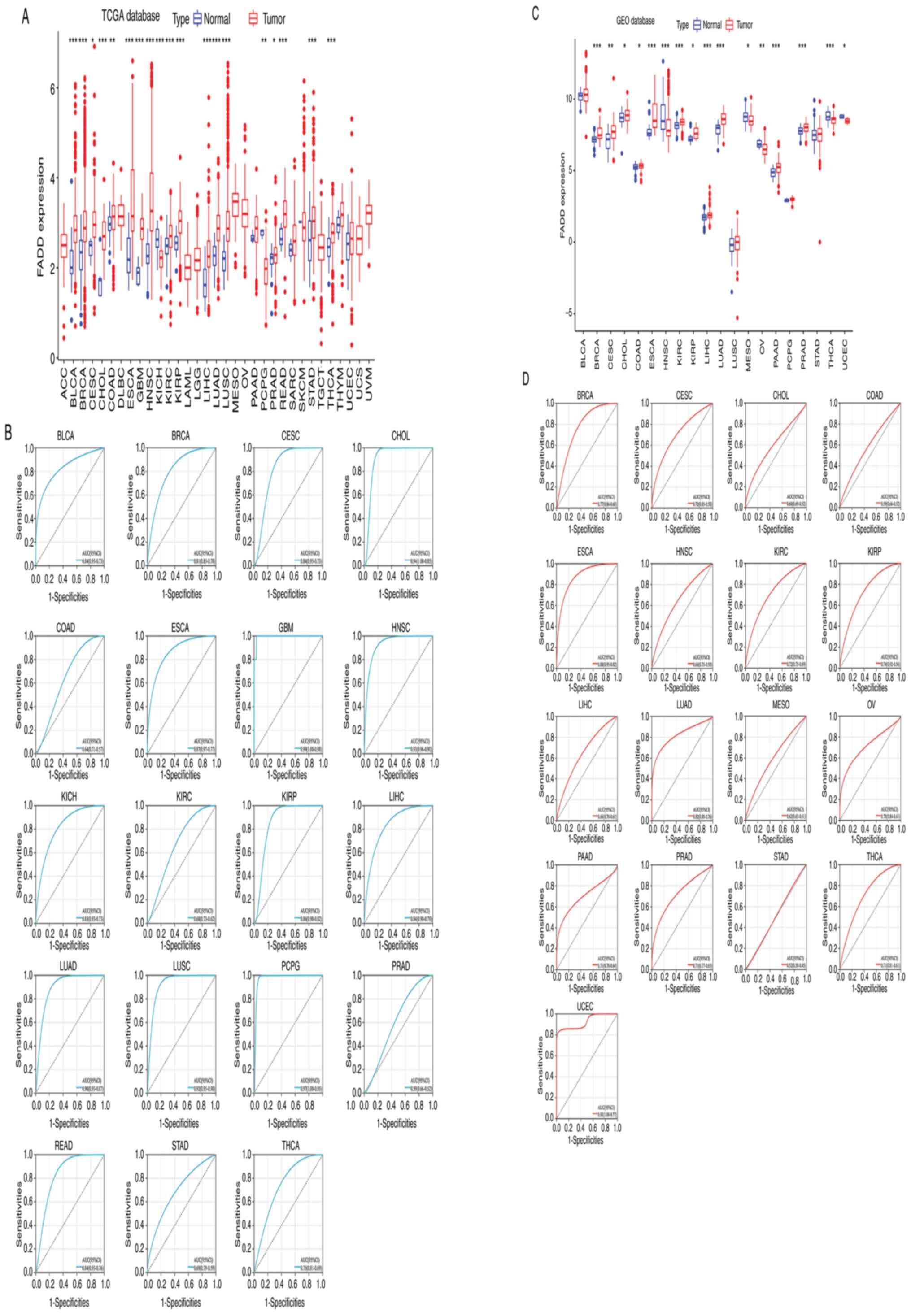
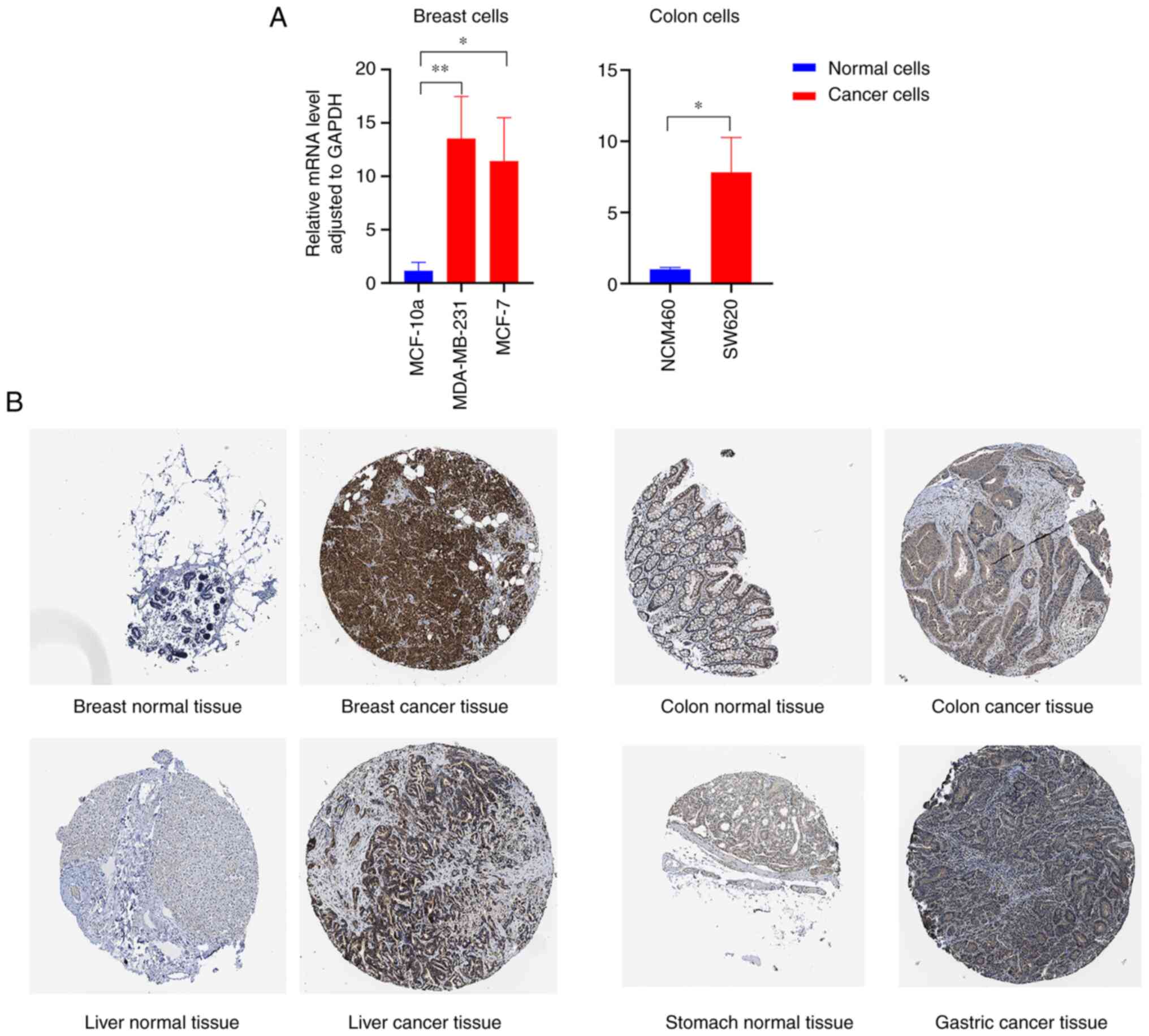
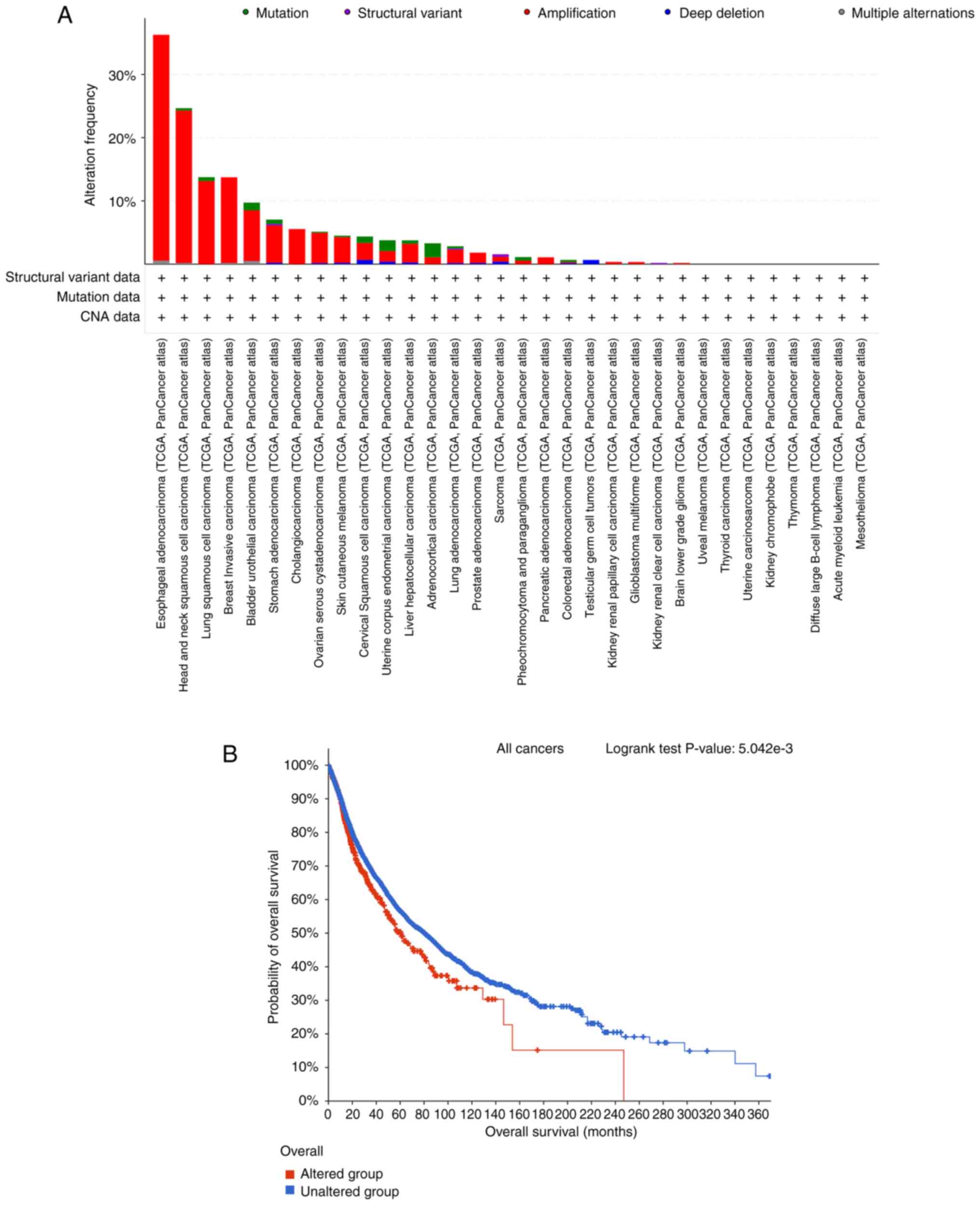
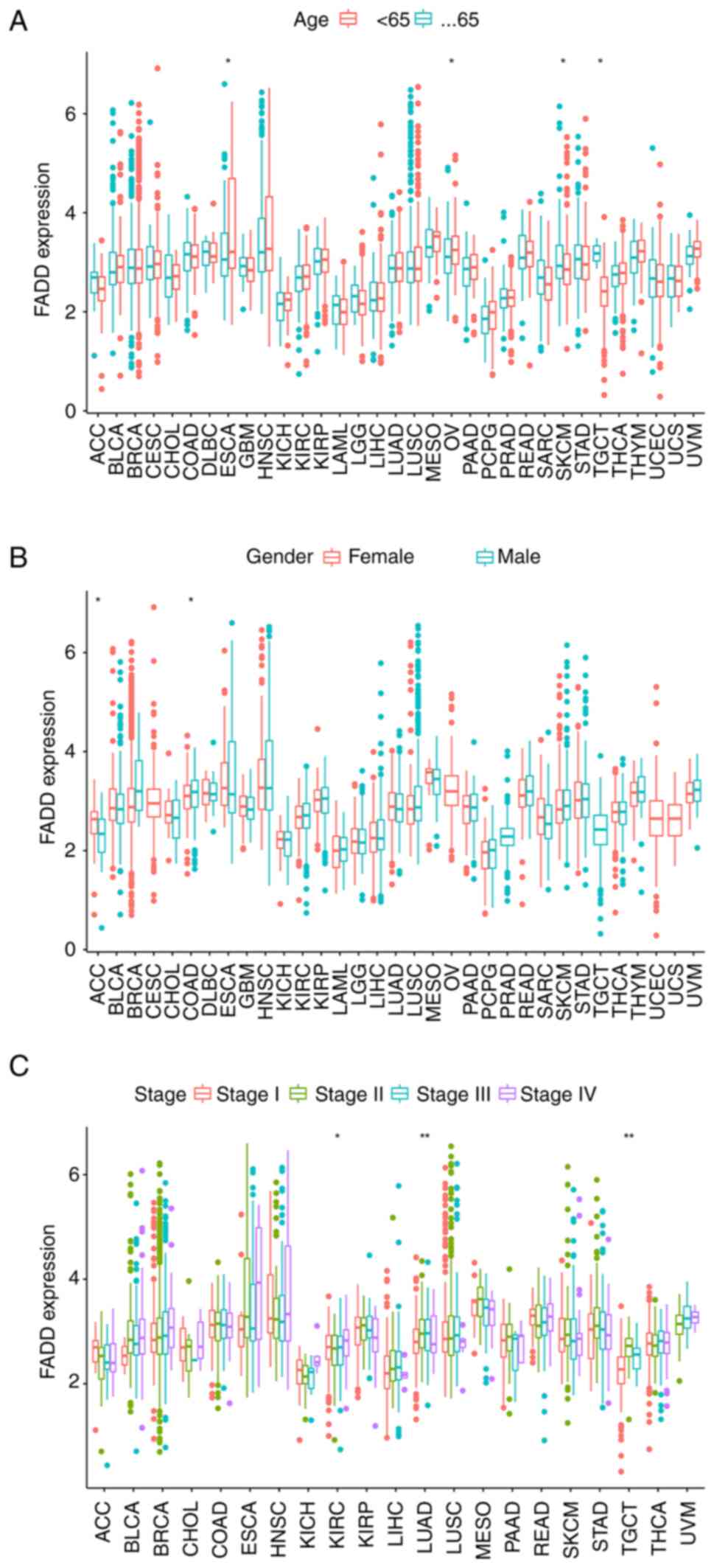
|
1
|
Mouasni S and Tourneur L: FADD at the
crossroads between cancer and inflammation. Trends Immunol.
39:1036–1053. 2018.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Wilkerson PM and Reis-Filho JS: The
11q13-q14 amplicon: Clinicopathological correlations and potential
drivers. Genes Chromosomes Cancer. 52:333–355. 2013.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Saggioro FP, Neder L, Stávale JN,
Paixão-Becker AN, Malheiros SM, Soares FA, Pittella JE, Matias CC,
Colli BO, Carlotti CG Jr and Franco M: Fas, FasL, and cleaved
caspases 8 and 3 in glioblastomas: A tissue microarray-based study.
Pathol Res Pract. 210:267–273. 2014.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Marín-Rubio JL, Vela-Martín L,
Fernández-Piqueras J and Villa-Morales M: FADD in cancer:
Mechanisms of altered expression and function, and clinical
implications. Cancers (Basel). 11(1462)2019.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Tourneur L and Chiocchia G: FADD: A
regulator of life and death. Trends Immunol. 31:260–269.
2010.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Zhuang H, Gan Z, Jiang W, Zhang X and Hua
ZC: Functional specific roles of FADD: Comparative proteomic
analyses from knockout cell lines. Mol Biosyst. 9:2063–2078.
