Introduction
Fas-associated death domain (FADD) is a ubiquitous
adaptor protein (1). The human
FADD gene consists of two exons and one intron and has been
mapped to chromosome 11q13.3, a region strongly associated with
breast invasive carcinoma (BRCA), lung cancer and esophageal
carcinoma (ESCA) (2). As an
important receptor protein in the tumor necrosis factor receptor
family-mediated apoptosis pathway, FADD modulates its binding to
death receptors of the tumor necrosis factor receptor family to
transmit apoptosis initiation signals (3-5).
In addition, FADD is involved in the regulation of cell
proliferation, gene expression and immunity (1,5-8).
As a universal adaptor molecule, abnormal expression of FADD
protein is associated with the occurrence and development of tumors
in both mature and embryonic tissues.
In the last few years, major breakthroughs have been
made in the treatment of cancer, including immunotherapy, which has
achieved remarkable results in clinical practice (9,10). As
the most important defense system of the human organism, the immune
system does not only eliminate pathogenic microorganisms, but also
destroys abnormal cancer cells, thus actively inhibits tumor growth
(11). However, the composition of
tumors and their related tumor microenvironment (TME) is relatively
complex, requiring precise immune responses (11,12).
Therefore, cancer immunotherapy can only achieve favorable results
in specific cancer types and patients (13,14).
Research to find potential targets for cancer immunotherapy and
predict its efficacy is critical to achieve specificity in cancer
treatment. Previous studies have demonstrated that FADD is involved
in and regulates signaling complexes, including necrosomes,
endosomes and inflammasomes (1,15,16).
Thus, FADD plays an indispensable role in innate immunity,
inflammation and cancer development (1). However, the role of FADD in
tumorigenesis is not fully understood, and whether it can be used
as a prognostic biomarker as well as its potential value for
clinical treatment require to be further explored. In the present
study, the differential expression, gene alteration, prognostic
value, tumor progression and promoter methylation level of FADD in
pan-cancer extent were evaluated based on The Cancer Genome Atlas
(TCGA) dataset. Subsequently, the expression level of FADD in
related cell lines and databases as well as its relationship with
immune cell infiltration, immune checkpoint, tumor mutation burden
(TMB) and microsatellite instability (MSI) were analyzed.
Materials and methods
Cell culture
Near diploid and normal human mammary epithelial
cells (MCF 10A), triple-negative breast cancer cells (MDA-MB-231)
and breast cancer cell (MCF-7) were purchased from Procell Life
Science & Technology Co., Ltd. (https://www.procell.com.cn/). Normal human colon
mucosal epithelial cell (NCM460) and human colon carcinoma cell
line (SW620) were purchased from MINGZHOUBIO Co., Ltd. (https://www.mingzhoubio.com/). All cells were cultured
with Dulbecco's modified eagle medium (Biological Industries)
containing 10% fetal bovine serum (Biological Industries) and
incubated at 37˚C in a thermostatic cell incubator containing 5%
CO2. Roswell Park Memorial Institute 1640 (Biological
Industries) was used to maintain cell growth. All cells retained
their original morphology throughout the study period.
Reverse-transcription quantitative PCR
(RT-qPCR)
Total RNA from MCF 10A, MDA-MB-231, MCF-7, NCM460
and SW620 was extracted using TRIzol reagent (Mei5 Biotechnology,
Co., Ltd., https://mei5bio.com/) according to the
manufacturer's instructions and converted into cDNA using M5 Sprint
qPCR RT kit with gDNA remover (Mei5 Biotechnology, Co., Ltd.)
according to the manufacturer's instructions. The extraction and
reverse transcription were performed in an enzyme-free environment.
AceQ qPCR SYBR Green Master Mix (Vazyme Biotech Co., Ltd.,
https://www.vazyme.com/) was used to quantify the
relative expression of FADD (Sangon Biotech Co., Ltd.) in mRNA. The
primers used were as follows: GAPDH forward,
5'-CAGGAGGCATTGCTGATGAT-3' and reverse, 5'-GAAGGCTGGGGCTCATTT-3';
and FADD forward, 5'-GACCGAGCTCAAGTTCCTATG-3' and reverse,
5'-GAGCATGGAGAAGAGGTCTAG-3'. The thermocycling conditions are
provided in Table I.
 | Table IThermocycling conditions of reverse
transcription-quantitative PCR. |
Table I
Thermocycling conditions of reverse
transcription-quantitative PCR.
| Stage 1 |
Pre-denaturation | Repeats: 1 | 95˚C | 5 min |
| Stage 2 | Amplification | Repeats: 40 | 95˚C | 10 sec |
| | | | 60˚C | 30 sec |
| Stage 3 | Melting Curve | Repeats: 1 | 95˚C | 15 sec |
| | | | 60˚C | 60 sec |
| | | | 95˚C | 15 sec |
Data acquisition and differential
expression of FADD in cancer tissues
Transcriptome data and patient clinical data of 33
human cancers were obtained from TCGA database on the UCSC Xena
website (xena.ucsc.edu). All gene names in the
expression matrix were transformed from Ensembl ID to the Symbol
format. In total, 20 datasets (GSE13057, GSE9750, GSE26566,
GSE44076, GSE23400, GSE30784, GSE167093, GSE15641, GSE25097,
GSE40791, GSE19188, GSE51024, GSE26712, GSE71729, GSE10927,
GSE70770, GSE26253, GSE33630 and GSE63678) containing 2,778 tumor
tissues and 1,821 non-tumor tissues were included from the Gene
Expression Omnibus (GEO) repository (17-36).
The R packages ‘plyr’ (version, 1.8.8; http://cran.ma.ic.ac.uk/web/packages/plyr/plyr.pdf),
‘reshap2’ (version, 1.4.4 http://cran.ma.ic.ac.uk/web/packages/reshape/reshape.pdf)
and ‘ggpubr’ (version, 0.6.0; http://cran.ma.ic.ac.uk/web/packages/ggpubr/ggpubr.pdf)
were used to create a box plot demonstrating FADD expression
differences. Furthermore, the immunohistochemical images of FADD
protein in different cancer tissues and normal tissues were
obtained from the Human Protein Atlas (HPA; https://www.proteinatlas.org).
FADD alteration and promoter
methylation in cancer
FADD alteration data were collected from the
cBioPortal website (https://www.cbioportal.org/) for a total of 10,953
patients with cancer, including the corresponding 10,967 samples of
mutation and CNA data, for analysis (37). Mutation, structural variant,
amplification, deep deletion and multiple alterations of FADD were
analyzed in different cancers. The University of Alabama at
Birmingham Cancer (UALCAN) data analysis portal (http://ualcan.path.uab.edu) was used to explore
differences in promoter methylation levels of FADD between tumor
and non-tumor samples in TCGA (38). P<0.05 was considered to indicate
a statistically significant difference.
Analysis of survival rate and clinical
association of patients with different expressions of FADD
The Kaplan-Meier plotter website (https://kmplot.com) was used to perform overall
survival (OS) and relapse-free survival (RFS) prognostic analysis.
According to the expression level of FADD, samples were divided
into high- and low-expression groups (39). Kaplan Meier analysis was used to
compare the differences between OS and RFS between high- and
low-expression groups, and values with P<0.05 were considered
statistically significant. The Cox proportional hazards model
method was used to compare FADD as a continuous variable with
survival status and survival time and to calculate the hazard ratio
(HR) value and P-value. Values with P<0.05 were considered to
indicate a statistically significant difference. A HR value >1
indicated that the expression of FADD was a high-risk factor in the
tumor, whereas a value <1 indicated that the expression of FADD
was considered. Based on these results, a forest map was
created.
Analysis of FADD expression, TME and
immune cell infiltration
TME encompasses the internal and external
environment in which tumors and tumor cells proliferate, develop
and metastasize (40). Changes in
TME contribute to the generation of tumor resistance (including
immune checkpoint inhibitors resistance) and the metabolic changes
in physiological processes (41).
The immune infiltration in TME is highly associated with the
occurrence and development of tumors and the clinical treatment
outcome of patients (42,43). The Spearman correlation test between
FADD expression and TME score was performed using the R packages
‘ggplot2’ (version, 3.4.3; https://cran.r-project.org/web/packages/ggplot2/index.html),
‘ggpubr’ and ‘ggExtra’, and the results satisfying the condition
(P<0.05, correlation coefficient >0.2) were plotted for
visualization. The relative content of immune cells in each sample
was determined using the Sangerbox website (http://vip.sangerbox.com/home.html). The relative
expression of FADD in the samples and the infiltration of immune
cells [B cells, CD4 cells, CD8 cells, neutrophils, macrophages and
dendritic cells (DCs)] were analyzed using TIMER2.0 tool
(http://timer.cistrome.org/) (44,45).
Correlation of FADD expression with
TMB and MSI
Although TMB and MSI (46) are not perfect indicators of cancer
immunotherapy response, they are still important biomarkers for
predicting the effect of immunotherapy (47,48).
The R package ‘fmsb’ (version, 0.7.5; http://cran.ma.ic.ac.uk/web/packages/fmsb/fmsb.pdf)
was used to analyze the correlation of the FADD expression with TMB
and MSI in all cancer samples (49). P<0.05 was considered to indicate
a statistically significant difference. These correlation analysis
results were illustrated in a radar map. A correlation coefficient
>0 indicated that FADD expression was positively correlated with
TMB and MSI, whereas a correlation coefficient <0 indicated that
FADD expression was negatively correlated with TMB and MSI.
Gene set enrichment analysis
(GSEA)
The GSEA method is useful for the discovery of genes
with no significant difference in expression but key biological
function (49). Using GSEA website
(http://www.gsea-msigdb.org/gsea), data
sets were obtained from the Kyoto Encyclopedia of Genes and Genomes
(KEGG) (https://www.kegg.jp/) and Gene Ontology
(GO) (http://www.geneology.org) databases. The
R packages ‘limma’ (version, 3.56.2; https://bioconductor.org/packages/release/bioc/html/limma.html),
‘org.Hs.eg.db’ (version, 3.17.0; https://bioconductor.org/packages/release/data/annotation/html/org.Hs.eg.db.html),
‘enrichmentplot’ and ‘clusterProfiler’ were used to perform KEGG
pathway analysis and GO function annotation analysis on genes
differentially expressed between high- and low-expression groups of
FADD (49,50). With P<0.05 as the threshold for
statistical significance, the top five most significant pathways
and biological processes were displayed.
