Introduction
Breast cancer (BC) is the most common malignancy
among women, with 2.3 million new cases and 685,000 cancer-related
deaths reported globally in 2020(1). According to the World Health
Organization (WHO), BC is the second most prevalent cancer
diagnosed in Saudi Arabia, with 14.2% new cases in 2020. Although
the 5-year survival rate has improved over the years owing to
developments in cancer treatment, a significant number of patients
with BC still succumb to this disease, due to regional and distant
metastases when diagnosed at later stages. A burden is associated
with BC owing to its heterogeneity, resulting in increased
incidence and low survival rates (2). Breast tissues contain heterogeneous
groups of carcinoma cells that exhibit tumorlike characteristics.
Breast carcinomas appear as elongated spindle cells in the presence
or absence of elongated or ovoid nuclei in pale cytoplasm. An
extensive loss of epithelial morphology and a gain in the
mesenchymal spindle cell profile occur due to biological plasticity
processes, such as epithelial-mesenchymal transition. Therefore,
different types of breast carcinomas can appear, such as
epithelial, mesenchymal, or biphasic carcinomas, which contain both
cell types (3).
The tumor microenvironment (TME) is highly
immunosuppressive and can constrain antitumor immune responses and
promote tumor progression. It has recently been reported that
molecules such as macroH2A1, a histone variant with a large
C-terminal portion, are associated with bone metastasis in BC.
MacroH2A1 expression is higher in metastatic BC than in
nonmetastatic BC (4). The release
of soluble molecules can shape the bone microenvironment and
promote tumor progression. Various molecules in a soluble secreted
form, but also membrane-bound, contribute to the suppression of
antitumor immunity. Membrane-bound molecules are shed from the cell
surface and secreted into the TME (5). Targeting of these molecules has shown
clinical benefits in cancer immunotherapy.
Immune checkpoint inhibitors with a role in
maintaining immunosurveillance against tumors, including two
antibodies against cytotoxic T-lymphocyte-associated antigen 4
(CTLA4) and five antibodies against programmed death 1 (PD-1) and
its ligands PDL-1 and PDL-2, have been used in several clinical
trials on BC treatment (6). These
immune checkpoint inhibitors are effective in patients with
CTLA4-positive and PD-1-positive BC, where differential expression
levels are associated with the effectiveness of related blockades
as tumor suppressor agents. Hence, only a small proportion of
patients with BC benefit from these cancer immunotherapies
(7,8).
In addition, BC is considered as a weakly
immunogenic cancer (7). Along with
CTLA4, PD-1, PDL-1 and PDL-2, B- and T-lymphocyte attenuator (BTLA)
has immune inhibitory effects. BTLA is induced in Th1 cells after
activation of T cells but is not expressed by naive T cells. The
interaction of BTLA with its ligand attenuates the production of
interleukin-2 (IL-2), thus affecting T-cell proliferation and
suggesting that it acts as a third immune checkpoint (9). BTLA binds to the herpesvirus entry
mediator (HVEM), a tumor necrosis factor (TNF) receptor. Cytotoxic
T-lymphocyte 8 (CD8-T)-cell suppression has been shown to be
mediated by aberrant expression of BTLA and HVEM (7), which has been linked to leukemia
(9) and B-cell malignancy (10). HVEM was first detected in
hematopoietic cells but later was also observed in cancer cells
such as BC (11), colorectal
carcinoma (12), hepatocellular
carcinoma (13), and renal
carcinoma (14). A negative
correlation between HVEM expression and survival rate was reported
in human esophageal squamous cell carcinoma, in which high levels
of HVEM were associated with lower SR (15). HVEM+ melanomas suppressed
the proliferation of BTLA+ tumor-specific
CD8+ T cells and inhibited the production of
interferon-γ (IFN-γ). This is due to the phosphorylation of
immunoreceptor tyrosine-based inhibition motifs (ITIMs) and Src
homology 2 (SH2) domain-containing phosphatase 1 (SHP-1)/SHP-2,
resulting in the reduction of both T-cell proliferation and
cytokine production (9). In
addition, HVEM expression in melanoma samples has been correlated
with an aggressive gene signature, such as MITF (16). These results suggest that the
interaction between HVEM and BTLA contributes to tumor evasion from
immunosurveillance. Indeed, antagonizing both BTLA and PD-1 has
been shown to restore NY-ESO-1-specific CD8+ T-cell
proliferation and cytokine production in melanoma compared with the
use of anti-PD-1 alone (17).
However, other studies have shown that HVEM overexpression in
pancreatic and bladder cancers improves the survical rate (18,19).
In addition, in melanoma treatment, the transfer of CD8+
BTLA+ tumor-infiltrating lymphocytes (TILs) in adoptive
cell therapy was revealed to be associated with improved clinical
results (20). Owing to the
contrasting results of these studies, further HVEM investigations
are required.
Furthermore, three inhibitory receptors in
dysfunctional T cells in cancer have been found to maintain tumor
tolerance including lymphocyte activating gene 3 (LAG-3), T-cell
immunoreceptor with immunoglobulin (Ig) and ITIM domains (TIGIT),
and T-cell immunoglobulin and mucin-3 (TIM-3) (21). Indoleamine 2,3-dioxygenase (IDO) is
a modulatory enzyme that interferes with T-cell survival (10) and has been detected in BC,
especially in the triple-negative subtype, resulting in cancer
escaping antitumor immunity (21).
Unlike previously described molecules, the glucocorticoid-induced
tumor necrosis factor (TNF) receptor-related (GITR) gene provides
co-stimulatory signals to CD4+ and CD8+ T
cells through the activation of the NF-κB pathway, resulting in the
production of inflammatory cytokines. GITR agonists induce effector
antitumor cells and overcome self-tolerance (22).
In the present study polymerase chain reaction (PCR)
was used to assess the expression levels of 12 genes with
immune-inhibitory effects. The clinicopathological characteristics
of patients with BC were then linked to genes that showed
substantially different values. Furthermore, the serum levels of
the differentially expressed genes were determined and linked to
clinicopathological features. The present study may lead to
enhanced understanding of the expression status of immune
inhibitory-related genes, which may be used as biomarkers for BC
prognosis.
Materials and methods
Study subjects
Ethical approval (approval no. HA-02-J-008) was
granted by the Biomedical Ethics Research Committee of King
Abdulaziz University Hospital (KAUH), Jeddah, Saudi Arabia.
Patients with recurrent BC who had begun treatment were excluded
from the study, and only those diagnosed with BC for the first time
were included. A total of 32 age-matched female patients, 16 with
malignant BC and 16 non-malignant control subjects (mean age ± SEM,
47.03±1.141 and 46.56±2.1, respectively), were recruited between
October 2016 and September 2017 from the Department of Radiology,
Mammogram Section, KAUH. The baseline characteristics of the
participants, obtained from the completed questionnaires, are
presented in Table I. The
participants were provided with information concerning the study
and requested to sign a consent form. Clinicopathological data were
acquired from the Department of Laboratory Medicine and Pathology,
KAUH (23).
 | Table IBaseline characteristics of studied
subjects. |
Table I
Baseline characteristics of studied
subjects.