2013.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Screaton RA, Kiessling S, Sansom OJ,
Millar CB, Maddison K, Bird A, Clarke AR and Frisch SM:
Fas-associated death domain protein interacts with methyl-CpG
binding domain protein 4: A potential link between genome
surveillance and apoptosis. Proc Natl Acad Sci USA. 100:5211–5216.
2003.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Gómez-Angelats M and Cidlowski JA:
Molecular evidence for the nuclear localization of FADD. Cell Death
Differ. 10:791–797. 2003.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Zhang Y and Zhang Z: The history and
advances in cancer immunotherapy: Understanding the characteristics
of tumor-infiltrating immune cells and their therapeutic
implications. Cell Mol Immunol. 17:807–821. 2020.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Jiang Y, Chen M, Nie H and Yuan Y: PD-1
and PD-L1 in cancer immunotherapy: Clinical implications and future
considerations. Hum Vaccin Immunother. 15:1111–1122.
2019.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Gajewski TF, Schreiber H and Fu YX: Innate
and adaptive immune cells in the tumor microenvironment. Nat
Immunol. 14:1014–1022. 2013.PubMed/NCBI View
Article : Google Scholar
|
|
12
|
Balkwill FR, Capasso M and Hagemann T: The
tumor microenvironment at a glance. J Cell Sci. 125:5591–5596.
2012.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Mellman I, Coukos G and Dranoff G: Cancer
immunotherapy comes of age. Nature. 480:480–489. 2011.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Esfahani K, Roudaia L, Buhlaiga N, Del
Rincon SV, Papneja N and Miller WH*Jr: A review of cancer
immunotherapy: From the past, to the present, to the future. Curr
Oncol. 27 (Suppl 2):S87–S97. 2020.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Ranjan K, Waghela BN, Vaidya FU and Pathak
C: Cell-penetrable peptide-conjugated FADD induces apoptosis and
regulates inflammatory signaling in cancer cells. Int J Mol Sci.
21(6890)2020.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Xiang R, Liu Y, Zhu L, Dong W and Qi Y:
Adaptor FADD is recruited by RTN3/HAP in ER-bound signaling
complexes. Apoptosis. 11:1923–1932. 2006.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Kim WJ, Kim EJ, Kim SK, Kim YJ, Ha YS,
Jeong P, Kim MJ, Yun SJ, Lee KM, Moon SK, et al: Predictive value
of progression-related gene classifier in primary non-muscle
invasive bladder cancer. Mol Cancer. 9(3)2010.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Wang J, Zhang X, Beck AH, Collins LC,
Collins LC, Chen WY, Tamimi RM, Hazra A, Brown M, Rosner B and
Hankinson SE: Alcohol consumption and risk of breast cancer by
tumor receptor expression. Horm Cancer. 6:237–246. 2015.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Scotto L, Narayan G, Nandula SV,
Arias-Pulido H, Subramaniyam S, Schneider A, Kaufmann AM, Wright
JD, Pothuri B, Mansukhani M and Murty VV: Identification of copy
number gain and overexpressed genes on chromosome arm 20q by an
integrative genomic approach in cervical cancer: Potential role in
progression. Genes Chromosomes Cancer. 47:755–765. 2008.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Andersen JB, Spee B, Blechacz BR, Avital
I, Komuta M, Barbour A, Conner EA, Gillen MC, Roskams T, Roberts
LR, et al: Genomic and genetic characterization of
cholangiocarcinoma identifies therapeutic targets for tyrosine
kinase inhibitors. Gastroenterology. 142:1021–1031.e15.
2012.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Solé X, Crous-Bou M, Cordero D, Olivares
D, Guinó E, Sanz-Pamplona R, Rodriguez-Moranta F, Sanjuan X, de Oca
J, Salazar R and Moreno V: Discovery and validation of new
potential biomarkers for early detection of colon cancer. PLoS One.
9(e106748)2014.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Su H, Hu N, Yang HH, Wang C, Takikita M,
Wang QH, Giffen C, Clifford R, Hewitt SM, Shou JZ, et al: Global
gene expression profiling and validation in esophageal squamous
cell carcinoma and its association with clinical phenotypes. Clin
Cancer Res. 17:2955–2966. 2011.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Chen C, Méndez E, Houck J, Fan W,
Lohavanichbutr P, Doody D, Yueh B, Futran ND, Upton M, Farwell DG,
et al: Gene expression profiling identifies genes predictive of
oral squamous cell carcinoma. Cancer Epidemiol Biomarkers Prev.