Statistical analysis
FADD expression levels in all cancer tissues and
adjacent tissue samples were determined using The R Project for
Statistical Computing 4.2.1 (R Foundation and R Core Team,
https://www.r-project.org/). The
Wilcoxon rank sum test was used to calculate the difference in FADD
expression between tumor and non-tumor tissuesand the receiver
operating characteristic (ROC) curve was drawn (Sangerbox website,
http://vip.sangerbox.com/home.html).
A hypothesis test probability (P<0.05) was considered
statistically significant. With GAPDH as the internal reference
gene, the 2-ΔΔCq method was used to calculate the
expression of FADD. Unpaired t-test was used to calculate the
significance of the relative expression of FADD between normal
breast cells and breast cancer cells. And an unpaired t test with
Welch's correction was used to calculate the significance of the
relative expression of FADD between colon mucosal epithelial cell
and colon carcinoma cell line. Statistical Calculation and Bar
Chart Drawing by GraphPad Prism 8.3.0 (Dotmatics).
Results
Expression of FADD in different
cancers
The analysis results of the expression of FADD mRNA
in tumor and non-tumor tissues collected from the TCGA database
revealed that FADD was significantly differentially expressed in 19
cancer types. FADD was relatively highly expressed in bladder
urothelial carcinoma (BLCA), BRCA, cervical squamous cell carcinoma
and endocervical adenocarcinoma (CESC), cholangiocarcinoma (CHOL),
colon adenocarcinoma (COAD), ESCA, glioblastoma multiforme (GBM),
head and neck squamous cell carcinoma (HNSC), kidney renal clear
cell carcinoma (KIRC), kidney renal papillary cell carcinoma
(KIRP), liver hepatocellular carcinoma (LIHC), lung adenocarcinoma
(LUAD), lung squamous cell carcinoma (LUSC), prostate
adenocarcinoma (PRAD), rectum adenocarcinoma (READ), stomach
adenocarcinoma STAD, and thyroid carcinoma (THCA) samples, but
showed relatively low expression in KICH, and pheochromocytoma and
paraganglioma (PCPG) samples (Fig.
1A and B). The results of mRNA
expression analysis of FADD in tumor and non-tumor samples
collected from the GEO database revealed that FADD was relatively
highly expressed in BRCA, CESC, CHOL, COAD, ESCA, KIRC, KIRP, LIHC,
LUAD, PAAD and PRAD samples, but showed relatively lower expression
in HNSC, mesothelioma (MESO), ovarian serous cystadenocarcinoma
(OV), THCA and uterine corpus endometrial carcinoma (UCEC) samples
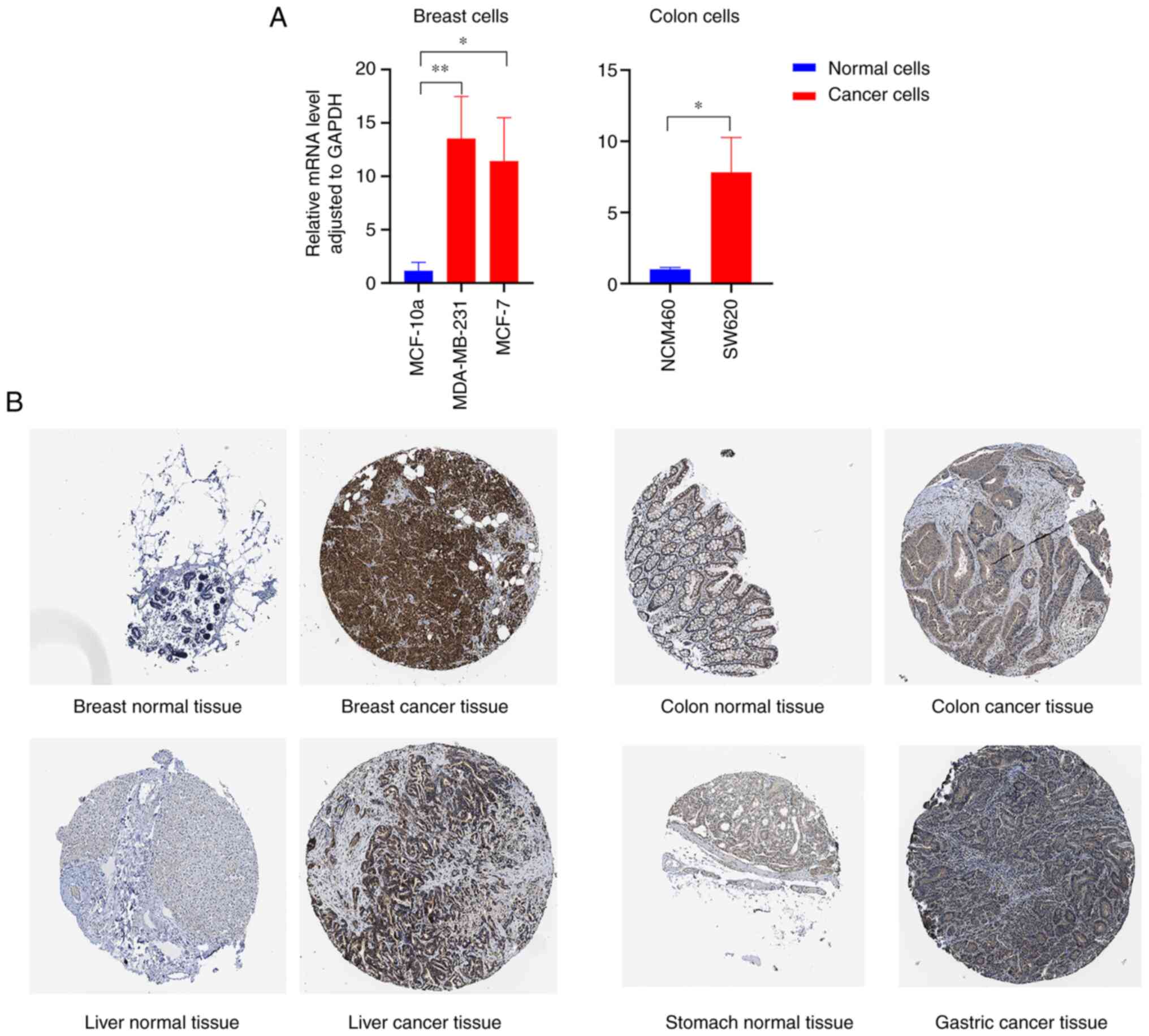
(Fig. 1C and D). RT-qPCR results revealed that FADD mRNA
was highly expressed in breast cancer cells and colon cancer cells
(Fig. 2A). The results of
immunohistochemical analysis of tumor and non-tumor samples in the
HPA database showed that FADD protein was relatively highly
expressed in breast, colon, liver and gastric cancer tissues
(Fig. 2B). Relative expression of
FADD in cancer cells is demonstrated in Table II.
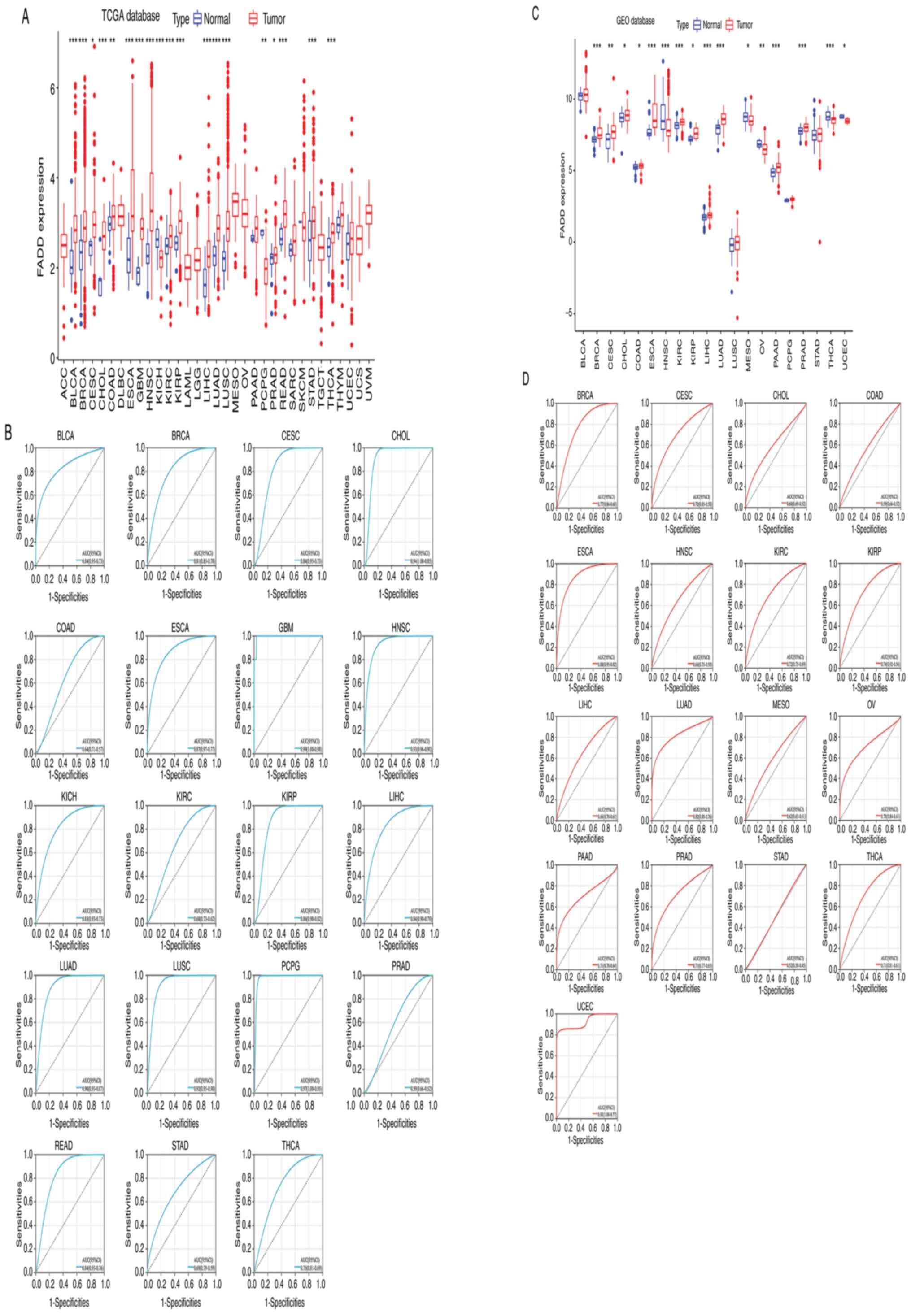 | Figure 1Expression in pan-cancer. (A) The
differential expression analysis of FADD in adjacent tissues and
cancer tissues from the TCGA database and (B) the ROC curve of
cancer species with statistical differences. (C) The differential
expression analysis of FADD in adjacent tissues and cancer tissues
from the GEO database and (D) the ROC curve of cancer species with
statistical differences. *P<0.05,
**P<0.01 and ***P<0.001. FADD,
fas-associated death domain; TCGA, The Cancer Genome Atlas; ROC,
receiver operating characteristic; GEO, Gene Expression Omnibus;
BLCA, bladder urothelial carcinoma; BRCA, breast invasive
carcinoma; CESC, cervical squamous cell carcinoma and endocervical
adenocarcinoma; CHOL, cholangiocarcinoma; COAD, colon
adenocarcinoma; ESCA, esophageal carcinoma; GBM, glioblastoma
multiforme; HNSC, head and neck squamous cell carcinoma; KICH,
kidney chromophobe; KIRC, kidney renal clear cell carcinoma; KIRP,
kidney renal papillary cell carcinoma; LIHC, liver hepatocellular
carcinoma; LUAD, lung adenocarcinoma; LUSC, lung squamous cell
carcinoma; PCPG, pheochromocytoma and paraganglioma; PRAD, prostate
adenocarcinoma; READ, rectum adenocarcinoma; STAD, stomach
adenocarcinoma; THCA, thyroid carcinoma; MESO, mesothelioma; OV,
ovarian serous cystadenocarcinoma; PAAD, pancreatic adenocarcinoma;
UCEC, uterine corpus endometrial carcinoma. |
 | Table IIRelative expression of fas-associated
death domain in cancer cells. |
Table II
Relative expression of fas-associated
death domain in cancer cells.