| | Total | Non-malignant
BC | Malignant BC |
|---|
| Parameters | Mean ± SEM | Median | IQR | Mean ± SEM | Median | IQR | Mean ± SEM | Median | IQR |
|---|
| Number of patients,
n (%) | 32(100) | | | 16 (50.0) | | | 16 (50.0) | | |
| Age, years | 47.03±1.41 | 48.00 | 12.25 | 47.5±1.92 | 49.00 | 11.75 | 46.56±2.13 | 45.00 | 10.50 |
| BMI,
kg/m2 | 29.56±0.96 | 28.85 | 6.17 | 30.4±1.40 | 29.80 | 8.50 | 28.71±1.31 | 28.10 | 5.87 |
| Waist
circumference, cm | 89.09±3.12 | 92.00 | 19.00 | 93.0±3.53 | 85.50 | 30.00 | 85.19±5.08 | 93.00 | 19.00 |
| Hip circumference,
cm | 103.8±3.50 | 106.00 | 16.8 | 107.2±3.57 | 105.00 | 14.50 | 100.4±6.03 | 106.00 | 18.55 |
| W/H ratio | 0.86±0.02 | 0.865 | 0.105 | 0.87±0.03 | 0.870 | 0.085 | 0.85±0.02 | 0.865 | 0.15 |
| Age of first
menstruation, years | 13.41±0.31 | 13.00 | 3.00 | 13.19±0.41 | 13.00 | 2.75 | 13.63±0.47 | 13.50 | 3.00 |
| Age since
menopause, years | 50.91±1.37 | 50.00 | 9.00 | 51.80±2.11 | 52.00 | 7.50 | 50.17±1.92 | 50.00 | 10.25 |
| Age of first
pregnancy, years | 21.89±0.80 | 21.00 | 6.00 | 20.92±1.19 | 20.00 | 7.25 | 22.67±1.06 | 22.00 | 6.00 |
Collection and storage of blood
samples
Peripheral blood samples were collected in two
different tubes. First, 5 ml of whole blood was collected in
PAXgene™ Blood RNA Tubes (BRT; Qiagen, Inc.) according to the
manufacturer's instructions. The samples were stored at -80˚C until
RNA extraction. Second, serum was separated from the DB
Vacutainer® SSTTM tube by centrifugation at 2,000 x g
for 20 min at room temperature. The serum was then aliquoted and
stored at -80˚C for multiplex immunoassays.
RNA extraction
Total RNA was extracted from the samples using the
PAXgene™ blood RNA kit (Qiagen, Inc.) following the manufacturer's
instructions. In brief, the BRT was centrifuged for 10 min at 4,800
x g and the pellet was washed twice with RNase-free water before
adding 40 µl of proteinase K (PK). The lysate was then directly
pipetted into a PAX gene shredder spin column. DNase I was added
directly to the spin column membrane, incubated for 15 min, and
centrifuged at 16,000 x g for 1 min. The elution step was repeated
twice, and the quality and quantity of the extracted RNA were
confirmed using a DeNovix DS-11 spectrophotometer (DeNovix, Inc.)
and gel electrophoresis on 1.2% agarose gel. The isolated RNA was
aliquoted and stored at -80˚C until further analysis. All steps of
this protocol were carried out at room temperature.
Complementary DNA (cDNA)
synthesis
Using a QuantiTect Reverse Transcription kit
(Qiagen, Inc.), 400 ng of extracted RNA was reverse-transcribed
according to the manufacturer's instructions. The cDNA produced was
maintained at -20˚C until analysis.
Reverse transcription-quantitative PCR
(RT-qPCR)
RT-qPCR was used to evaluate the expression levels
of 12 genes, selected based on their previously reported
association with cancer and their involvement in impairing
antitumor immunity, in malignant BC and non-malignant controls
(Table II) (24-35).
Primers targeting these genes were designed using the Primer3web
tool (version 4.1.0; https://bioinfo.ut.ee/primer3/) and were evaluated
using the In-Silico PCR tool of the University of California, Santa
Cruz Genome Browser for Human Gene Assembly GRCh38 (http://genome.ucsc.edu/index.html). Relative gene
expression levels were adjusted using the internal reference
housekeeping gene, glyceraldehyde 3-phosphate dehydrogenase
(GAPDH). The primer sequences used are listed in Table III. Samples were processed in
duplicate in 96-well plates using Bio-Rad IQ SYBR Green mix and a
CFX Connect™ Real-Time PCR device (Bio-Rad Laboratories, Inc.)
according to the manufacturer's instructions. RT-qPCR was conducted
using a single initial cycle of 30 sec at 95˚C, followed by 40
amplification cycles of 15 sec at 98˚C, and 30 sec at 60˚C. The
amplified products were verified at the end of each cycle and their
purity was determined by analyzing the melting curves. Relative
expression was quantified using the comparative Cq method
(2-ΔΔCq) (36) and the
REST 2009 software version 2.0.13(37).
 | Table IISelected genes associated with
BC. |
Table II
Selected genes associated with
BC.
| Target gene | Full name | Biological function
on immune cells | Expression status
in literature |
|---|
| BTLA | B- and T-lymphocyte
attenuator | Inhibitory
receptor, expressed by T and B lymphocytes and dendritic cells,
able to suppress lymphocyte activation | Highly expressed in
B cell malignancy (24,25) and gastric cancer and gene
polymorphisms in BC (26) |
| GITR |
Glucocorticoid-induced TNF receptor
related gene | Co-stimulatory
protein for T cells, highly expressed in regulatory T cells and
lower in Th and CTLs | Overexpressed in
regulatory T cells in peripheral blood mononuclear cells of
patients with BC (27) |
| GITRL | GITR ligand | Associated with
worse relapse-free survival | Expressed in
platelets of BC patients (28) |
| CTLA4 | Cytotoxic
T-lymphocyte-associated protein 4 | Inhibitory receptor
suppresses antitumor immunity through binding to B7 molecules | Overexpression
detected in BC (8,29) |
| HVEM | Herpesvirus entry
mediator | HVEM expression
decreases lymphocyte infiltration, perforin, and IFN-γ, suggesting
its suppressor effects | Expression detected
in BC tissues (30) |
| LAG3 |
Lymphocyte-activation gene 3 | Inhibitory receptor
suppresses antitumor immunity | Upregulation of
LAG-3 observed in BC (31) |
| PD1 | Programmed cell
death protein 1 | Inhibitory receptor
suppresses antitumor immunity | Upregulated in BC
(32) |
| PDL-1 | Programmed death
ligand 1 | Inhibitory receptor
suppresses antitumor immunity | Upregulated in BC
(32) |
| PDL-2 | Programmed death
ligand 2 | Inhibitory ligand
suppresses antitumor immunity | Increased
expression in BC (33) |
| TIM3 | T-cell
immunoglobulin and mucin-domain containing 3 | Inhibitory receptor
suppresses anti-tumor immunity | Upregulated in BC
(34) |
| TIGIT | T cell
immunoreceptor with Ig and ITIM domains | Inhibitory receptor
suppresses anti-tumor immunity | Upregulated in BC
(5) |
| IDO1 | Indoleamine
2,3-dioxygenase 1 | Suppresses immune
surveillance by catalyzing tryptophan to kynurenine, resulting in
lack of essential amino acids for immune cells | Increased in BC
(35) |
 | Table IIIPCR primer sequences of target
genes. |
Table III
PCR primer sequences of target
genes.
| Gene symbol | Gene name | Accession
number | Forward primer
(5'-3') | Reverse primer
(5'-3') |
|---|
| BTLA | B- and T-lymphocyte
attenuator | NM_001085357 |
GTCATACCGCTGTTCTGCAA |
CTGCTTGCCATTTCGTCCTT |
| CTLA4 | Cytotoxic
T-lymphocyte-associated protein 4 | NM_005214 |
ATCCCTGTCTTCTGCAAAGC |
TACTCACACACAAAGCTGGC |
| GITRL | Ligand for receptor
TNFRSF18/AITR/GITR | NM_005092 |
GATCATCCTGGAAGCTGTGG |
CTCCTTAGCAGTCTCTAATTGGA |
| GITR |
Glucocorticoid-induced TNFR-related
protein | NM_004195 |
TGTCCAGCCTGAATTCCACT |
AGCCAAAACTGAATTTCCCCT |
| HVEM | Herpesvirus entry
mediator | NM_003820 |
ACCTACATTGCCCACCTCAA |
CGTTCTCTGTCCTGGAGCA |
| LAG3 |
Lymphocyte-activation gene 3 | NM_002286 |
CCAGGCCTCGATGACTGC |
TCAGGGCGGCTGAAGGAG |
| PD-L1 | Programmed death
ligand 1 | NM_014143 |
CCTGAGGAAAACCATACAGCTG |
TGGCTCCCAGAATTACCAAGT |
| PD-L2 | Programmed death
ligand 2 | NM_025239 |
ATCATCTATGGGGTCGCCTG |
GCTCTACCTCATCTGTTTCTGG |
| PD1 | Programmed cell
death protein 1 | NM_005018 |
CTCTGTGGGGCCATCTCC |
TCTGCCCTTCTCTCTGTCAC |
| TIM3 | T-cell
immunoglobulin and mucin-domain containing 3 | NM_032782.5 |
CAGACACTGGGGAGCCTC |
TTGCTCCAGAGTCCCGTAAG |
| TIGIT | T cell
immunoreceptor with Ig and ITIM domains | NM_173799 |
GGGACGTACACTGGGAGAAT |
CACCACGATGACTGCTGTG |
| IDO1 | Indoleamine
2,3-dioxygenase 1 | NM_002164 |
TTCGTGATGGAGACTGCAGT |
CAAAGTGTCCCGTTCTTGCA |
| GAPDH | Glyceraldehyde
3-phosphate dehydrogenase | NM_002046 |
TCACCAGGGCTGCTTTTAAC |
GATGATCTTGAGGCTGTTGTCA |
Multiplex ELISA
The MILLIPLEX® Human Immuno-Oncology
Checkpoint Protein premixed immunoassay (cat. no. HCKP1-11KPX17;
Merck Millipore, Inc.) was used to determine serum levels of HVEM
and CTL4. The assay kit included all of the reagents required for
the analysis. Serum samples were thawed to room temperature before
analysis and no other pretreatment was required. Fluorescent
color-coded magnetic beads (Merck Millipore, Inc.) were read after
the assay was completed using a Luminex MAGPIX™ reader, according
to the manufacturer's instructions. For validation, 25 µl of sample
was added to the polystyrene beads on a microtiter plate and
incubated overnight at 4˚C. After adding the serum samples and
standards, incubating, and washing the plate, the detection
antibody and streptavidin-phycoerythrin solution were set up to
react with the beads independently before being washed again.
