantly affected by
Resveratrol treatment relative to the others, perhaps indicating
more resistance to changes in expression compared with the other
transcripts. As specific changes to DLEU2 exon expression have been
confirmed in esophageal adenocarcinoma (26) and pediatric acute myeloid leukemia
(27), exon expression changes are
definitely a possibility in altering liver Dleu2 transcript
expression. However, more specific testing is required to determine
these alternative splicing changes in future experiments, perhaps
using RNASeq to sidestep the limitations of overlapping targeted
primer sets on the nested mouse Dleu2 transcripts. Additionally,
several of these initial results need be assessed and confirmed
in vivo or even using human liver tissues to identify the
effects of alternative splicing.
A previous study looked at a novel alternative
splicing subset within an overlapping DLEU2/LEU5/RFP2 cluster,
containing multiple alternative splicing sites to produce
monocistronic transcripts or a bicistronic transcript (34). Primers were designed to individually
target exons of the LEU5/RFP2 cluster as well of that of DLEU2 and
the overlapping sequences between the two genes. Whilst this
previous study involved artificial constructs of areas surrounding
exons from DLEU2/LEU5/RFP2, they were able to confirm specific
intronic sequences between exons using RT-qPCR. The present study
only investigated Dleu2 sequences within specific unique exons,
whereas the previous study was searching for sequences spanning
multiple exons, although RT-qPCR was used in both studies to build
a larger exon-exon map of alternative splicing regions within and
surrounding lncRNA Dleu2.
In other studies involving pediatric AML (27) and esophageal adenocarcinoma (26), alternative splicing of DLEU2 was
investigated, and hypermethylation or expression of specific exons
was shown to be associated with cancer progression. As such, there
may be numerous other mechanisms available, in addition to simple
alternative splicing, by which Dleu2 promotes the development
and/or progression of cancer.
The way in which both mouse and human Dleu2 and
DLEU2, respectively, are organized in their promoter regions and
location/number of alternative transcription sites may exert an
effect on how DLEU2 is modulated between the two species (24). Mouse Dleu2 is primarily layered such
that most of the functional transcripts reside nested within one
another, thus it is difficult to find specific primer sets that
will only target one transcript and does not overlap multiple
transcripts. As such, there are more promoter sites, and a more
spread out area for human DLEU2 alternative splicing sites
(Fig. S3A and B). Whereas the miR-15a/16 locus is also
present for a simple majority of the human DLEU2 alternative
splicing sites, there are multiple DLEU2 alternative splicing sites
coded outside of that region in humans (24). It is probable that with the
additional human DLEU2 transcripts compared to mouse Dleu2
regulation, there may be additional modulatory factors present in
the regulation of human Dleu2, particularly through different
alternative splicing transcription patterns, or through other
dysfunctional regulatory factors. Finding these transcriptional
changes within transcriptome populations could ultimately lead to
novel diagnostic tools and aid in prevention of disease.
In conclusion, using the splicing map of Dleu2, the
present study was the first to show the modulation of Dleu2
alternative spliced region expression was shown through knockdown
and overexpression. Furthermore, a nutraceutical, Resveratrol, was
shown to increase the expression of Dleu2. This may assist in
identifying a means of cancer prevention for certain types of
cancer. Recently, Soreq et al (22) used wide annotations of alternate
promoters, splicing and alternative poly-A sites to identify and
quantify both disease- and treatment-induced splicing shifts, miRNA
binding site modifications, putatively changed protein-protein
interactions and other transcript structural changes in Parkinson's
leukocytes. Future studies in in vivo mouse and human
samples should expand in the above directions to investigate Dleu2
and alternative splicing in depth.
Supplementary Material
Mouse lncRNA Dleu2 transcript map.