| Cell line | Mean ± SD | P-value |
|---|
| MCF 10A | 1.1651±0.7826 | |
| MDA-MB-231 | 13.5447±3.9375 | 0.0059 |
| MCF-7 | 11.4218±4.0622 | 0.0127 |
| NCM460 | 1.0059±0.1364 | |
| SW620 | 7.8246±2.4492 | 0.0400 |
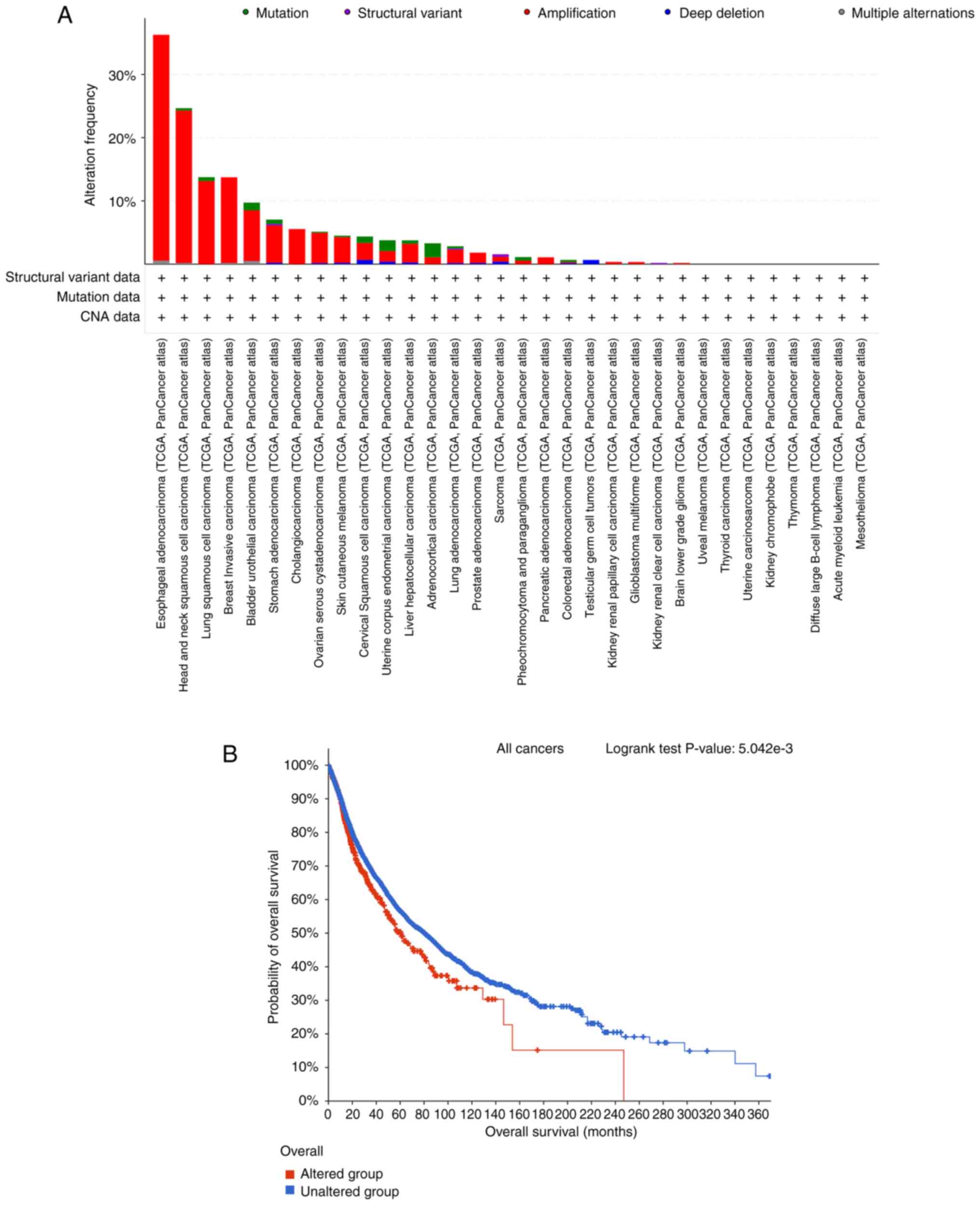
FADD alteration in cancer
The analysis results of the types of FADD alteration
in 32 cancers in the TCGA database demonstrated that FADD changes
were most frequent in patients with ESCA (Fig. 3A). Kaplan-Meier prognosis result of
OS in FADD-altered and non-altered patients showed that FADD
alteration was significantly associated with shorter OS in cancer
patients (Fig. 3B).
FADD promoter methylation level
The analysis of tumor samples and non-tumor samples
in the UALCAN database revealed that FADD promoter methylation
level was relatively high in CESC (Fig.
4B), ESCA (Fig. 4C), KIRC
(Fig. 4D), LUSC (Fig. 4F) and PAAD (Fig. 4G) samples, but lower in BLCA
(Fig. 4A), LIHC (Fig. 4E), PRAD (Fig. 4H), sarcoma (SARC; Fig. 4I), testicular germ cell tumors
(TGCT; Fig. 4J), THCA (Fig. 4K) and UCEC (Fig. 4L) samples.
 | Figure 4Promoter methylation levels.
Difference analysis of the methylation levels of FADD promoter in
(A) BLCA, (B) CESC, (C) ESCA, (D) KIRC, (E) LIHC, (F) LUSC, (G)
PAAD, (H) PRAD, (I) SARC, (J) TGCT, (K) THCA and (L) UCEC. FADD,
fas-associated death domain; TCGA, The Cancer Genome Atlas; BLCA,
bladder urothelial carcinoma; CESC, cervical squamous cell
carcinoma and endocervical adenocarcinoma; ESCA, esophageal
carcinoma; KIRC, kidney renal clear cell carcinoma; LIHC, liver
hepatocellular carcinoma; LUSC, lung squamous cell carcinoma; PAAD,
pancreatic adenocarcinoma; PRAD, prostate adenocarcinoma; SARC,
sarcoma; TGCT, testicular germ cell tumors; THCA, thyroid
carcinoma; UCEC, uterine corpus endometrial carcinoma. |
Correlation between FADD expression
and clinical characteristics of various cancers
To analyze the correlation between the expression of
FADD and age, patients were divided into two cohorts: i) Patients
aged <65 years and ii) patients aged 65 years or older. The
expression level of FADD was relatively higher in elderly cancer
patients with ESCA and OV and patients younger than 65 years with
skin cutaneous melanoma and TGCT (Fig.
5A). In addition, FADD was highly expressed in female patients
with adrenocortical carcinoma and male patients with COAD (Fig. 5B). Notably, the difference in the
expression of FADD among different stages of KIRC, LUAD and TGCT
patients was statistically significant (Fig. 5C).