Individual bead types were determined, and the fluorescence signal
of the immunoassay sandwich was quantified using a MAGPIX™ analyzer
(Luminex Corporation, Inc.), which was correlated with a set of
standards (standard curve) measured using MAGPIX (38,39).
Statistical analysis
GraphPad Prism version 8.0.1 was used for
statistical analysis of the collected data (GraphPad Software,
Inc.). Significant changes in gene expression between malignant BC
and non-malignant controls were detected using an unpaired
two-tailed t-test. In addition, a two-tailed P-value was utilized
to examine the distribution of clinicopathological features in
malignant BC patients, using one way ANOVA (two-tailed and
Kruskal-Wallis tests) for certain parameters as the comparison
between three groups were applied. The results are displayed as the
mean ± standard error of the mean (SEM). The median and
interquartile range (IQR) were calculated using Excel. Using the
easyROC web tool (ver.1.3.1; https://www.biosoft.hacettepe.edu.tr/easyROC/),
receiver operating characteristic (ROC) curves were created to
examine the sensitivity and specificity of HVEM as a possible
biomarker compared to CTLA4 using their expression values
(2-ΔΔCq) in malignant BC and non-malignant controls.
P≤0.05 was considered to indicate a statistically significant
difference. The G-power software to calculate post-hoc power was
used to verify the small sample size.
Results
Evaluation of selected gene expression
in malignant BC and non-malignant control samples
The expression levels of the selected immune
checkpoint protein genes (BTLA, GITR, GITRL, CTLA4, HVEM, LAG3,
PD1, PDL-1, PDL-2, TIM3, TIGIT, and IDO1) were evaluated. Among
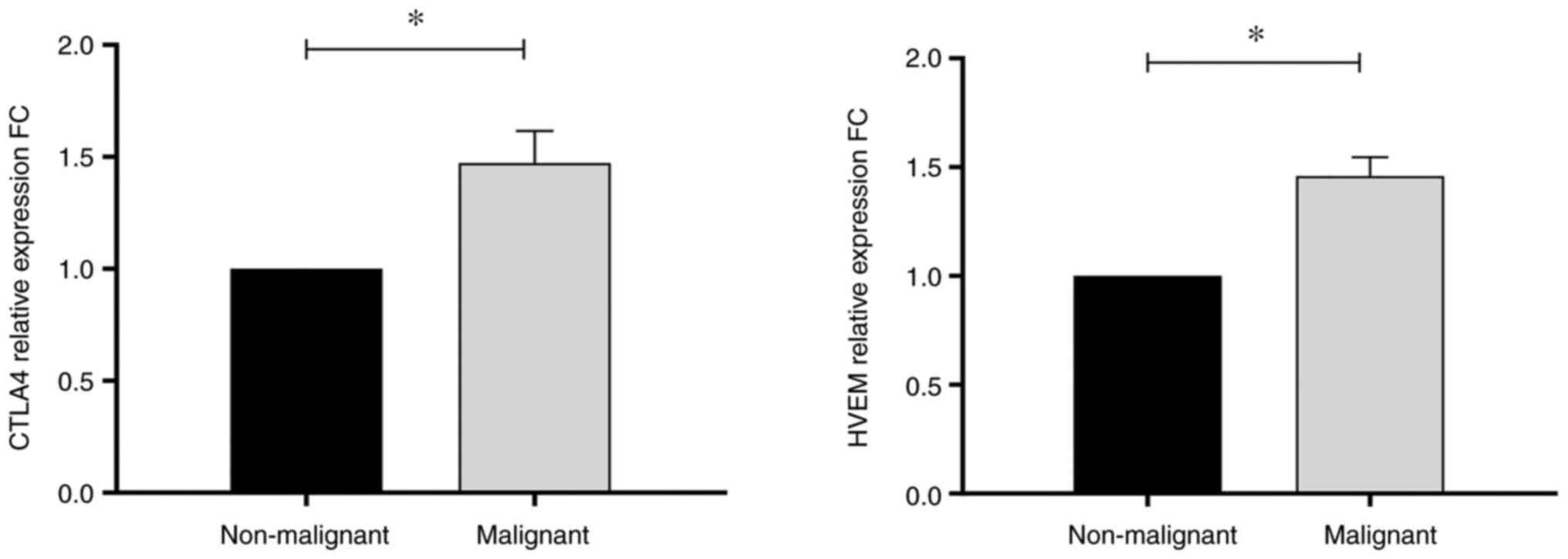
these 12 genes, CTLA4 and HVEM showed significantly different
expression levels between malignant BC and control subjects, with
fold changes (FC) of 1.47 (P=0.036) and 1.46 (P=0.012),
respectively (Table IV and
Fig. 1). ROC curves were created
using gene expression values in patients with malignant BC and
non-malignant controls to evaluate the sensitivity and specificity
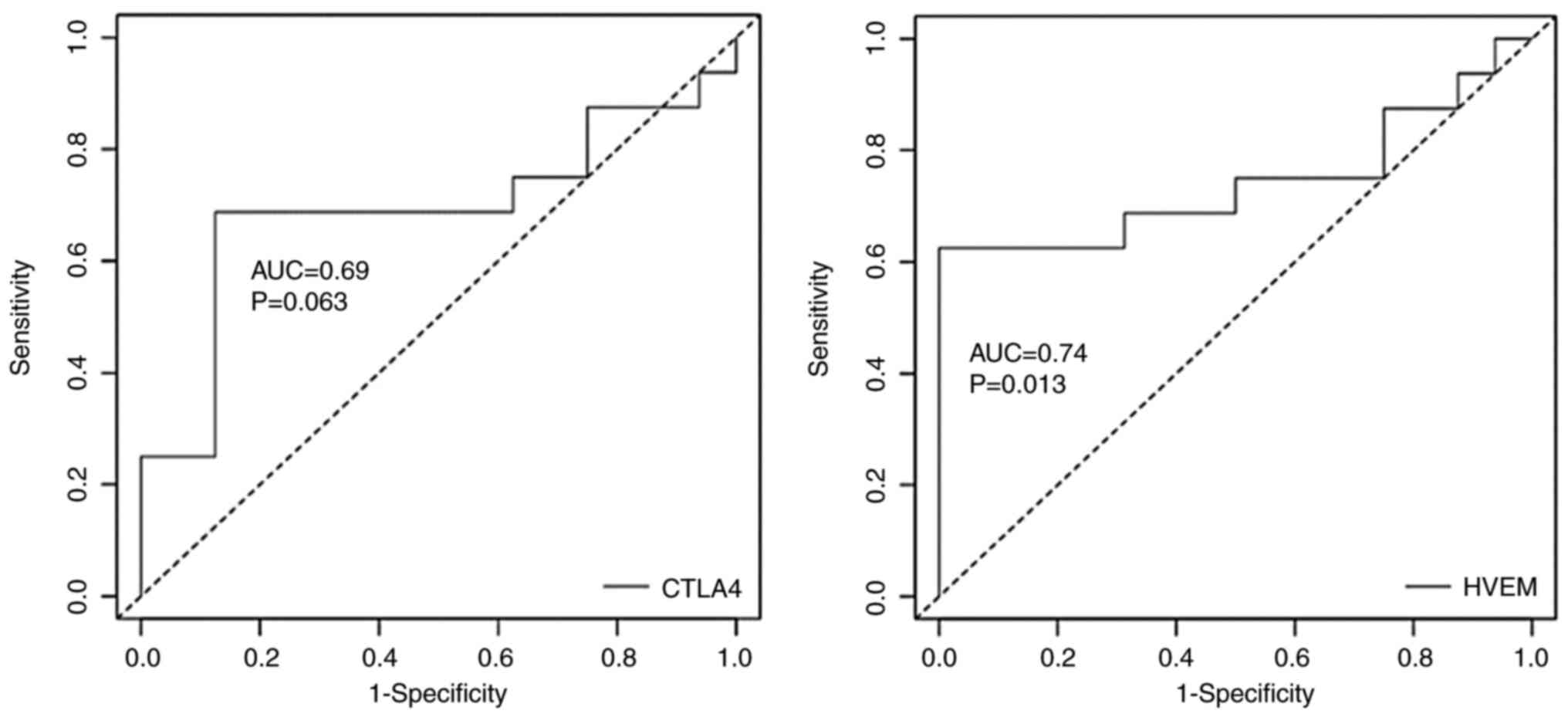
of HVEM as a potential BC biomarker. ROC curve analysis revealed
that HVEM expression allows significant differentiation between
patients with malignant BC and controls, with an area under the
curve (AUC) equal to 0.74 (P=0.013). The same was observed for
CTLA4, with an AUC equal to 0.69 (P=0.063). Therefore, at the
genetic level, HVEM may act as a potential biomarker for malignant
BC (Fig. 2 and Table SI).