17:2152–2162. 2008.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Laskar RS, Li P, Ecsedi S, Abedi-Ardekani
B, Durand G, Robinot N, Hubert JN, Janout V, Zaridze D, Mukeria A,
et al: Sexual dimorphism in cancer: Insights from transcriptional
signatures in kidney tissue and renal cell carcinoma. Hum Mol
Genet. 30:343–355. 2021.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Jones J, Otu H, Spentzos D, Kolia S, Inan
M, Beecken WD, Fellbaum C, Gu X, Joseph M, Pantuck AJ, et al: Gene
signatures of progression and metastasis in renal cell cancer. Clin
Cancer Res. 11:5730–5739. 2005.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Ivanovska I, Zhang C, Liu AM, Wong KF, Lee
NP, Lewis P, Philippar U, Bansal D, Buser C, Scott M, et al: Gene
signatures derived from a c-MET-driven liver cancer mouse model
predict survival of patients with hepatocellular carcinoma. PLoS
One. 6(e24582)2011.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Zhang Y, Foreman O, Wigle DA, Kosari F,
Vasmatzis G, Salisbury JL, van Deursen J and Galardy PJ: USP44
regulates centrosome positioning to prevent aneuploidy and suppress
tumorigenesis. J Clin Invest. 122:4362–4374. 2012.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Hou J, Aerts J, den Hamer B, van Ijcken W,
den Bakker M, Riegman P, van der Leest C, van der Spek P, Foekens
JA, Hoogsteden HC, et al: Gene expression-based classification of
non-small cell lung carcinomas and survival prediction. PLoS One.
5(e10312)2010.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Suraokar MB, Nunez MI, Diao L, Chow CW,
Kim D, Behrens C, Lin H, Lee S, Raso G, Moran C, et al: Expression
profiling stratifies mesothelioma tumors and signifies deregulation
of spindle checkpoint pathway and microtubule network with
therapeutic implications. Ann Oncol. 25:1184–1192. 2014.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Vathipadiekal V, Wang V, Wei W, Waldron L,
Drapkin R, Gillette M, Skates S and Birrer M: Creation of a human
secretome: A novel composite library of human secreted proteins:
Validation using ovarian cancer gene expression data and a virtual
secretome array. Clin Cancer Res. 21:4960–4969. 2015.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Moffitt RA, Marayati R, Flate EL, Volmar
KE, Loeza SG, Hoadley KA, Rashid NU, Williams LA, Eaton SC, Chung
AH, et al: Virtual microdissection identifies distinct tumor- and
stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat
Genet. 47:1168–1178. 2015.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Giordano TJ, Kuick R, Else T, Gauger PG,
Vinco M, Bauersfeld J, Sanders D, Thomas DG, Doherty G and Hammer
G: Molecular classification and prognostication of adrenocortical
tumors by transcriptome profiling. Clin Cancer Res. 15:668–676.
2009.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Whitington T, Gao P, Song W, Ross-Adams H,
Lamb AD, Yang Y, Svezia I, Klevebring D, Mills IG, Karlsson R, et
al: Gene regulatory mechanisms underpinning prostate cancer
susceptibility. Nat Genet. 48:387–397. 2016.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Oh SC, Sohn BH, Cheong JH, Kim SB, Lee JE,
Park KC, Lee SH, Park JL, Park YY, Lee HS, et al: Clinical and
genomic landscape of gastric cancer with a mesenchymal phenotype.
Nat Commun. 9(1777)2018.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Dom G, Tarabichi M, Unger K, Thomas G,
Oczko-Wojciechowska M, Bogdanova T, Jarzab B, Dumont JE, Detours V
and Maenhaut C: A gene expression signature distinguishes normal
tissues of sporadic and radiation-induced papillary thyroid
carcinomas. Br J Cancer. 107:994–1000. 2012.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Pappa KI, Polyzos A, Jacob-Hirsch J,
Amariglio N, Vlachos GD, Loutradis D and Anagnou NP: Profiling of
discrete gynecological cancers reveals novel transcriptional
modules and common features shared by other cancer types and
embryonic stem cells. PLoS One. 10(e0142229)2015.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Cerami E, Gao J, Dogrusoz U, Gross BE,
Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et
al: The cBio cancer genomics portal: An open platform for exploring
multidimensional cancer genomics data. Cancer Discov. 2:401–404.