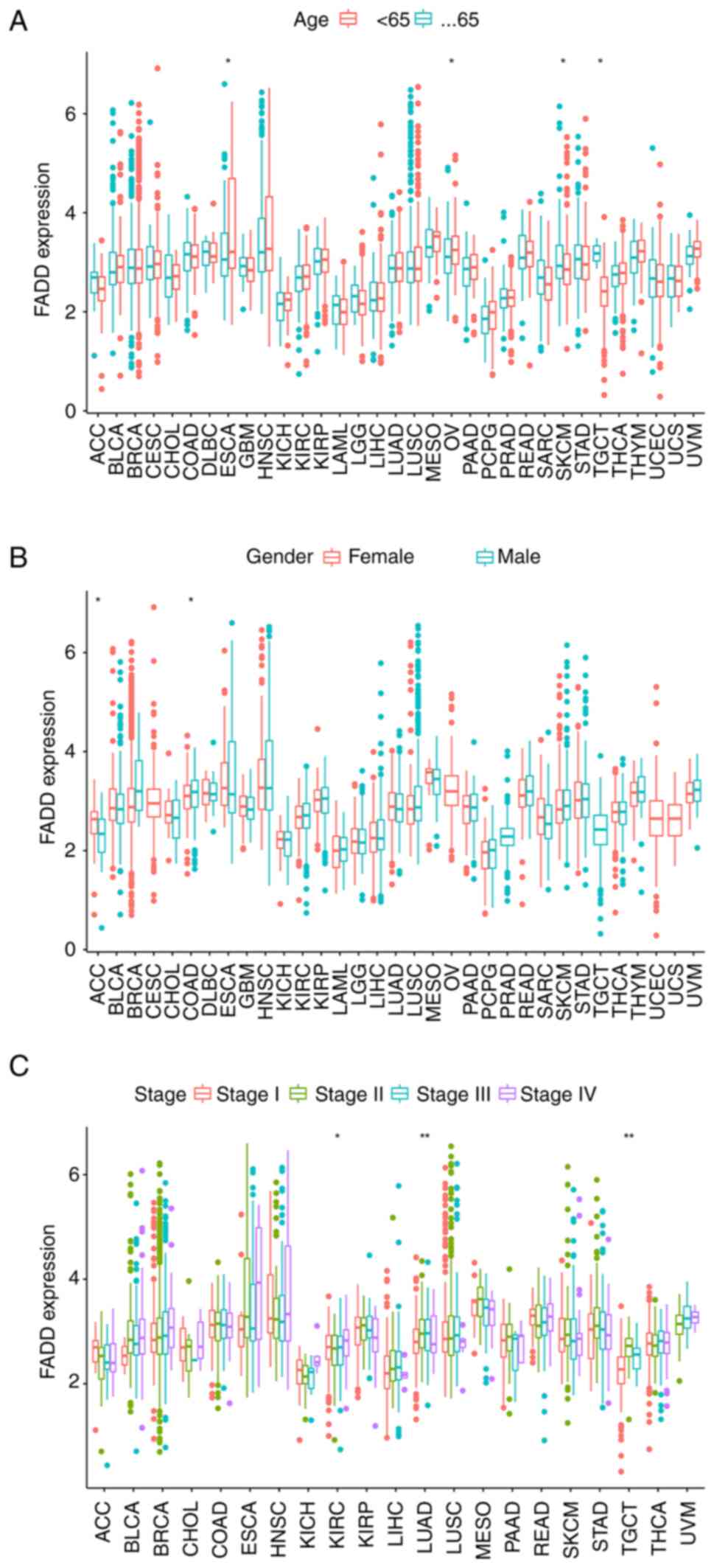 | Figure 5Clinical correlation. FADD expression
correlates with (A) age, (B) sex and (C) cancer stage in patients
with cancer. *P<0.05 and **P<0.01.
FADD, fas-associated death domain. ACC, adrenocortical carcinoma;
BLCA, bladder urothelial carcinoma; BRCA, breast invasive
carcinoma; CESC, cervical squamous cell carcinoma; CHOL,
cholangiocarcinoma; COAD, colon adenocarcinoma; DLBC, diffuse large
B cell lymphoma; ESCA, esophageal carcinoma; GBM, glioblastoma
multiforme; HNSC, head and neck squamous cell carcinoma; KICH,
kidney chromophobe; KIRC, kidney renal clear cell carcinoma; KIRP,
kidney renal papillary cell carcinoma; LAML, acute myeloid
leukemia; LGG, brain lower grade glioma; LIHC, liver hepatocellular
carcinoma; LUAD, lung adenocarcinoma; LUSC, lung squamous cell
carcinoma; MESO, mesothelioma; OV, ovarian serous
cystadenocarcinoma; PAAD, pancreatic adenocarcinoma; PCPG,
pheochromocytoma and paraganglioma; PRAD, prostate adenocarcinoma;
READ, rectum adenocarcinoma; SARC, sarcoma; SKCM, skin cutaneous
melanoma; STAD, stomach adenocarcinoma; TGCT, testicular germ cell
tumors; THCA, thyroid carcinoma; THYM, thymoma; UCEC, uterine
corpus endometrial carcinoma; UCS, uterine carcinosarcoma; UVM,
uveal melanoma. |
Correlation between FADD expression
and prognosis of various cancers
The Kaplan-Meier-plot website was used to
investigate the relationship between FADD expression and the
prognosis of patients with cancer. The results demonstrated that
the high expression of FADD was significantly associated with
shorter OS in CESC (Fig. 6B), HNSC
(Fig. 6C), LIHC (Fig. 6D), LUAD (Fig. 6E), LUSC (Fig. 6F) and PAAD (Fig. 6G), but not in STAD (Fig. 6H), and significantly longer OS in
thymoma (THYM) (Fig. 6I) and THCA
(Fig. 6J). The low expression of
FADD in CESC (Fig. 6L), LUAD
(Fig. 6M), PAAD (Fig. 6N) and TGCT (Fig. 6P) was significantly associated with
shorter RFS, whereas the high expression of FADD in BRCA (Fig. 6K) and STAD (Fig. 6O) was associated with longer RFS.
Cox regression analysis exhibited that FADD was a poor prognostic
factor for HNSC, acute myeloid leukemia, brain lower grade glioma
(LGG), LIHC, LUAD and PAAD, but a protective factor for MESO and
THCA (Fig. 6A).
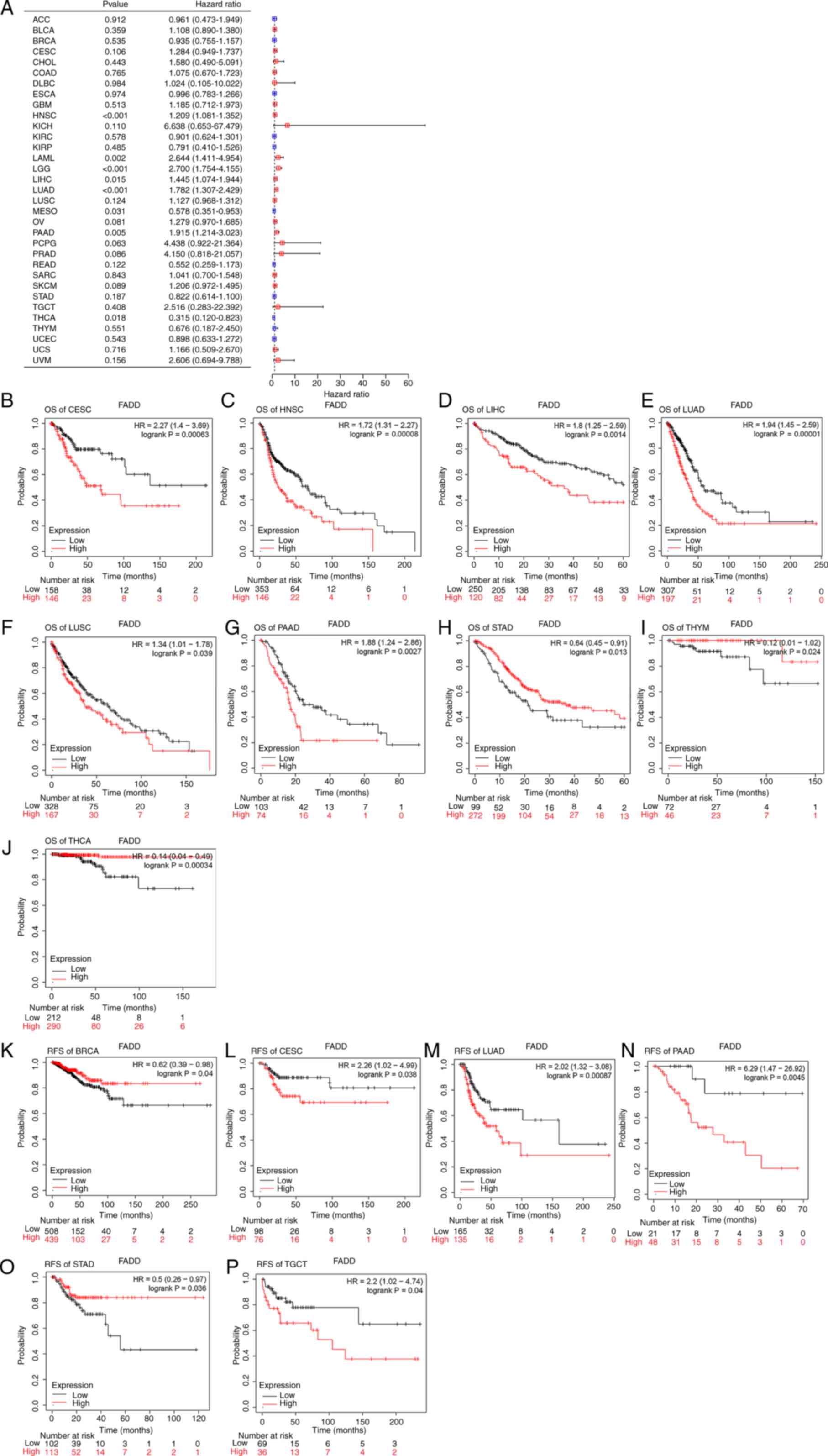 | Figure 6Prognosis analysis. (A) Cox
regression analysis of 33 types of cancer. FADD expression and
Kaplan-Meier prognosis analysis (based on OS) of (B) CESC, (C)
HNSC, (D) LIHC, (E) LUAD, (F) LUSC, (G) PAAD, (H) STAD, (I) THYM
and (J) THCA. FADD expression and Kaplan-Meier prognosis analysis
(based on RFS) of (K) BRCA, (L) CESC, (M) LUAD, (N) PAAD, (O) STAD
and (P) TGCT. FADD, fas-associated death domain; OS, overall
survival; CESC, cervical squamous cell carcinoma and endocervical
adenocarcinoma; HNSC, head and neck squamous cell carcinoma; LIHC,
liver hepatocellular carcinoma; LUAD, lung adenocarcinoma; LUSC,
lung squamous cell carcinoma; PAAD, pancreatic adenocarcinoma;
STAD, stomach adenocarcinoma; THYM, thymoma; THCA, thyroid
carcinoma; RFS, relapse-free survival; BRCA, breast invasive
carcinoma; TGCT, testicular germ cell tumors. |
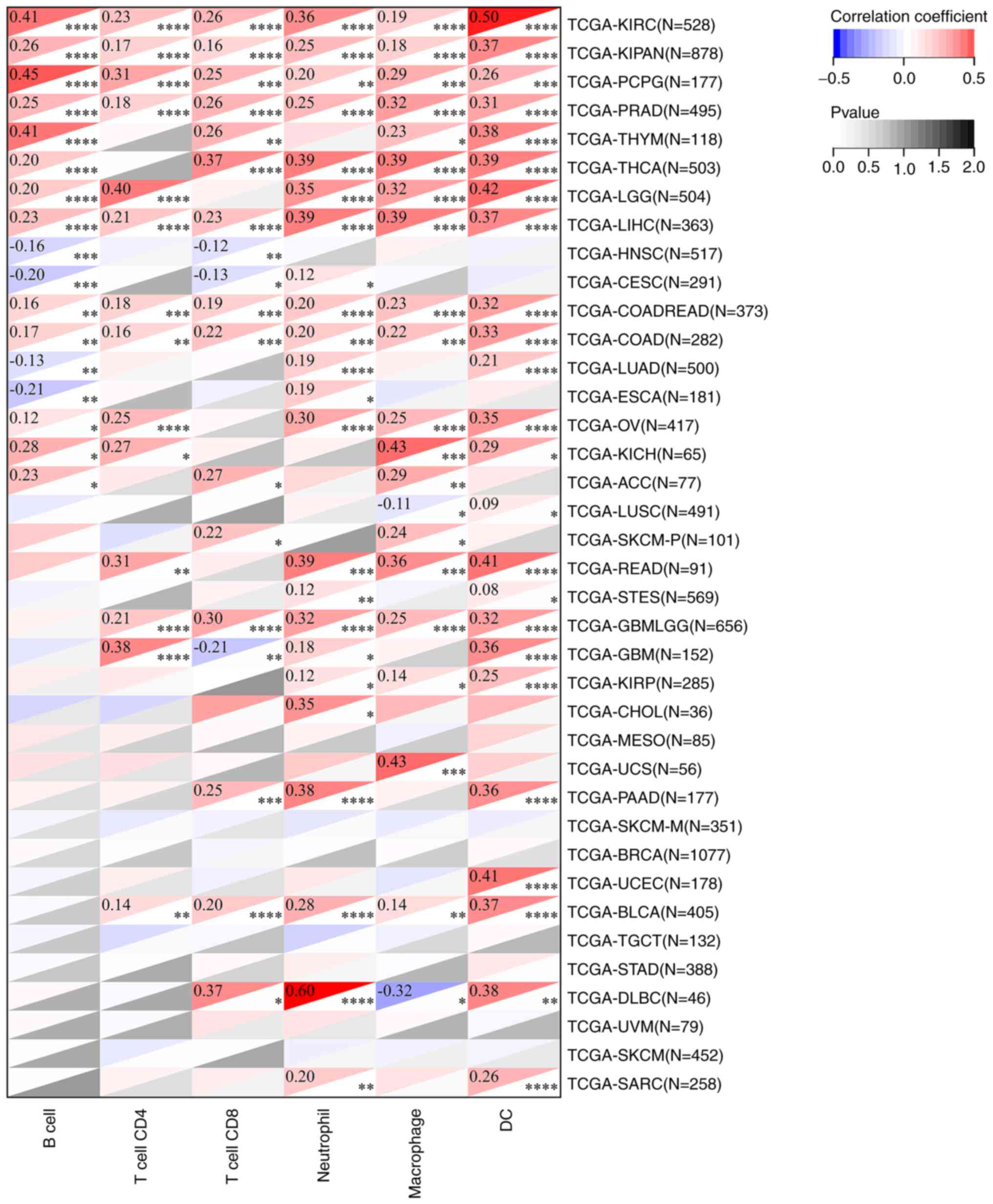
Effects of FADD on TME and immune cell
infiltration
The results of TME analysis showed that the
expression of FADD was positively correlated with the ImmuneScore
of LGG (Fig. 7A), SARC (Fig. 7B), THCA (Fig. 7D), uterine carcinosarcoma (UCS;
Fig. 7E) and uveal melanoma (UVM;
Fig. 7F), and negatively correlated
with the ImmuneScore of TGCT (Fig.
7C). The expression of FADD was positively correlated with the
StromalScore of LGG (Fig. 7G) and
negatively correlated with the StromalScore of MESO (Fig. 7H) and THYM (Fig. 7I). Analysis of the data obtained
from the TCGA database using the TIMER method revealed that the
expression of FADD was positively correlated with B cell
infiltration in KIRC, KIPAN, PCPG, PRAD, THYM, THCA, LGG, LIHC,
COADREAD, COAD, OV, KICH and ACC, but negatively correlated with B
cell infiltration in HNSC, CESC, LUAD and ESCA. Moreover, the
expression of FADD was positively correlated with CD4 cell
infiltration in KIRC, KIPAN, PCPG, PRAD, LGG, LIHC, COADREAD, COAD,