 | Table IVRelative change in expression of
selected genes in patients with malignant breast cancer compared
with non-malignant control. |
Table IV
Relative change in expression of
selected genes in patients with malignant breast cancer compared
with non-malignant control.
| Gene name | Relative expression
fold change | SEM | Median | IQR | P-value |
|---|
| BTLA | 0.853 | 0.161 | 1.156 | 0.962 | 0.636 |
| GITR | 1.573 | 0.178 | 2.145 | 1.269 | 0.062 |
| GITRL | 1.861 | 0.396 | 1.450 | 3.990 | 0.147 |
| CTLA4 | 1.473 | 0.143 | 1.657 | 1.232 | 0.036a |
| HVEM | 1.459 | 0.087 | 1.543 | 0.708 | 0.012a |
| LAG3 | 1.372 | 0.164 | 1.668 | 1.353 | 0.107 |
| PD1 | 1.490 | 0.177 | 1.643 | 1.430 | 0.083 |
| PDL-1 | 1.616 | 0.259 | 1.845 | 1.098 | 0.076 |
| PDL-2 | 1.449 | 0.297 | 1.456 | 1.826 | 0.193 |
| TIM3 | 1.345 | 0.158 | 1.194 | 0.797 | 0.122 |
| TIGIT | 1.292 | 0.130 | 1.402 | 1.645 | 0.170 |
| IDO1 | 1.249 | 0.264 | 1.539 | 0.995 | 0.384 |
Association between gene expression
levels of CTLA4 and HVEM with clinicopathological characteristics
of patients with BC
The clinicopathological characteristics of the
patients under investigation, including hormone receptor phenotype,
estrogen receptor (ER), progesterone receptor (PR), human epidermal
growth factor receptor-2 (HER2), lymph node involvement, tumor
size, BC histotype, vascular invasion, and margin invasion, are
presented in Table SII.
Significant associations between CTLA4 and HVEM expression levels
and some of these clinicopathological parameters were detected
among patients with BC (P≤0.05; Fig.
3). Association with hormone receptor phenotype in Fig. 3A, revealed significantly higher
expression of CTLA4 (FC=5.98, P=0.002) and HVEM (FC=3.53, P=0.009)
in the HER2-enriched phenotype of patients with malignant BC
compared with the non-malignant control reference baseline levels;
whereas a significantly high expression of HVEM only was observed
in the triple-negative phenotype (FC=2.07, P=0.016) and no
significant differences were found in the expression of CTLA4 and
HVEM in the luminal-like phenotype. Furthermore, significantly high
expression levels of both CTLA4 and HVEM were detected in negative
ER/PR (FC=1.94, P=0.030 and FC=1.79, P=0.012, respectively;
Fig. 3B and C); while, a significantly high expression
of HVEM only was detected in HER2- (FC=2.07, P=0.016;
Fig. 3D). Significantly different
levels of CTLA4 and HVEM were also associated with negative lymph
node involvement (FC=1.94, P=0.018 and FC=1.69, P=0.028,
respectively; Fig. 3E).
Furthermore, HVEM was significantly higher in patients with grade
III tumors compared with non-malignant controls (FC=1.89, P=0.025),
in contrast to CTLA4 that exhibited no significant differences in
tumor grades (Fig. 3F). Moreover,
no significant differences were found in the expression of CTLA4
and HVEM with regard to tumor size or BC histotype (Fig. 3G and H). A significant difference of HVEM
expression levels was detected in negative vascular invasion of
patients with malignant BC and non-malignant controls (FC=1.68;
P=0.046; Fig. 3I); as well as, of
both CTLA4 and HVEM in negative margin invasion (FC=1.73, P=0.046
and FC=1.68, P=0.036, respectively; Fig. 3J).