Location of mDleu2 which can encode 15 different transcripts; the
spliced-in exons are shown by vertical bars, and the spliced-out
introns are shown by horizontal lines. Promoter regions are shown
in red at the bottom. Introns coding for miR-15/16 denoted by
yellow star. Figure obtained from ENSEMBL (uswest.ensembl.org/Mus_musculus/Gene/Summary?db=core;g=ENSMUSG00000097589;r=14:61602839-61682373).
lncRNA, long non-coding RNA; miRNA/miR, microRNA; misc,
miscellaneous; lincRNA, long intervening noncoding RNA; Dleu2,
deleted in leukemia 2 gene.
Expression of genes associated with
proliferation. (A) PCNA, (B) TGF-β and (C) EGFR expression in the
negative control and Dleu2-silenced AML-12 cells. n=6. Data are
presented as the mean ± the standard error of the mean of at least
6 independent experiments. siRNA, small interfering RNA; Dleu2,
deleted in leukemia 2 gene; PCNA, proliferating cell nuclear
antique; TGF-β, transforming growth factor-β; EGFR, epidermal
growth factor receptor.
Differences between the mouse Dleu2
splicing organization compared with Human DLEU2 splicing
organization. (A) Mouse Dleu2 transcripts are more nested compared
with (B) human DLEU2, which have a longer range away from the
miR15a/16 loci and are more spread out across different promoter
sites. Figures obtained from ENSEMBL (uswest.ensembl.org/Mus_musculus/Gene/Summary?db=core;g=ENSMUSG00000097589;r=14:61602839-61682373
and uswest.ensembl.org/Homo_sapiens/Gene/Summary?db=core;g=ENSG00000231607;r=13:49956670-50125720).
Dleu2/ DLEU2, deleted in leukemia 2 gene; lncRNA, long non-coding
RNA; miRNA/miR, microRNA; misc, miscellaneous; lincRNA, long
intervening noncoding RNA.
Primer sets used for identifying
mDleu2 transcripts and genes associated with
proliferationa.
Acknowledgements
We would like to thank the undergraduate lab
assistants Miss Sumiya Wahab, Miss Jazmin Diaz, Miss Victoria
Tilson and Miss Faith Upton (Texas A&M University) for their
contributions to this work.
Funding
Funding: The present study was supported by funding from the
Morris L Lichtenstein Jr. Medical Research Foundation (grant no.
M1500307).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
MC and MKK designed the study and wrote the
manuscript. MKK performed the experiments. JZ analyzed and
interpreted the data. All authors read and approved the final
manuscript. MKK, JZ and MC confirmed the authenticity of all the
raw data.
Ethics approval and consent to
participate
Not applicable
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Klein UL, Lia M, Shen Q, Smith PM, Tang H,
Mo T, Crespo M, Siegel R, Bhagat G and Dalla-Favera R: The
DLEU2/Mir-15a/Mir-16-1 locus, commonly deleted in B-cell chronic
Lymphocytic leukemia (CLL), controls B-cell compartment expansion
and its deletion leads to CLL in mice. Blood. 112(25)2008.
|
|
2
|
Klein U, Lia M, Crespo M, Siegel R, Shen
Q, Mo T, Ambesi-Impiombato A, Califano A, Migliazza A, Bhagat G, et
al: The DLEU2/miR-15a/16-1 cluster controls B cell proliferation
and its deletion leads to chronic lymphocytic leukemia. Cancer
Cell. 17:28–40. 2010.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Döhner H, Stilgenbauer S, Benner A,
Leupolt E, Kröber A, Bullinger L, Döhner K, Bentz M and Lichter P:
Genomic aberrations and survival in chronic lymphocytic leukemia. N
Engl J Med. 343:1910–1916. 2000.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Kalachikov S, Migliazza A, Cayanis E,
Fracchiolla NS, Bonaldo MF, Lawton L, Jelenc P, Ye X, Qu X, Chien
M, et al: Cloning and gene mapping of the chromosome 13q14 region
deleted in chronic lymphocytic leukemia. Genomics. 42:369–377.