OV, KICH, READ, GBMLGG, GBM and BLCA. The expression of FADD was
positively correlated with T cell CD8 infiltration in LIRC, KIPAN,
PCPG, PRAD, THYM, THCA, LIHC, COADREAD, COAD, ACC, SKCM-P, GBMLGG,
PAAD, BLCA and DLBC, and negatively correlated with T cell CD8
infiltration in HNSC, CESC and GBM. In addition, the expression of
FADD was positively correlated with neutrophil and macrophage
infiltration in all cancer types except LUSC and DLBC in which the
expression of FADD was negatively correlated with macrophage
infiltration. Furthermore, the expression of FADD was positively
correlated with dendritic cells (DC) infiltration in KIRC, KIPAN,
PCPG, PRAD, THYM, THCA, LGG, LIHC, COADREAD, COAD, LUAD, OV, KICH,
LUSC, READ, STES, GBMLGG, GBM, KIRP, PAAD, UCEC, BLCA, DLBC and
SARC (Fig. 8).
 | Figure 7Tumor microenvironment. Correlation
between FADD expression and ImmuneScore of (A) LGG, (B) SARC, (C)
TGCT, (D) THCA, (E) UCS and (F) UVM; the correlation between FADD
expression and StromalScore of LGG (G), MESO (H) and THYM (I).
FADD, fas-associated death domain; LGG, brain lower grade glioma;
SARC, sarcoma; TGCT, testicular germ cell tumors; THCA, thyroid
carcinoma; UCS, uterine carcinosarcoma; UVM, uveal melanoma; MESO,
mesothelioma; THYM, thymoma. |
Correlation of FADD expression with
MSI and TMB in cancer
FADD was observed to be positively correlated with
MSI in LGG, LUAD, PAAD, SARC and UCEC, and negatively correlated
with MSI in COAD, PCPG, READ and THYM (Fig. 9A). By contrast, FADD expression was
positively correlated with HNSC, KIRC, KIRP, PRAD, SARC, THCA, UCEC
and UVM, while it was revealed as negatively correlated with TMB in
LUAD and READ (Fig. 9B).
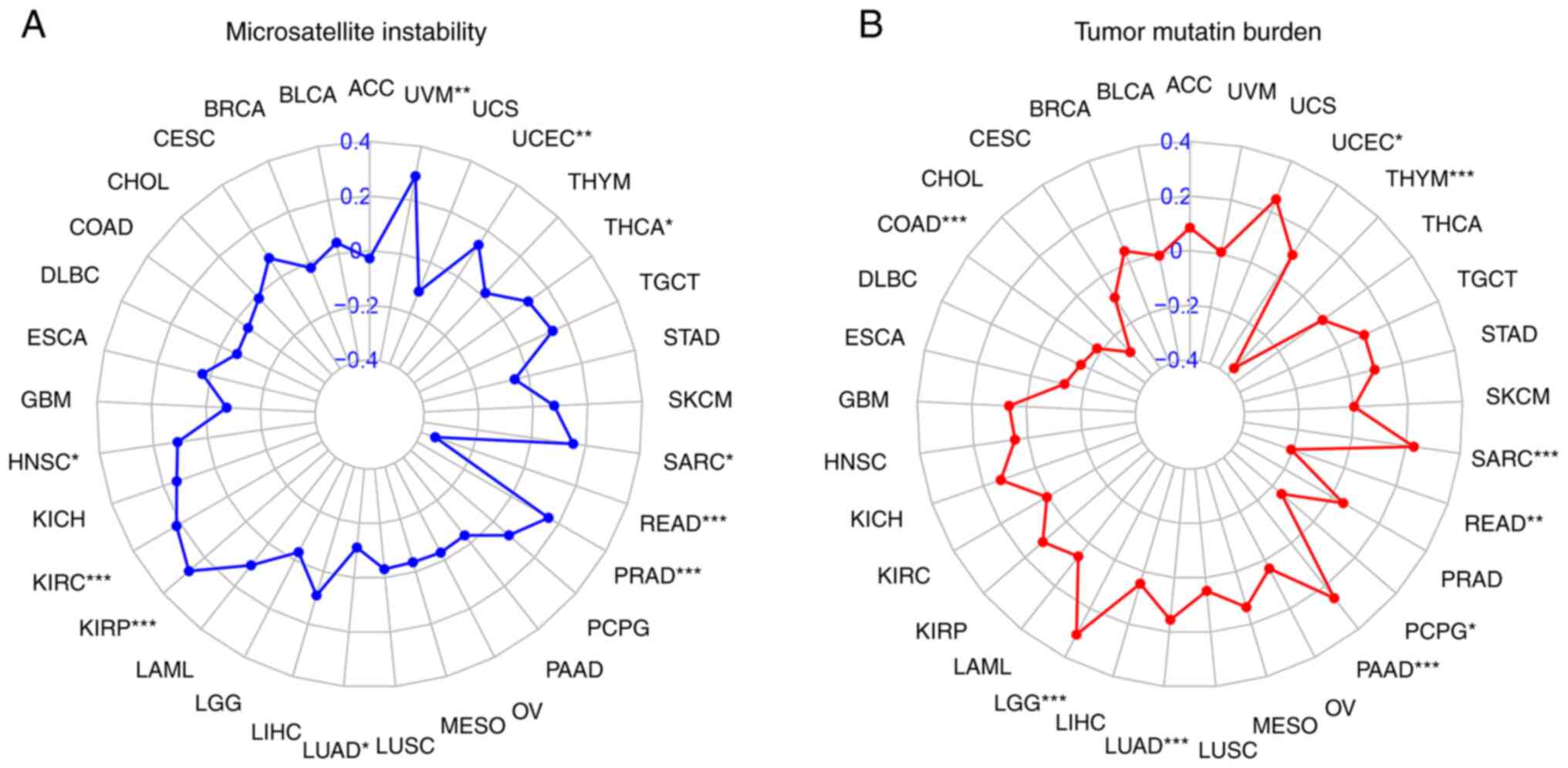 | Figure 9MSI and TMB. Correlation of FADD
expression with (A) MSI and (B) TMB in cancer
*P<0.05, **P<0.01 and
***P<0.001. FADD, fas-associated death domain; MSI,
microsatellite instability; TMB, tumor mutation burden. ACC,
adrenocortical carcinoma; BLCA, bladder urothelial carcinoma; BRCA,
breast invasive carcinoma; CESC, cervical squamous cell carcinoma;
CHOL, cholangiocarcinoma; COAD, colon adenocarcinoma; DLBC, diffuse
large B cell lymphoma; ESCA, esophageal carcinoma; GBM,
glioblastoma multiforme; HNSC, head and neck squamous cell
carcinoma; KICH, kidney chromophobe; KIRC, kidney renal clear cell
carcinoma; KIRP, kidney renal papillary cell carcinoma; LAML, acute
myeloid leukemia; LGG, brain lower grade glioma; LIHC, liver
hepatocellular carcinoma; LUAD, lung adenocarcinoma; LUSC, lung
squamous cell carcinoma; MESO, mesothelioma; OV, ovarian serous
cystadenocarcinoma; PAAD, pancreatic adenocarcinoma; PCPG,
pheochromocytoma and paraganglioma; PRAD, prostate adenocarcinoma;
READ, rectum adenocarcinoma; SARC, sarcoma; SKCM, skin cutaneous
melanoma; STAD, stomach adenocarcinoma; TGCT, testicular germ cell
tumors; THCA, thyroid carcinoma; THYM, thymoma; UCEC, uterine
corpus endometrial carcinoma; UCS, uterine carcinosarcoma; UVM,
uveal melanoma. |
Analysis of different FADD expressions
(GSEA)
The expression of FADD was divided into two groups
and GSEA analysis was performed. Furthermore, the results
demonstrated that FADD was involved in different signaling pathways
and biological processes in various cancers. GO enrichment results
(Fig. S1) revealed that the main
active biological processes associated with the high expression
FADD group were the detection of chemical stimulus (in three
cancers), epidermal cell differentiation (in three cancers), and
epidermis development (in three cancers as well). On the contrary,
the main active biological processes associated with the low
expression FADD group were the detection of chemical stimulus (in
18 cancers) and the detection of stimulus involved in sensory
perception (in 15 cancers). KEGG analysis results (Fig. S2) indicated that the main active
signaling pathways in the high expression FADD group were olfactory
transduction (in 4 cancers) and systemic lupus atherosclerosis (in
3 cancers). By contrast, the main active signaling pathways
associated with the low expression FADD group were olfactory
transduction (in 19 cancers) and neuroactive ligand-receptor
interaction (in 10 cancers).