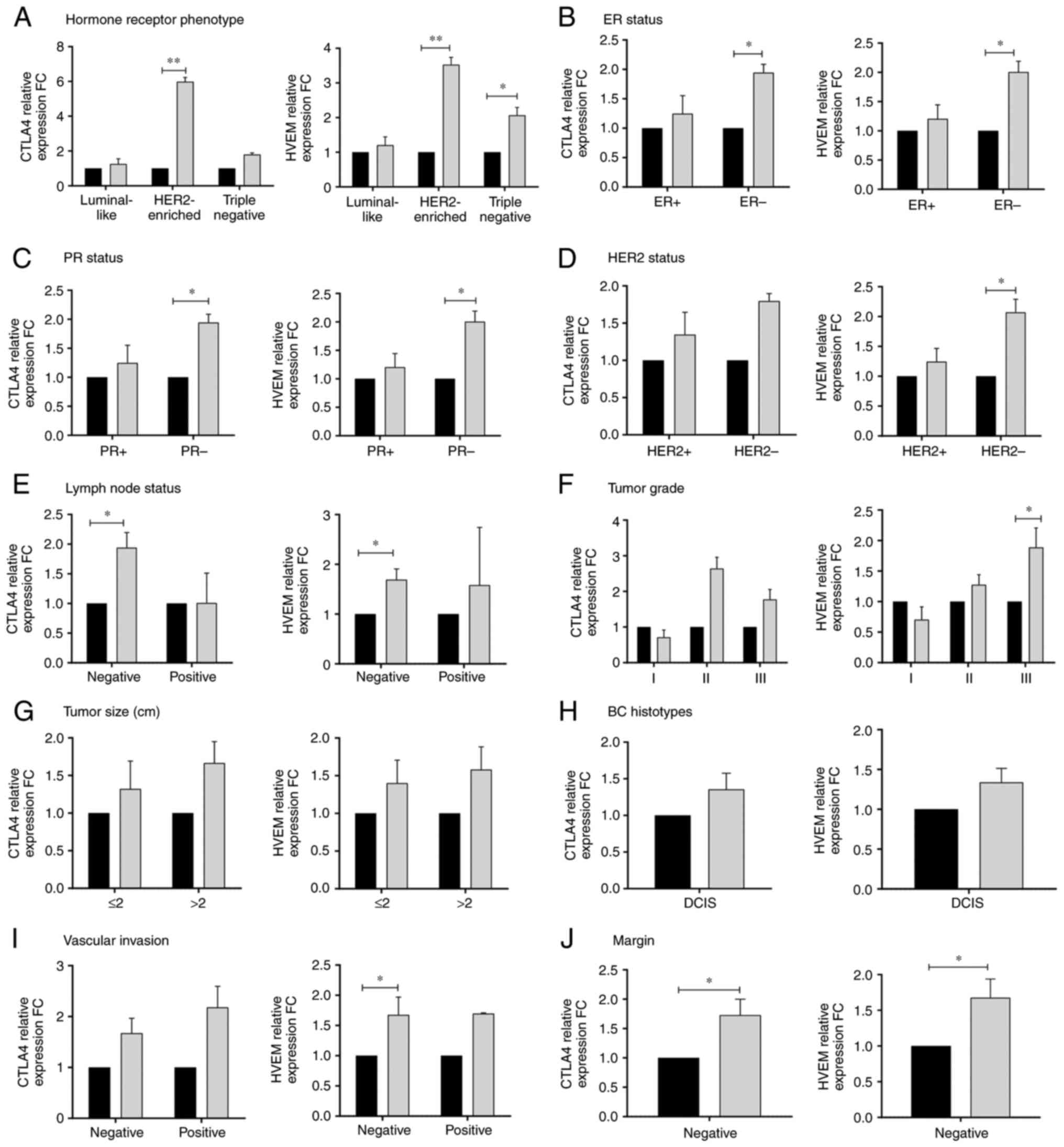 | Figure 3Relative expression fold change of
T-lymphocyte-associated antigen 4 (left) and herpesvirus entry
mediator (right) in association with clinicopathologic features
including (A) hormone receptor phenotype, (B) estrogen receptor
status, (C) progesterone status, (D) human epidermal growth factor
receptor-2 status, (E) lymph node status, (F) tumor grade, (G)
tumor size (cm), (H) BC histotypes, (I) vascular invasion, (J)
margins, in patients with malignant BC (gray bars) compared with
controls (black bars). Error bars represent the standard error of
the mean; *P≤0.05 and **P≤0.01. BC, breast
cancer; CTLA4, T-lymphocyte-associated antigen 4; HVEM, herpesvirus
entry mediator; ER, estrogen receptor; PR, progesterone receptor;
HER2, human epidermal growth factor receptor-2. |
Evaluation of CTLA4 and HVEM serum
levels in patients with malignant BC compared with non-malignant
controls associated with clinicopathological characteristics of
patients
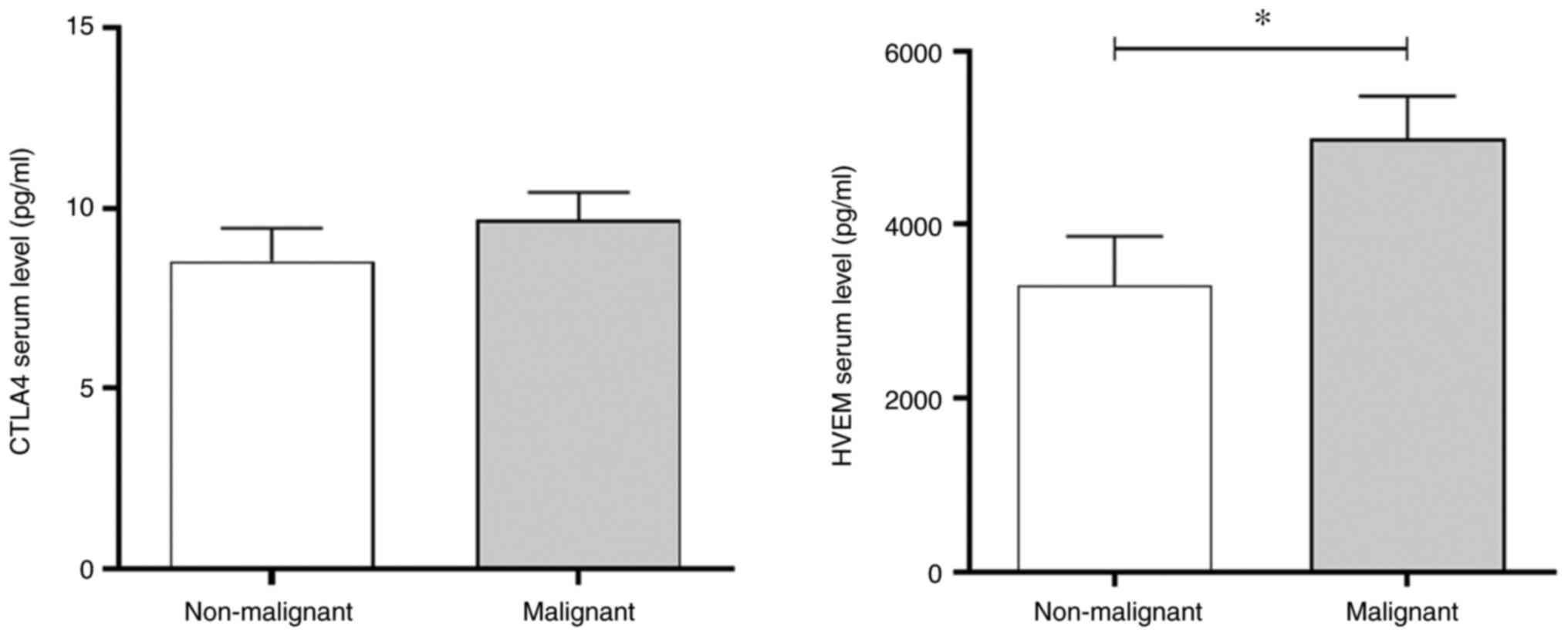
A positive association was observed between the
expression levels of CTLA4 and HVEM in patients with malignant BC.
To assess whether the expression of these genes could induce the
production of related proteins, the serum levels were measured
using a multiplex immunoassay. In healthy subjects, CTLA4 serum
levels were measured at an average of 10 pg/ml, whereas HVEM
exhibited much higher levels of ~3,000 pg/ml. The results in
Fig. 4 revealed that patients with
malignant BC exhibited a significant increase in the serum levels
of HVEM compared with non-malignant control subjects, whereas CTLA4
levels remain unaltered. In addition, the increase in HVEM serum
levels was not associated with any specific clinicopathological
characteristic in the patients with malignant BC (Table V).
 | Table VDistribution of HVEM serum levels
associated with the clinicopathologic data of the patients. |
Table V
Distribution of HVEM serum levels
associated with the clinicopathologic data of the patients.
| | HVEM serum
level | |
|---|
| Parameters | Categories | Mean, pg/ml | SEM | Median | IQR | P-value |
|---|
| Hormone receptor
phenotype | Luminal | 4470 | 664.7 | 4025 | 2074 | 0.6499 |
| | HER2-enriched | 6171 | 0.000 | 6171 | 0 | |
| | Triple
negative | 5107 | 913.7 | 4628 | 3354 | |
| ER status | ER- | 5320 | 739.0 | 5507 | 3155 | 0.0959 |
| | ER+ | 3899 | 393.5 | 3872 | 2019 | |
| PR status | PR- | 5320 | 739.0 | 5507 | 3155 | 0.0959 |
| | PR+ | 3899 | 393.5 | 3872 | 2019 | |
| HER2 status |
HER2- | 5107 | 913.7 | 4628 | 3354 | 0.3226 |
| |
HER2+ | 4183 | 443.6 | 4025 | 2074 | |
| Lymph node
involvement | Negative | 4701 | 795.8 | 4450 | 3548 | 0.9273 |
| | Positive | 4843 | 664.5 | 4843 | 1329 | |
| Size of tumor,
cm | ≤2 | 4354 | 750.3 | 3748 | 2019 | 0.0873 |
| | >2 | 6580 | 910.7 | 6171 | 3678 | |
| Tumor grade | I | 4178 | 0.000 | 4178 | 0 | 0.08998 |
| | II | 4703 | 875.3 | 4445 | 3095 | |
| | III | 4994 | 688.8 | 4690 | 2968 | |
| Histotype | DCIS | 5103 | 525.8 | 5142 | 2527 | - |
| | LCIS | No samples | | | | |
| Vascular
invasion | Negative | 4622 | 668.4 | 3963 | 2499 | 0.2513 |
| | Positive | 6171 | 0.000 | 6171 | 0 | |
| Margin | Negative | 5026 | 684.7 | 4843 | 2997 | - |
| | Positive | No samples | | | | |
Discussion
Although cancer immunotherapy is still emerging, it
is considered a promising cancer treatment (6). Clinical trials of several immune
checkpoint inhibitors have shown positive disease outcomes in
different types of cancer, such as melanoma, lung, kidney, and
bladder cancers, as well as Hodgkin's lymphoma (5). However, immune checkpoint inhibitors
are considered controversial in BC, and only a minority of patients
with BC have benefited from them (40). Combining immunotherapy with
chemotherapy results in improved overall survival but causes severe
adverse effects (41).