1997.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Bullrich F, Fujii H, Calin G, Mabuchi H,
Negrini M, Pekarsky Y, Rassenti L, Alder H, Reed JC, Keating MJ, et
al: Characterization of the 13q14 tumor suppressor locus in CLL:
Identification of ALT1, an alternative splice variant of the LEU2
gene. Cancer Res. 61:6640–6648. 2001.PubMed/NCBI
|
|
6
|
Corcoran MM, Rasool O, Liu Y, Iyengar A,
Grander D, Ibbotson RE, Merup M, Wu X, Brodyansky V, Gardiner AC,
et al: Detailed molecular delineation of 13q14.3 loss in B-cell
chronic lymphocytic leukemia. Blood. 91:1382–1390. 1998.PubMed/NCBI
|
|
7
|
Liu Y, Corcoran M, Rasool O, Ivanova G,
Ibbotson R, Grandér D, Iyengar A, Baranova A, Kashuba V, Merup M,
et al: Cloning of two candidate tumor suppressor genes within a 10
kb region on chromosome 13q14, frequently deleted in chronic
lymphocytic leukemia. Oncogene. 15:2463–2473. 1997.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Li Y, Sun N, Lu Z, Sun S, Huang J, Chen Z
and He J: Prognostic alternative mRNA splicing signature in
non-small cell lung cancer. Cancer Lett. 393:40–51. 2017.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Lerner M, Harada M, Lovén J, Castro J,
Davis Z, Oscier D, Henriksson M, Sangfelt O, Grandér D and Corcoran
MM: DLEU2, frequently deleted in malignancy, functions as a
critical host gene of the cell cycle inhibitory microRNAs miR-15a
and miR-16-1. Exp Cell Res. 315:2941–2952. 2009.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Jiang XP, Ai WB, Wan LY, Zhang YQ and Wu
JF: The roles of microRNA families in hepatic fibrosis. Cell
Biosci. 7(34)2017.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Chen Z, Zhang J, Zhang Z, Feng Z, Wei J,
Lu J, Fang Y, Liang Y, Cen J, Pan Y, et al: The putative tumor
suppressor microRNA-30a-5p modulates clear cell renal cell
carcinoma aggressiveness through repression of ZEB2. Cell Death
Dis. 8(e2859)2017.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Xu B, Gong X, Zi L, Li G, Dong S, Chen X
and Li Y: Silencing of DLEU2 suppresses pancreatic cancer cell
proliferation and invasion by upregulating microRNA-455. Cancer
Sci. 110:1676–1685. 2019.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Boue S, Letunic I and Bork P: Alternative
splicing and evolution. BioEssays. 25:1031–1034. 2003.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Modrek B and Lee C: A genomic view of
alternative splicing. Nat Genet. 30:13–19. 2002.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Pan Q, Shai O, Lee LJ, Frey BJ and
Blencowe BJ: Deep surveying of alternative splicing complexity in
the human transcriptome by high-throughput sequencing. Nat Genet.
40:1413–1415. 2008.PubMed/NCBI View
Article : Google Scholar
|
|
16
|
Soergel DAW, Lareau LF and Brenner SE:
Regulation of gene expression by coupling of alternative splicing
and NMD. In: Madame Curie Bioscience Database. Landes Bioscience,
Austin, TX, 2000-2013.
|
|
17
|
Edwalds-Gilbert G: Regulation of mRNA
Splicing by Signal Transduction. Nat Educ. 3(43)2010.
|
|
18
|
Tress ML, Abascal F and Valencia A:
Alternative splicing may not be the key to proteome complexity.
Trends Biochem Sci. 42:98–110. 2017.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Ankö ML and Neugebauer KM: Long noncoding
RNAs add another layer to pre-mRNA splicing regulation. Mol Cell.
39:833–834. 2010.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Romero-Barrios N, Legascue MF, Benhamed M,
Ariel F and Crespi M: Splicing regulation by long noncoding RNAs.