Discussion
Analysis of the differential expression of FADD
between cancer and normal samples in the TCGA database revealed
that FADD was substantially expressed in 18 of the 33 malignancies
analyzed and in 11 of the 20 GSE datasets selected. The area under
the ROC curve of 6 cancer types (CHOL, GBM, HNSC, LUAD, LUSC and
PCPG) from TCGA database had a value of >0.9, and the area under
the ROC curve of 4 cancers (ESCA, HNSC, LUAD and STAD) from GEO
database had a value of >0.8. RT-qPCR exhibited that FADD mRNA
was relatively significantly expressed in breast, colon, liver and
stomach cancer cells, which was consistent with immunohistochemical
images obtained from the HPA database. These findings showed that
FADD may have diagnostic utility as a biomarker for cancer. FADD is
a ubiquitous adapter protein that not only conveys apoptotic
signals mediated by death receptors but also mediates inflammation
and cancer (51-53).
Inflammation is a hallmark of a substantial percentage of cancers,
which may explain the relatively high expression of FADD in the
vast majority of cancers (54,55),
including oral squamous cell cancer (56). FADD alteration in the cBioPortal
database demonstrated that FADD is more likely to change in more
than 30, 20, 10 and 10% of patients with ESCA, HNSC, LUSC and BRCA,
respectively, and amplification is the predominant FADD alteration.
The human FADD gene is located on chromosome 11q13.3,
11q13-q14 amplification has a relatively high incidence in breast,
ovary, head and neck, esophageal, melanoma and bladder tumors,
which is consistent with the expression patterns of FADD in cancer
(2). This suggests that FADD
amplification may cause certain cancers. Methylation of FADD is
also linked to cancer. A previous study has identified an
association between FADD methylation and oral squamous cell
carcinoma (57). The UALCAN
database analysis revealed that abnormal FADD promoter methylation
was associated with 12 tumor samples, indicating that both FADD
mutation and promoter methylation are associated with malignancy.
Compared with normal tissue, FADD promoter methylation levels were
significantly reduced in primary tumors of BLCA, LIHC, PRAD and
THCA, and differential analysis revealed that FADD mRNA was
significantly highly expressed in these cancer tissues. The high
expression of FADD mRNA in BLCA, LIHC, PRAD and THCA may be related
to the decrease of the promoter methylation level. FADD mRNA was
also highly expressed in CESC, KIRC and LUSC, but FADD promoter
methylation levels were significantly lower in primary tumor
tissues than in corresponding normal tissues in these cancers. This
may be due to the low expression or no expression of FADD mRNA in
the corresponding normal tissue. Even if the methylation level of
the promoter is increased, the expression of FADD in primary tissue
remains significantly higher than that in normal tissue. This
suggests that FADD is reliable as a biomarker for the diagnosis of
these cancers.
High expression of FADD was significantly associated
with shorter OS in six types of cancer patients and RFS in four
types of cancer patients, as exhibited by Kaplan-Meier analysis.
FADD expression was a risk factor for numerous cancers (6 types)
and a protective factor for fewer cancers (2 types), according to
Cox regression analysis. For instance, Kaplan-Meier prognostic
analysis and Cox regression analysis of patients with HNSC, LIHC
and LUAD revealed that FADD expression was a risk factor for these
malignancies. A recent study has demonstrated the predictive
utility of FADD gene can in the prognosis of lung
adenocarcinoma in women (58).
Because FADD amplification occurs in high frequency in HNSC,
numerous studies have investigated its potential as a biomarker of
HNSC (59,60). Additionally, the immunohistochemical
results of FADD overexpression were substantially linked with poor
OS in patients with HNSC, according to a previous meta-analysis
(61). As one of the
apoptosis-related factors, FADD is associated with the occurrence
of LIHC, but further research is needed to confirm its prognostic
value for patients with LIHC (62-64).
Different prognostic analysis approaches have demonstrated that
FADD predicts poor OS in LIHC patients, indicating that FADD may be
employed as both a diagnostic and prognostic biomarker for patients
with LIHC. Analysis of TME and immune cell infiltration revealed
that FADD expression influences tumor immunity in a number of
malignancies, particularly various tumors where neutrophil and DC
infiltration are strongly positively associated. The neutrophil is
a crucial cell that regulates inflammation and immune response,
whereas DC is an antigen-presenting cell with a significant effect
on tumor immunity (65-68).
This suggests that FADD may influence tumor immunity by boosting
neutrophil and DC infiltration into tumors. FADD expression was
substantially related to MSI in 9 malignancies and TMB in 10
tumors, suggesting its potential as an immunotherapy marker. Bowman
et al (69) revealed that
the phosphorylation of FADD promoted the proliferation of lung
cancer cells, suggesting that FADD indeed plays a role in
tumorigenesis and development, and it is necessary to conduct
in-depth research on it in the future.
Because of inconsistencies between the GEO and TCGA
databases, expression data for 33 tumors was not gathered to verify
the differential expression of FADD. The present study is limited
to the expression and clinical relevance of FADD in different
cancers, and lacks clarification of the specific role of FADD in
tumorigenesis and progression, which is necessary to explore FADD
as a therapeutic target. Although FADD expression was identified as
a potential diagnostic and prognostic biomarker for specific
cancers, its clinical application and applicability in clinical
practice need to be rigorously evaluated and verified by
large-scale clinical trials.
The present study carefully evaluated the expression
of FADD in various malignancies and its effect on the prognoses of
patients with cancer. Analysis of various databases revealed that
FADD was highly expressed in BRCA, CESC, CHOL, COAD, ESCA, KIRC,
KIRP, LIHC, LUAD and PRAD. Moreover, the high expression of FADD
was confirmed in BRCA, COAD, LIHC and STAD using RT-qPCR,
supporting the potential utility of FADD as a prognostic biomarker
for patients with LIHC. In conclusion, FADD is highly expressed in
numerous malignancies and can be utilized as a diagnostic biomarker
for BRCA, COAD, LIHC, and STAD. FADD expression is a predictive
risk factor for HNSC, LIHC, and LUAD patients and has potential
value as a prognostic marker for these tumors.
Supplementary Material
GO enrichment analysis of differential
genes in high and low expression groups of FADD. GO, Gene Ontology;
FADD, fas-associated death domain; GOBP, GO biological process;
DLBC, lymphoid neoplasm diffuse large B-cell lymphoma; ESCA,
esophageal carcinoma; GBM, glioblastoma multiforme; HNSC, head and
neck squamous cell carcinoma; KICH, kidney chromophobe; KIRC,
kidney renal clear cell carcinoma; KIRP, kidney renal papillary
cell carcinoma; LAML, acute myeloid leukemia; LGG, brain lower
grade glioma; LIHC, liver hepatocellular carcinoma; LUAD, lung
adenocarcinoma; LUSC, lung squamous cell carcinoma; MESO,
mesothelioma; OV, ovarian serous cystadenocarcinoma; PAAD,
pancreatic adenocarcinoma; PCPG, pheochromocytoma and
paraganglioma; PRAD, prostate adenocarcinoma; READ, rectum
adenocarcinoma; SARC, sarcoma; SKCM, skin cutaneous melanoma; STAD,
stomach adenocarcinoma; TGCT, testicular germ cell tumors; THCA,
thyroid carcinoma; THYM, thymoma; UCEC, uterine corpus endometrial
carcinoma; UCS, uterine carcinosarcoma; UVM, uveal melanoma; AAC,
adenoid cystic carcinoma; BLCA, bladder urothelial carcinoma; BRCA,
breast invasive carcinoma; CESC, cervical squamous cell carcinoma
and endocervical adenocarcinoma; CHOL, cholangiocarcinoma; COAD,
colon adenocarcinoma.