Chemoresistant patients with PDL-1-positive metastatic BC treated
with PD-1 blockade pembrolizumab exhibited an 18% objective
response rate (ORR) compared with the 15% ORR when using a
combination of PD-1 and CTLA4 blockade (40). Numerous trials have investigated
combinations of other targeted immune checkpoints (42). Therefore, there is still a need to
investigate additional immune checkpoint molecules that may play
potential roles in BC diagnosis and treatment. Numerous studies
have focused mainly on PD-1 and CTLA4, but there are a few other
promising immune checkpoints, such as TIGIT, GAT-3, BTLA, and HVEM
(43), which require further
investigation.
In the present study, the gene expression of 12
selected immune checkpoint molecules, namely BTLA, GITR, GITRL,
CTLA4, HVEM, LAG3, PD1, PDL-1, PDL-2, TIM3, TIGIT, and IDO1, was
measured in the blood samples of patients with BC to identify a
systematic BC biomarker. The gene expression patterns in BC were
distinct; among the 12 selected genes, only CTLA4 and HVEM were
significantly upregulated in the blood of patients with malignant
BC compared with control subjects. This result is consistent with
that of a recent study by Fang et al on 50 immune
checkpoints, which found that both CTLA4 and HVEM gene expression
levels were upregulated in BC tissues (5); however, upregulation was also detected
in TIGIT, PD1, IDO, and LAG3. The differences between the two
studies may be due to differences in sample types. In the present
study, blood samples were used to identify a systematic BC
biomarker, whereas this aforementioned study (33) investigated biomarkers within the TME
of BC tissues. Although this may indicate that blood signatures
differ from those of tumor tissues in BC, blood samples from cancer
patients differ from those of non-malignant control samples.
Therefore, the blood may serve as a potential hub for systematic BC
biomarkers.
CTLA4 has been approved for cancer treatment, but
HVEM is still under study. It has been reported that HVEM is
inducible by the TME and has a broader expression than the other
immune checkpoints, such as PD-L1. The overexpression of HVEM was
revealed to be directly associated with aggressiveness and a poorer
cancer prognosis. A previous study concluded that HVEM has a
significant oncogenic role in breast carcinogenesis and suggests
HVEM as a tumor-specific target (30). In addition, in melanoma, HVEM was
revealed to be a negative prognostic marker with potential as a
treatment target (16). Therefore,
a comparative study of CTLA4 and HVEM gene expression levels
associated with clinicopathological data was conducted. A
significant association between HVEM expression and tumor grade
(grade III) was detected, consistent with the findings of Tsang
et al indicating that HVEM gene expression in BC tissues and
blood is associated with higher tumor grades (30). In contrast to HVEM, no significant
correlation was found between CTLA4 expression and tumor size or
grade. Moreover, the findings of the present study showed no link
between HVEM gene expression in the blood of patients with BC and
tumor size. This suggests that when tumors grow, HVEM is
overexpressed in BC tissue, and this expression level is maintained
in cancer cells and is not secreted into the HVEM soluble form in
the blood. Consistent with this previous study (30), significant differences in HVEM
expression were associated with the triple-negative phenotype,
which is considered the most aggressive type of BC. It was also
reported that the presence of HER2 is associated with positive
outcomes by increasing levels of TILs (30). In the present study, it was observed
that HVEM expression, in contrast to CTLA4 expression, was
significantly associated with negative HER2 status. This suggests
that HVEM expression in the blood of patients with HER2-negative BC
may reduce infiltration, resulting in worse outcomes by
downregulating the immune response. In addition, the present study
demonstrated that HVEM expression may directly be associated with
tumor grade. HVEM was significantly higher in the blood samples of
patients with malignant BC with tumor grade III, compared with
non-malignant controls, in contrast to CTLA4 which exhibited no
significant differences between patients with malignant BC and
non-malignant controls in all tumor grades. Similar results have
been previously observed in BC tissues, as HVEM was revealed to be
associated with aggressive forms of BC with high grade (30). Effective antitumor immunity is
evidenced by the production of perforin, granzyme B, and IFN-γ
(44), which is reduced by HVEM in
hepatocellular carcinomas (13).
HVEM expression in BC tissues has been demonstrated to be
associated with shorter overall survival. Significantly higher HVEM
expression has been reported in tissue samples from early recurrent
BC than in those from later cancer recurrence (30). This suggests the involvement of HVEM
shedding in tumor tissues. Moreover, the integration of gene
expression and metabolites as serum proteins may provide unique
insights into the pathways associated with patients with malignant
BC (45). Therefore, the serum
levels of HVEM were assessed in the present study. Notably,
significant differences in serum HVEM levels were observed in
patients with malignant BC compared with control subjects, whereas
no difference in CTLA4 levels was detected. HVEM may act as an
agonist that interacts with BTLA in its circulating, soluble form.
BTLA is strongly expressed on naïve and effector antitumor cells,
in contrast to PD-1, which is strongly expressed on activated T
cells. Therefore, circulating soluble HVEM can affect naive T
cells, resulting in their arrest in an inactive state and
preventing them from infiltrating the TME. HVEM has also been
demonstrated to promote tumor cell progression. Furthermore, it has
been shown to act as an oncogene to promote the colorectal cancer
cell cycle as its silencing induces cell cycle arrest (15).
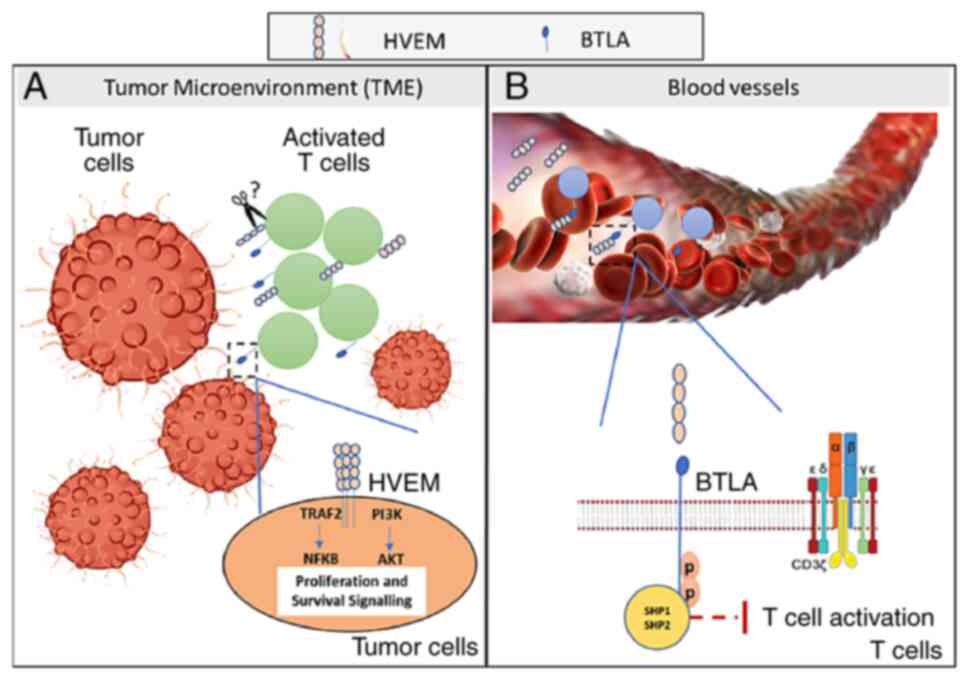
The model presented in Fig. 5 is based on the results of the
present study. The TME plays a role in immune editing, resulting in
the shedding of HVEM from immune cells. Activated T cells express
both HVEM and its ligand BTLA. Following T-cell activation, HVEM
can be shed from the surface of immune cells within the TME, which
then circulate in the blood. However, HVEM in tumor cells remains
bound to cells, providing them with survival signals. Ligation of
circulating HVEM with BTLA on T cells in the blood provides an
inhibitory signal to T cells, preventing their activation. This
suggests that the TME accelerates shedding of HVEM into its soluble
form (Fig. 5).