Nucleic Acids Res. 46:2169–2184. 2018.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Gil N and Ulitsky I: Production of spliced
long noncoding RNAs specifies regions with increased enhancer
activity. Cell Syst. 7:537–547.e3. 2018.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Soreq L, Guffanti A, Salomonis N,
Simchovitz A, Israe Z, Bergman H and Soreq H: Long non-coding RNA
and alternative splicing modulations in Parkinson's leukocytes
identified by RNA sequencing. PLOS Comput Biol.
10(e1003517)2014.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Kiegle EA, Garden A, Lacchini E and Kater
MM: A genomic view of alternative splicing of long non-coding RNAs
during rice seed development reveals extensive splicing and lncRNA
gene families. Front Plant Sci. 9(115)2018.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Yates AD, Achuthan P, Akanni W, Allen J,
Allen J, Alvarez-Jarreta J, Amode MR, Armean IM, Azov AG, Bennett
R, et al: Ensembl 2020. Nucleic Acids Res. 48:D682–D688.
2020.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Williams M, Cheng YY, Kirschner MB, Sarun
KH, Schelch K, Winata P, McCaughan B, Kao S, Van Zandwijk N and
Reid G: Transcriptional suppression of the miR-15/16 family by
c-Myc in malignant pleural mesothelioma. Oncotarget. 10:4125–4138.
2019.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Ma W, Zhang CQ, Dang CX, Cai HY, Li HL,
Miao GY, Wang JK and Zhang LJ: Upregulated long-non-coding RNA
DLEU2 exon 9 expression was an independent indicator of unfavorable
overall survival in patients with esophageal adenocarcinoma. Biomed
Pharmacother. 113(108655)2019.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Morenos L, Chatterton Z, Ng JL, Halemba
MS, Parkinso-Bates M, Mechinaud F, Elwood N, Saffery R and Wong NC:
Hypermethylation and down-regulation of DLEU2 in paediatric acute
myeloid leukaemia independent of embedded tumour suppressor
miR-15a/16-1. Mol Cancer. 13(123)2014.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Park MH, Gutiérrez-García AK and Choudhury
M: Mono-(2-ethylhexyl) phthalate aggravates inflammatory response
via sirtuin regulation and inflammasome activation in RAW 264.7
cells. Chem Res Toxicol. 32:935–942. 2019.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Guo Y, Bai M, Lin L, Huang J, An Y, Liang
L, Liu Y and Huang W: LncRNA DLEU2 aggravates the progression of
hepatocellular carcinoma through binding to EZH2. Biomed
Pharmacother. 118(109272)2019.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Wu W, Zhao Y, Gao E, Li Y, Guo X, Zhao T,
He W and Zhang H: LncRNA DLEU2 accelerates the tumorigenesis and
invasion of non-small cell lung cancer by sponging miR-30a-5p. J
Cell Mol Med. 24:441–450. 2020.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Zhou Y, Shi H, Du Y, Zhao G, Wang X, Li Q,
Liu J, Ye L, Shen Z, Guo Y, et al: lncRNA DLEU2 modulates cell
proliferation and invasion of non-small cell lung cancer by
regulating miR-30c-5p/SOX9 axis. Aging (Albany NY). 11:7386–7401.
2019.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Lu T, Wang R, Cai H and Cui Y: Long
non-coding RNA DLEU2 promotes the progression of esophageal cancer
through miR-30e-5p/E2F7 axis. Biomed Pharmacother.
123(109650)2020.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Zheng ZM and Wang X: Regulation of
cellular miRNA expression by human papillomaviruses. Biochim
Biophys Acta. 1809:668–677. 2011.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Corcoran MM, Hammarsund M, Zhu C, Lerner
M, Kapanadze B, Wilson B, Larsson C, Forsberg L, Ibbotson RE,
Einhorn S, et al: DLEU2 encodes an antisense RNA for the putative
bicistronic RFP2/LEU5 gene in humans and mouse. Genes Chromosomes
Cancer. 40:285–297. 2004.PubMed/NCBI View Article : Google Scholar
|