KEGG enrichment analysis of
differential genes in high and low expression groups of FADD. KEGG,
Kyoto Encyclopedia of Genes and Genomes; FADD, fas-associated death
domain; PRAD, prostate adenocarcinoma; PCPG, pheochromocytoma and
paraganglioma; PAAD, pancreatic adenocarcinoma; OV, ovarian serous
cystadenocarcinoma; LUSC, lung squamous cell carcinoma; LUAD, lung
adenocarcinoma; LIHC, liver hepatocellular carcinoma; LGG, brain
lower grade glioma; READ, rectum adenocarcinoma; SARC, sarcoma;
SKCM, skin cutaneous melanoma; STAD, stomach adenocarcinoma; TGCT,
testicular germ cell tumors; THCA, thyroid carcinoma; THYM,
thymoma; UCEC, uterine corpus endometrial carcinoma; BRCA, breast
invasive carcinoma; CESC, cervical squamous cell carcinoma and
endocervical adenocarcinoma; CHOL, cholangiocarcinoma; COAD, colon
adenocarcinoma; DLBC, lymphoid neoplasm diffuse large B-cell
lymphoma; ESCA, esophageal carcinoma; GBM, glioblastoma multiforme;
HNSC, head and neck squamous cell carcinoma; KICH, kidney
chromophobe; KIRC, kidney renal clear cell carcinoma; KIRP, kidney
renal papillary cell carcinoma; LAML, acute myeloid leukemia.
Acknowledgements
The authors wish to thank the Key Laboratory of
Molecular Diagnostics and Precision Medicine for Surgical Oncology
in Gansu Province and the DaVinci Surgery System Database
(www.davincisurgery.com) for their help
and support in the methodology.
Funding
Funding: The present study was supported by the 2021 Central to
guide local scientific and Technological Development (grant no.
ZYYDDFFZZJ-1), the Natural Science Foundation of Gansu Province,
China (grant no. 18JR3RA052), the Lanzhou Talent Innovation and
Entrepreneurship Project Task Contract (grant no. 2016-RC-56), the
Gansu Da Vinci robot high-end diagnosis and treatment team
construction project, the Gansu Provincial Youth Science and
Technology Fund Program (grant no. 20JR10RA415) and the National
Key Research and Development Program (grant no.
2018YFC1311500).
Availability of data and materials
The datasets generated and/or analyzed during the
current study are available in TCGA (https://portal.gdc.cancer.gov/), UCSC Xena website
(xena.ucsc.edu/), Kaplan Meier plotter portal
(https://kmplot.com/), GEO database (https://www.ncbi.nlm.nih.gov/), The Molecular
Signatures Database (https://www.gsea-msigdb.org/gsea/msigdb), The Human
Protein Atlas (https://www.proteinatlas.org/), cBioPortal database
(http://www.cbioportal.org/), UALCAN
portal (ualcan.path.uab.edu/) and sangerbox
website (http://vip.sangerbox.com/home.html).
Authors' contributions
XJ and CW conceived the study. ZX and QZ
comprehensively collected relevant data. XJ and CW contributed in
data analysis and in drafting the manuscript. XJ and CW confirm the
authenticity of all the raw data. CW revised the manuscript and HC
reviewed the manuscript. All authors read and approved the final
version of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Mouasni S and Tourneur L: FADD at the
crossroads between cancer and inflammation. Trends Immunol.
39:1036–1053. 2018.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Wilkerson PM and Reis-Filho JS: The
11q13-q14 amplicon: Clinicopathological correlations and potential
drivers. Genes Chromosomes Cancer. 52:333–355. 2013.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Saggioro FP, Neder L, Stávale JN,
Paixão-Becker AN, Malheiros SM, Soares FA, Pittella JE, Matias CC,
Colli BO, Carlotti CG Jr and Franco M: Fas, FasL, and cleaved
caspases 8 and 3 in glioblastomas: A tissue microarray-based study.
Pathol Res Pract. 210:267–273. 2014.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Marín-Rubio JL, Vela-Martín L,
Fernández-Piqueras J and Villa-Morales M: FADD in cancer:
Mechanisms of altered expression and function, and clinical
implications. Cancers (Basel). 11(1462)2019.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Tourneur L and Chiocchia G: FADD: A
regulator of life and death. Trends Immunol. 31:260–269.
2010.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Zhuang H, Gan Z, Jiang W, Zhang X and Hua
ZC: Functional specific roles of FADD: Comparative proteomic
analyses from knockout cell lines. Mol Biosyst. 9:2063–2078.
2013.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Screaton RA, Kiessling S, Sansom OJ,
Millar CB, Maddison K, Bird A, Clarke AR and Frisch SM:
Fas-associated death domain protein interacts with methyl-CpG
binding domain protein 4: A potential link between genome
surveillance and apoptosis. Proc Natl Acad Sci USA. 100:5211–5216.
2003.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Gómez-Angelats M and Cidlowski JA:
Molecular evidence for the nuclear localization of FADD. Cell Death
Differ. 10:791–797. 2003.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Zhang Y and Zhang Z: The history and
advances in cancer immunotherapy: Understanding the characteristics
of tumor-infiltrating immune cells and their therapeutic
implications. Cell Mol Immunol. 17:807–821. 2020.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Jiang Y, Chen M, Nie H and Yuan Y: PD-1
and PD-L1 in cancer immunotherapy: Clinical implications and future
considerations. Hum Vaccin Immunother. 15:1111–1122.
2019.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Gajewski TF, Schreiber H and Fu YX: Innate
and adaptive immune cells in the tumor microenvironment. Nat
Immunol. 14:1014–1022. 2013.PubMed/NCBI View
Article : Google Scholar
|
|
12
|
Balkwill FR, Capasso M and Hagemann T: The
tumor microenvironment at a glance. J Cell Sci. 125:5591–5596.
2012.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Mellman I, Coukos G and Dranoff G: Cancer
immunotherapy comes of age. Nature. 480:480–489. 2011.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Esfahani K, Roudaia L, Buhlaiga N, Del
Rincon SV, Papneja N and Miller WH*Jr: A review of cancer
immunotherapy: From the past, to the present, to the future. Curr
Oncol. 27 (Suppl 2):S87–S97. 2020.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Ranjan K, Waghela BN, Vaidya FU and Pathak
C: Cell-penetrable peptide-conjugated FADD induces apoptosis and
regulates inflammatory signaling in cancer cells. Int J Mol Sci.
21(6890)2020.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Xiang R, Liu Y, Zhu L, Dong W and Qi Y:
Adaptor FADD is recruited by RTN3/HAP in ER-bound signaling
complexes. Apoptosis. 11:1923–1932. 2006.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Kim WJ, Kim EJ, Kim SK, Kim YJ, Ha YS,
Jeong P, Kim MJ, Yun SJ, Lee KM, Moon SK, et al: Predictive value
of progression-related gene classifier in primary non-muscle
invasive bladder cancer. Mol Cancer. 9(3)2010.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Wang J, Zhang X, Beck AH, Collins LC,
Collins LC, Chen WY, Tamimi RM, Hazra A, Brown M, Rosner B and
Hankinson SE: Alcohol consumption and risk of breast cancer by
tumor receptor expression. Horm Cancer. 6:237–246. 2015.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Scotto L, Narayan G, Nandula SV,
Arias-Pulido H, Subramaniyam S, Schneider A, Kaufmann AM, Wright
JD, Pothuri B, Mansukhani M and Murty VV: Identification of copy
number gain and overexpressed genes on chromosome arm 20q by an
integrative genomic approach in cervical cancer: Potential role in
progression. Genes Chromosomes Cancer. 47:755–765. 2008.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Andersen JB, Spee B, Blechacz BR, Avital
I, Komuta M, Barbour A, Conner EA, Gillen MC, Roskams T, Roberts
LR, et al: Genomic and genetic characterization of
cholangiocarcinoma identifies therapeutic targets for tyrosine
kinase inhibitors. Gastroenterology. 142:1021–1031.e15.
2012.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Solé X, Crous-Bou M, Cordero D, Olivares
D, Guinó E, Sanz-Pamplona R, Rodriguez-Moranta F, Sanjuan X, de Oca
J, Salazar R and Moreno V: Discovery and validation of new
potential biomarkers for early detection of colon cancer. PLoS One.
9(e106748)2014.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Su H, Hu N, Yang HH, Wang C, Takikita M,
Wang QH, Giffen C, Clifford R, Hewitt SM, Shou JZ, et al: Global
gene expression profiling and validation in esophageal squamous
cell carcinoma and its association with clinical phenotypes. Clin
Cancer Res. 17:2955–2966. 2011.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Chen C, Méndez E, Houck J, Fan W,
Lohavanichbutr P, Doody D, Yueh B, Futran ND, Upton M, Farwell DG,
et al: Gene expression profiling identifies genes predictive of
oral squamous cell carcinoma. Cancer Epidemiol Biomarkers Prev.
17:2152–2162. 2008.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Laskar RS, Li P, Ecsedi S, Abedi-Ardekani
B, Durand G, Robinot N, Hubert JN, Janout V, Zaridze D, Mukeria A,
et al: Sexual dimorphism in cancer: Insights from transcriptional
signatures in kidney tissue and renal cell carcinoma. Hum Mol
Genet. 30:343–355. 2021.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Jones J, Otu H, Spentzos D, Kolia S, Inan
M, Beecken WD, Fellbaum C, Gu X, Joseph M, Pantuck AJ, et al: Gene
signatures of progression and metastasis in renal cell cancer. Clin
Cancer Res. 11:5730–5739. 2005.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Ivanovska I, Zhang C, Liu AM, Wong KF, Lee
NP, Lewis P, Philippar U, Bansal D, Buser C, Scott M, et al: Gene
signatures derived from a c-MET-driven liver cancer mouse model
predict survival of patients with hepatocellular carcinoma. PLoS
One. 6(e24582)2011.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Zhang Y, Foreman O, Wigle DA, Kosari F,
Vasmatzis G, Salisbury JL, van Deursen J and Galardy PJ: USP44
regulates centrosome positioning to prevent aneuploidy and suppress
tumorigenesis. J Clin Invest. 122:4362–4374. 2012.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Hou J, Aerts J, den Hamer B, van Ijcken W,
den Bakker M, Riegman P, van der Leest C, van der Spek P, Foekens
JA, Hoogsteden HC, et al: Gene expression-based classification of
non-small cell lung carcinomas and survival prediction. PLoS One.