In conclusion, the present study demonstrated the
upregulated expression of HVEM and CTLA4 genes in the blood of
patients with malignant BC, which suggests that the upregulated
HVEM gene translates into HVEM protein, which could then be
secreted in a soluble form, as indicated by increased HVEM serum
levels, in contrast to CTLA4, which remains bound to cells. HVEM at
both genetic and protein levels may serve as a prognostic and
diagnostic BC biomarker that can be easily measured in blood
samples. It may also serve as an effective target for immunotherapy
in patients with the most aggressive phenotype and histological
high-grade BC.
Although the small sample size of the present study
is a limitation, HVEM expression in the 16 samples for each group
was significant, where the power of the sample was shown to be 0.81
(data not shown). The most obvious limitation is that the study
used a small cohort of patients with BC without predicting the
power of the samples. This is because patient samples had to be
obtained before treatment and at short notice, which, together with
the excessive cost of the experiment, made it difficult to expand
the number of patients in this study. Although the calculated
post-hoc power of the HVEM expression was strong (0.81), it can
only be associated with this experimental design and should
generally be considered as suggestive power, which may also be
biased as it is entirely determined by the P-value. Therefore, the
present study should be followed-up with future studies, taking
care to use larger samples to obtain statistically significant
results. Further studies are underway to elucidate the underlying
HVEM mechanism and to determine the diagnostic and prognostic value
of HVEM in BC. This will involve comparing HVEM expression in BC
tissues and the blood of patients with BC, which could provide
insights into the poor prognosis. Furthermore, conducting
phenotypic analysis of blood cells using flow cytometry may provide
insights into the use of markers and their mechanisms in BC for the
development of more effective therapies.
Supplementary Material
ROC curve analysis of CTLA4 and HVEM
gene expression.
Expression level of CTLA4 and HVEM in
patients with malignant BC compared with non-malignant controls in
association with the clinicopathological characteristics of
patients.
Acknowledgements
Not applicable.
Funding
Funding: The present study was funded by the Deputyship for
Research and Innovation, Ministry of Education, Saudi Arabia
(project no. IFPRC-042-290-2020).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding authors.
Authors' contributions
MH, AA, KAS, and FB designed and coordinated the
experiments. KAS and AA obtained ethical approval, patient consent,
and blood sample collection. MH performed the molecular experiments
and JA, KZ and PNP performed the Multiplex immunoassay. MH, PNP and
FB analyzed the data. AA, JA, PNP, and KAS contributed to
laboratory facilities and project funding. FB, MH and KAS drafted
the manuscript. KAS, AA, PNP, and KZ edited the manuscript. AA, MH,
FB, and KAS confirm the authenticity of all the raw data. All
authors have read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved (approval no.
HA-02-J-008) by the Biomedical Ethics Research Committee of King
Abdulaziz University Hospital (KAUH; Jeddah, Saudi Arabia). All
patients signed a consent form to participate in this study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing of
interests.
References
|
1
|
Lei S, Zheng R, Zhang S, Wang S, Chen R,
Sun K, Zeng H, Zhou J and Wei W: Global patterns of breast cancer
incidence and mortality: A population-based cancer registry data
analysis from 2000 to 2020. Cancer Commun (Lond). 41:1183–1194.
2021.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Sharma R: Breast cancer incidence,
mortality and mortality-to-incidence ratio (MIR) are associated
with human development, 1990-2016: Evidence from Global Burden of
Disease Study 2016. Breast Cancer. 26:428–445. 2019.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Magro G, Salvatorelli L, Puzzo L, Piombino
E, Bartoloni G, Broggi G and Vecchio GM: Practical approach to
diagnosis of bland-looking spindle cell lesions of the breast.
Pathologica. 111:344–360. 2019.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Broggi G, Filetti V, Ieni A, Rapisarda V,
Ledda C, Vitale E, Varricchio S, Russo D, Lombardo C, Tuccari G, et
al: MacroH2A1 immunoexpression in breast cancer. Front Oncol.
10(1519)2020.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Fang J, Chen F, Liu D, Gu F, Chen Z and
Wang Y: Prognostic value of immune checkpoint molecules in breast
cancer. Biosci Rep. 40(BSR20201054)2020.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Yi H, Li Y, Tan Y, Fu S, Tang F and Deng
X: Immune checkpoint inhibition for Triple-negative breast cancer:
Current landscape and future perspectives. Front Oncol.
11(648139)2021.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Planes-Laine G, Rochigneux P, Bertucci F,
Chrétien AS, Viens P, Sabatier R and Gonçalves A: PD-1/PD-L1
targeting in breast cancer: The first clinical evidences are
emerging. A literature review. Cancers (Basel).
11(1033)2019.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Kern R and Panis C: CTLA-4 expression and
its clinical significance in breast cancer. Arch Immunol Ther Exp
(Warsz). 69(16)2021.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Watanabe N, Gavrieli M, Sedy JR, Yang J,
Fallarino F, Loftin SK, Hurchla MA, Zimmerman N, Sim J, Zang X, et
al: BTLA is a lymphocyte inhibitory receptor with similarities to
CTLA-4 and PD-1. Nat Immunol. 4:670–679. 2003.PubMed/NCBI View
Article : Google Scholar
|
|
10
|
Dill EA, Dillon PM, Bullock TN and Mills
AM: IDO expression in breast cancer: An assessment of 281 primary
and metastatic cases with comparison to PD-L1. Mod Pathol.
31:1513–1522. 2018.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Miselis NR, Linn D, Restaino C, Baral T,
Xia J, Ueda R, Sawant A, Baker J, Raghunathan G, Wang X, et al:
Abstract 577: Antagonism of the co-inhibitory receptor BTLA
enhances efficacy of anti-PD-1 treatment in murine syngeneic tumor
models. Cancer Res. 77(577)2017.
|
|
12
|
Inoue T, Sho M, Yasuda S, Nishiwada S,
Nakamura S, Ueda T, Nishigori N, Kawasaki K, Obara S, Nakamoto T,
et al: HVEM expression contributes to tumor progression and
prognosis in human colorectal cancer. Anticancer Res. 35:1361–1367.
2015.PubMed/NCBI
|
|
13
|
Hokuto D, Sho M, Yamato I, Yasuda S, Obara
S, Nomi T and Nakajima Y: Clinical impact of herpesvirus entry
mediator expression in human hepatocellular carcinoma. Eur J
Cancer. 51:157–165. 2015.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Tang M, Cao X, Li Y, Li GQ, He QH, Li SJ,
Chen J, Xu GL and Zhang KQ: High expression of herpes virus entry
mediator is associated with poor prognosis in clear cell renal cell
carcinoma. Am J Cancer Res. 9:975–987. 2019.PubMed/NCBI
|
|
15
|
Migita K, Sho M, Shimada K, Yasuda S,
Yamato I, Takayama T, Matsumoto S, Wakatsuki K, Hotta K, Tanaka T,
et al: Significant involvement of herpesvirus entry mediator in
human esophageal squamous cell carcinoma. Cancer. 120:808–817.