2012.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Zhang Y, Chen F, Chandrashekar DS,
Varambally S and Creighton CJ: Proteogenomic characterization of
2002 human cancers reveals pan-cancer molecular subtypes and
associated pathways. Nat Commun. 13(2669)2022.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Lánczky A and Győrffy B: Web-based
survival analysis tool tailored for medical research (KMplot):
Development and implementation. J Med Internet Res.
23(e27633)2021.PubMed/NCBI View
Article : Google Scholar
|
|
40
|
Whiteside TJ: The tumor microenvironment
and its role in promoting tumor growth. Oncogene. 27:5904–5912.
2008.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Arneth B: Tumor microenvironment. Medicina
(Kaunas). 56(15)2019.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Fridman WH, Galon J, Dieu-Nosjean MC,
Cremer I, Fisson S, Damotte D, Pagès F, Tartour E and
Sautès-Fridman C: Immune infiltration in human cancer: Prognostic
significance and disease control. Curr Top Microbiol Immunol.
344:1–24. 2011.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Pagès F, Galon J, Dieu-Nosjean MC, Tartour
E, Sautès-Fridman C and Fridman WH: Immune infiltration in human
tumors: A prognostic factor that should not be ignored. Oncogene.
29:1093–1102. 2010.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Shen W, Song Z, Zhong X, Huang M, Shen D,
Gao P, Qian X, Wang M, He X, Wang T, et al: Sangerbox: A
comprehensive, interaction-friendly clinical bioinformatics
analysis platform. iMeta. 1(e36)2022.
|
|
45
|
Li T, Fu J, Zeng Z, Cohen D, Li J, Chen Q,
Li B and Liu XS: TIMER2.0 for analysis of tumor-infiltrating immune
cells. Nucleic Acids Res. 48 (W1):W509–W514. 2020.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Niu B, Ye K, Zhang Q, Lu C, Xie M,
McLellan MD, Wendl MC and Ding L: MSIsensor: Microsatellite
instability detection using paired tumor-normal sequence data.
Bioinformatics. 30:1015–1016. 2014.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Goodman AM, Kato S, Bazhenova L, Patel SP,
Frampton GM, Miller V, Stephens PJ, Daniels GA and Kurzrock R:
Tumor mutational burden as an independent predictor of response to
immunotherapy in diverse cancers. Mol Cancer Ther. 16:2598–2608.
2017.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Chalmers ZR, Connelly CF, Fabrizio D, Gay
L, Ali SM, Ennis R, Schrock A, Campbell B, Shlien A, Chmielecki J,
et al: Analysis of 100,000 human cancer genomes reveals the
landscape of tumor mutational burden. Genome Med.
9(34)2017.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Mootha VK, Lindgren CM, Eriksson KF,
Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E,
Ridderstråle M, Laurila E, et al: PGC-1alpha-responsive genes
involved in oxidative phosphorylation are coordinately
downregulated in human diabetes. Nat Genet. 34:267–273.
2003.PubMed/NCBI View
Article : Google Scholar
|
|
50
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Harris MA, Clark J, Ireland A, Lomax J,
Ashburner M, Foulger R, Eilbeck K, Lewis S, Marshall B, Mungall C,
et al: The gene ontology (GO) database and informatics resource.
Nucleic Acids Res. 32 (Database Issue):D258–D261. 2004.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Sharma VK, Singh TG, Singh S, Garg N and
Dhiman S: Apoptotic pathways and Alzheimer's disease: Probing
therapeutic potential. Neurochem Res. 46:3103–3122. 2021.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Zhou W, Lai Y, Zhu J, Xu X, Yu W, Du Z, Wu
L, Zhang X and Hua Z: The classical apoptotic adaptor FADD
regulates glycolytic capacity in acute lymphoblastic leukemia. Int
J Biol Sci. 18:3137–3155. 2022.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Lee EW, Seo J, Jeong M, Lee S and Song J:
The roles of FADD in extrinsic apoptosis and necroptosis. BMB Rep.