5(e10312)2010.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Suraokar MB, Nunez MI, Diao L, Chow CW,
Kim D, Behrens C, Lin H, Lee S, Raso G, Moran C, et al: Expression
profiling stratifies mesothelioma tumors and signifies deregulation
of spindle checkpoint pathway and microtubule network with
therapeutic implications. Ann Oncol. 25:1184–1192. 2014.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Vathipadiekal V, Wang V, Wei W, Waldron L,
Drapkin R, Gillette M, Skates S and Birrer M: Creation of a human
secretome: A novel composite library of human secreted proteins:
Validation using ovarian cancer gene expression data and a virtual
secretome array. Clin Cancer Res. 21:4960–4969. 2015.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Moffitt RA, Marayati R, Flate EL, Volmar
KE, Loeza SG, Hoadley KA, Rashid NU, Williams LA, Eaton SC, Chung
AH, et al: Virtual microdissection identifies distinct tumor- and
stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat
Genet. 47:1168–1178. 2015.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Giordano TJ, Kuick R, Else T, Gauger PG,
Vinco M, Bauersfeld J, Sanders D, Thomas DG, Doherty G and Hammer
G: Molecular classification and prognostication of adrenocortical
tumors by transcriptome profiling. Clin Cancer Res. 15:668–676.
2009.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Whitington T, Gao P, Song W, Ross-Adams H,
Lamb AD, Yang Y, Svezia I, Klevebring D, Mills IG, Karlsson R, et
al: Gene regulatory mechanisms underpinning prostate cancer
susceptibility. Nat Genet. 48:387–397. 2016.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Oh SC, Sohn BH, Cheong JH, Kim SB, Lee JE,
Park KC, Lee SH, Park JL, Park YY, Lee HS, et al: Clinical and
genomic landscape of gastric cancer with a mesenchymal phenotype.
Nat Commun. 9(1777)2018.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Dom G, Tarabichi M, Unger K, Thomas G,
Oczko-Wojciechowska M, Bogdanova T, Jarzab B, Dumont JE, Detours V
and Maenhaut C: A gene expression signature distinguishes normal
tissues of sporadic and radiation-induced papillary thyroid
carcinomas. Br J Cancer. 107:994–1000. 2012.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Pappa KI, Polyzos A, Jacob-Hirsch J,
Amariglio N, Vlachos GD, Loutradis D and Anagnou NP: Profiling of
discrete gynecological cancers reveals novel transcriptional
modules and common features shared by other cancer types and
embryonic stem cells. PLoS One. 10(e0142229)2015.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Cerami E, Gao J, Dogrusoz U, Gross BE,
Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et
al: The cBio cancer genomics portal: An open platform for exploring
multidimensional cancer genomics data. Cancer Discov. 2:401–404.
2012.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Zhang Y, Chen F, Chandrashekar DS,
Varambally S and Creighton CJ: Proteogenomic characterization of
2002 human cancers reveals pan-cancer molecular subtypes and
associated pathways. Nat Commun. 13(2669)2022.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Lánczky A and Győrffy B: Web-based
survival analysis tool tailored for medical research (KMplot):
Development and implementation. J Med Internet Res.
23(e27633)2021.PubMed/NCBI View
Article : Google Scholar
|
|
40
|
Whiteside TJ: The tumor microenvironment
and its role in promoting tumor growth. Oncogene. 27:5904–5912.
2008.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Arneth B: Tumor microenvironment. Medicina
(Kaunas). 56(15)2019.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Fridman WH, Galon J, Dieu-Nosjean MC,
Cremer I, Fisson S, Damotte D, Pagès F, Tartour E and
Sautès-Fridman C: Immune infiltration in human cancer: Prognostic
significance and disease control. Curr Top Microbiol Immunol.
344:1–24. 2011.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Pagès F, Galon J, Dieu-Nosjean MC, Tartour
E, Sautès-Fridman C and Fridman WH: Immune infiltration in human
tumors: A prognostic factor that should not be ignored. Oncogene.
29:1093–1102. 2010.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Shen W, Song Z, Zhong X, Huang M, Shen D,
Gao P, Qian X, Wang M, He X, Wang T, et al: Sangerbox: A
comprehensive, interaction-friendly clinical bioinformatics
analysis platform. iMeta. 1(e36)2022.
|
|
45
|
Li T, Fu J, Zeng Z, Cohen D, Li J, Chen Q,
Li B and Liu XS: TIMER2.0 for analysis of tumor-infiltrating immune
cells. Nucleic Acids Res. 48 (W1):W509–W514. 2020.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Niu B, Ye K, Zhang Q, Lu C, Xie M,
McLellan MD, Wendl MC and Ding L: MSIsensor: Microsatellite
instability detection using paired tumor-normal sequence data.
Bioinformatics. 30:1015–1016. 2014.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Goodman AM, Kato S, Bazhenova L, Patel SP,
Frampton GM, Miller V, Stephens PJ, Daniels GA and Kurzrock R:
Tumor mutational burden as an independent predictor of response to
immunotherapy in diverse cancers. Mol Cancer Ther. 16:2598–2608.
2017.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Chalmers ZR, Connelly CF, Fabrizio D, Gay
L, Ali SM, Ennis R, Schrock A, Campbell B, Shlien A, Chmielecki J,
et al: Analysis of 100,000 human cancer genomes reveals the
landscape of tumor mutational burden. Genome Med.
9(34)2017.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Mootha VK, Lindgren CM, Eriksson KF,
Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E,
Ridderstråle M, Laurila E, et al: PGC-1alpha-responsive genes
involved in oxidative phosphorylation are coordinately
downregulated in human diabetes. Nat Genet. 34:267–273.
2003.PubMed/NCBI View
Article : Google Scholar
|
|
50
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Harris MA, Clark J, Ireland A, Lomax J,
Ashburner M, Foulger R, Eilbeck K, Lewis S, Marshall B, Mungall C,
et al: The gene ontology (GO) database and informatics resource.
Nucleic Acids Res. 32 (Database Issue):D258–D261. 2004.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Sharma VK, Singh TG, Singh S, Garg N and
Dhiman S: Apoptotic pathways and Alzheimer's disease: Probing
therapeutic potential. Neurochem Res. 46:3103–3122. 2021.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Zhou W, Lai Y, Zhu J, Xu X, Yu W, Du Z, Wu
L, Zhang X and Hua Z: The classical apoptotic adaptor FADD
regulates glycolytic capacity in acute lymphoblastic leukemia. Int
J Biol Sci. 18:3137–3155. 2022.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Lee EW, Seo J, Jeong M, Lee S and Song J:
The roles of FADD in extrinsic apoptosis and necroptosis. BMB Rep.
45:496–508. 2012.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Singh N, Baby D, Rajguru JP, Patil PB,
Thakkannavar SS and Pujari VB: Inflammation and cancer. Ann Afr
Med. 18:121–126. 2019.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Murata M: Inflammation and cancer. Environ
Health Prev Med. 23(50)2018.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Prapinjumrune C, Morita K, Kuribayashi Y,
Hanabata Y, Shi Q, Nakajima Y, Inazawa J and Omura K: DNA
amplification and expression of FADD in oral squamous cell
carcinoma. J Oral Pathol Med. 39:525–532. 2010.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Saberi E, Kordi-Tamandani DM, Jamali S and
Rigi-Ladiz MA: Analysis of methylation and mRNA expression status
of FADD and FAS genes in patients with oral squamous cell
carcinoma. Med Oral Patol Oral Cir Bucal. 19:e562–e568.
2014.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Liu Z, Zhang K, Zhao Z, Qin Z and Tang H:
Prognosis-related autophagy genes in female lung adenocarcinoma.
Medicine (Baltimore). 101(e28500)2022.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Jiang Y, Li Y, Ge H, Wu Y, Zhang Y, Guo S,
Zhang P, Cheng J and Wang Y: Identification of an autophagy-related
prognostic signature in head and neck squamous cell carcinoma. J
Oral Pathol Med. 50:1040–1049. 2021.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Fan S, Müller S, Chen ZG, Pan L,
Tighiouart M, Shin DM, Khuri FR and Sun SY: Prognostic impact of
Fas-associated death domain, a key component in death receptor
signaling, is dependent on the presence of lymph node metastasis in
head and neck squamous cell carcinoma. Cancer Biol Ther.
14:365–369. 2013.PubMed/NCBI View Article : Google Scholar
|
|
62
|
González-Moles MÁ, Ayén Á, González-Ruiz
I, de Porras-Carrique T, González-Ruiz L, Ruiz-Ávila I and
Ramos-García P: Prognostic and clinicopathological significance of
FADD upregulation in head and neck squamous cell carcinoma: A
systematic review and meta-analysis. Cancers (Basel).
12(2393)2020.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Verboom L, Martens A, Priem D, Hoste E,
Sze M, Vikkula H, Van Hove L, Voet S, Roels J, Maelfait J, et al:
OTULIN prevents liver inflammation and hepatocellular carcinoma by
inhibiting FADD- and RIPK1 kinase-mediated hepatocyte apoptosis.
Cell Rep. 30:2237–2247.e6. 2020.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Harari-Steinfeld R, Gefen M, Simerzin A,
Zorde-Khvalevsky E, Rivkin M, Ella E, Friehmann T, Gerlic M,
Zucman-Rossi J, Caruso S, et al: The lncRNA H19-derived
MicroRNA-675 promotes liver necroptosis by targeting FADD. Cancers
(Basel). 13(411)2021.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Liu W, Jing ZT, Xue CR, Wu SX, Chen WN,
Lin XJ and Lin X: PI3K/AKT inhibitors aggravate death
receptor-mediated hepatocyte apoptosis and liver injury. Toxicol
Appl Pharmacol. 381(114729)2019.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Euler M and Hoffmann MH: The double-edged
role of neutrophil extracellular traps in inflammation. Biochem Soc
Trans. 47:1921–1930. 2019.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Liew PX and Kubes P: The neutrophil's role
during health and disease. Physiol Rev. 99:1223–1248.
2019.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Murphy TL and Murphy KM: Dendritic cells
in cancer immunology. Cell Mol Immunol. 19:3–13. 2022.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Bowman BM, Sebolt KA, Hoff BA, Boes JL,
Daniels DL, Heist KA, Galbán CJ, Patel RM, Zhang J, Beer DG, et al:
Phosphorylation of FADD by the kinase CK1α promotes
KRASG12D-induced lung cancer. Sci Signal. 8(ra9)2015.PubMed/NCBI View Article : Google Scholar
|