2014.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Malissen N, Macagno N, Granjeaud S,
Granier C, Moutardier V, Gaudy-Marqueste C, Habel N, Mandavit M,
Guillot B, Pasero C, et al: HVEM has a broader expression than
PD-L1 and constitutes a negative prognostic marker and potential
treatment target for melanoma. Oncoimmunology.
8(e1665976)2019.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Fourcade J, Sun Z, Pagliano O, Guillaume
P, Luescher IF, Sander C, Kirkwood JM, Olive D, Kuchroo V and
Zarour HM: CD8(+) T cells specific for tumor antigens can be
rendered dysfunctional by the tumor microenvironment through
upregulation of the inhibitory receptors BTLA and PD-1. Cancer Res.
72:887–896. 2012.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Sideras K, Biermann K, Yap K, Mancham S,
Boor PPC, Hansen BE, Stoop HJA, Peppelenbosch MP, van Eijck CH,
Sleijfer S, et al: Tumor cell expression of immune inhibitory
molecules and tumor-infiltrating lymphocyte count predict
cancer-specific survival in pancreatic and ampullary cancer. Int J
Cancer. 141:572–582. 2017.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Zhu YD and Lu MY: Increased expression of
TNFRSF14 indicates good prognosis and inhibits bladder cancer
proliferation by promoting apoptosis. Mol Med Rep. 18:3403–3410.
2018.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Haymaker CL, Wu RC, Ritthipichai K,
Bernatchez C, Forget MA, Chen JQ, Liu H, Wang E, Marincola F, Hwu P
and Radvanyi LG: BTLA marks a less-differentiated
tumor-infiltrating lymphocyte subset in melanoma with enhanced
survival properties. Oncoimmunology. 4(e1014246)2015.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Kim S, Park S, Cho MS, Lim W, Moon BI and
Sung SH: Strong correlation of indoleamine 2,3-dioxygenase 1
expression with basal-like phenotype and increased lymphocytic
infiltration in triple-negative breast cancer. J Cancer. 8:124–130.
2017.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Schaer DA, Murphy JT and Wolchok JD:
Modulation of GITR for cancer immunotherapy. Curr Opin Immunol.
24:217–224. 2012.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Choudhry H, Hassan MA, Al-Malki AL and
Al-Sakkaf KA: Suppression of circulating AP001429.1 long non-coding
RNA in obese patients with breast cancer. Oncol Lett.
22(508)2021.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Lan X, Li S, Gao H, Nanding A, Quan L,
Yang C, Ding S and Xue Y: Increased BTLA and HVEM in gastric cancer
are associated with progression and poor prognosis. Onco Targets
Ther. 10:919–926. 2017.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Pasero C and Olive D: Interfering with
coinhibitory molecules: BTLA/HVEM as new targets to enhance
anti-tumor immunity. Immunol Lett. 151:71–75. 2013.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Fu Z and Li D, Jiang W, Wang L, Zhang J,
Xu F, Pang D and Li D: Association of BTLA gene polymorphisms with
the risk of malignant breast cancer in Chinese women of
Heilongjiang Province. Breast Cancer Res Treat. 120:195–202.
2010.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Khalife E, Khodadadi A, Talaeizadeh A,
Rahimian L, Nemati M and Jafarzadeh A: Overexpression of regulatory
T Cell-related markers (FOXP3, CTLA-4 and GITR) by peripheral blood
mononuclear cells from patients with breast cancer. Asian Pacific J
Cancer Prev. 19:3019–3025. 2018.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Zhou Y, Heitmann JS, Clar KL, Kropp KN,
Hinterleitner M, Engler T, Koch A, Hartkopf AD, Zender L, Salih HR,
et al: Platelet-expressed immune checkpoint regulator GITRL in
breast cancer. Cancer Immunol Immunother. 70:2483–2496.
2021.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Jaberipour M, Habibagahi M, Hosseini A,
Habibabad SR, Talei A and Ghaderi A: Increased CTLA-4 and FOXP3
transcripts in peripheral blood mononuclear cells of patients with
breast cancer. Pathol Oncol Res. 16:547–551. 2010.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Tsang JYS, Chan KW, Ni YB, Hlaing T, Hu J,
Chan SK, Cheung SY and Tse GM: Expression and clinical significance
of herpes virus entry mediator (HVEM) in breast cancer. Ann Surg
Oncol. 24:4042–4050. 2017.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Wu S, Shi X, Wang J, Wang X, Liu Y, Luo Y,
Mao F and Zeng X: Triple-negative breast cancer: Intact mismatch
repair and partial Co-expression of PD-L1 and LAG-3. Front Immunol.
12(561793)2021.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Kitano A, Ono M, Yoshida M, Noguchi E,
Shimomura A, Shimoi T, Kodaira M, Yunokawa M, Yonemori K, Shimizu
C, et al: Tumour-infiltrating lymphocytes are correlated with
higher expression levels of PD-1 and PD-L1 in early breast cancer.
ESMO Open. 2(e000150)2017.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Baptista MZ, Sarian LO, Derchain SF, Pinto
GA and Vassallo J: Prognostic significance of PD-L1 and PD-L2 in
breast cancer. Hum Pathol. 47:78–84. 2016.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Yasinska IM, Sakhnevych SS, Pavlova L, Teo
Hansen Selnø A, Teuscher Abeleira AM, Benlaouer O, Gonçalves Silva
I, Mosimann M, Varani L, Bardelli M, et al: The Tim-3-Galectin-9
pathway and its regulatory mechanisms in human breast cancer. Front
Immunol. 10(1594)2019.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Carvajal-Hausdorf DE, Mani N, Velcheti V,
Schalper KA and Rimm DL: Objective measurement and clinical
significance of IDO1 protein in hormone receptor-positive breast
cancer. J Immunother Cancer. 5(81)2017.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Pfaffl MW, Horgan GW and Dempfle L:
Relative expression software tool (REST) for group-wise comparison
and statistical analysis of relative expression results in
real-time PCR. Nucleic Acids Res. 30(e36)2002.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Hamed MH, Pushparaj PN, Rehman S, Al-Karim
S, Bazarah S and Qadri I: Deciphering the significance of plasma
chemokines as prognostic biomarkers in pegylated
IFN-Α-2a/Ribavirin-treated chronic Hepatitis C Genotype 4 patients.
Infect Disord Drug Targets. 22:58–62. 2022.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Huwaikem MAH, Kalamegam G, Alrefaei G,
Ahmed F, Kadam R, Qadah T, Sait KHW and Pushparaj PN: Human
Wharton's Jelly stem cell secretions inhibit human leukemic cell
line K562 in vitro by inducing cell cycle arrest and apoptosis.
Front Cell Dev Biol. 9(614988)2021.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Ledys F, Kalfeist L, Galland L, Limagne E
and Ladoire S: Therapeutic associations comprising Anti-PD-1/PD-L1
in breast cancer: Clinical challenges and perspectives. Cancers
(Basel). 13(5999)2021.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Ji Q, Ding J, Hao M, Luo N, Huang J and
Zhang W: Immune checkpoint inhibitors combined with chemotherapy
compared with chemotherapy alone for triple-negative breast cancer:
A systematic review and meta-analysis. Front Oncol.
11(795650)2021.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Miglietta F, Cona MS, Dieci MV, Guarneri V
and La Verde N: An overview of immune checkpoint inhibitors in
breast cancer. Explor Target Antitumor Ther. 1:452–472.
2020.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Manni M and Läubli H: Targeting
glyco-immune checkpoints for cancer therapy. Expert Opin Biol Ther.
21:1063–1071. 2021.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Basingab FS, Ahmadi M and Morgan DJ:
IFNγ-dependent interactions between ICAM-1 and LFA-1 counteract
prostaglandin E2-mediated inhibition of antitumor CTL responses.
Cancer Immunol Res. 4:400–411. 2016.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Hassan MA, Al-Sakkaf K, Shait Mohammed MR,
Dallol A, Al-Maghrabi J, Aldahlawi A, Ashoor S, Maamra M, Ragoussis
J, Wu W, et al: Integration of transcriptome and metabolome
provides unique insights to pathways associated with obese breast
cancer patients. Front Oncol. 10(804)2020.PubMed/NCBI View Article : Google Scholar
|