45:496–508. 2012.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Singh N, Baby D, Rajguru JP, Patil PB,
Thakkannavar SS and Pujari VB: Inflammation and cancer. Ann Afr
Med. 18:121–126. 2019.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Murata M: Inflammation and cancer. Environ
Health Prev Med. 23(50)2018.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Prapinjumrune C, Morita K, Kuribayashi Y,
Hanabata Y, Shi Q, Nakajima Y, Inazawa J and Omura K: DNA
amplification and expression of FADD in oral squamous cell
carcinoma. J Oral Pathol Med. 39:525–532. 2010.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Saberi E, Kordi-Tamandani DM, Jamali S and
Rigi-Ladiz MA: Analysis of methylation and mRNA expression status
of FADD and FAS genes in patients with oral squamous cell
carcinoma. Med Oral Patol Oral Cir Bucal. 19:e562–e568.
2014.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Liu Z, Zhang K, Zhao Z, Qin Z and Tang H:
Prognosis-related autophagy genes in female lung adenocarcinoma.
Medicine (Baltimore). 101(e28500)2022.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Jiang Y, Li Y, Ge H, Wu Y, Zhang Y, Guo S,
Zhang P, Cheng J and Wang Y: Identification of an autophagy-related
prognostic signature in head and neck squamous cell carcinoma. J
Oral Pathol Med. 50:1040–1049. 2021.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Fan S, Müller S, Chen ZG, Pan L,
Tighiouart M, Shin DM, Khuri FR and Sun SY: Prognostic impact of
Fas-associated death domain, a key component in death receptor
signaling, is dependent on the presence of lymph node metastasis in
head and neck squamous cell carcinoma. Cancer Biol Ther.
14:365–369. 2013.PubMed/NCBI View Article : Google Scholar
|
|
62
|
González-Moles MÁ, Ayén Á, González-Ruiz
I, de Porras-Carrique T, González-Ruiz L, Ruiz-Ávila I and
Ramos-García P: Prognostic and clinicopathological significance of
FADD upregulation in head and neck squamous cell carcinoma: A
systematic review and meta-analysis. Cancers (Basel).
12(2393)2020.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Verboom L, Martens A, Priem D, Hoste E,
Sze M, Vikkula H, Van Hove L, Voet S, Roels J, Maelfait J, et al:
OTULIN prevents liver inflammation and hepatocellular carcinoma by
inhibiting FADD- and RIPK1 kinase-mediated hepatocyte apoptosis.
Cell Rep. 30:2237–2247.e6. 2020.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Harari-Steinfeld R, Gefen M, Simerzin A,
Zorde-Khvalevsky E, Rivkin M, Ella E, Friehmann T, Gerlic M,
Zucman-Rossi J, Caruso S, et al: The lncRNA H19-derived
MicroRNA-675 promotes liver necroptosis by targeting FADD. Cancers
(Basel). 13(411)2021.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Liu W, Jing ZT, Xue CR, Wu SX, Chen WN,
Lin XJ and Lin X: PI3K/AKT inhibitors aggravate death
receptor-mediated hepatocyte apoptosis and liver injury. Toxicol
Appl Pharmacol. 381(114729)2019.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Euler M and Hoffmann MH: The double-edged
role of neutrophil extracellular traps in inflammation. Biochem Soc
Trans. 47:1921–1930. 2019.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Liew PX and Kubes P: The neutrophil's role
during health and disease. Physiol Rev. 99:1223–1248.
2019.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Murphy TL and Murphy KM: Dendritic cells
in cancer immunology. Cell Mol Immunol. 19:3–13. 2022.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Bowman BM, Sebolt KA, Hoff BA, Boes JL,
Daniels DL, Heist KA, Galbán CJ, Patel RM, Zhang J, Beer DG, et al:
Phosphorylation of FADD by the kinase CK1α promotes
KRASG12D-induced lung cancer. Sci Signal. 8(ra9)2015.PubMed/NCBI View Article : Google Scholar
|