|
1
|
Kinoshita T: Biosynthesis and biology of
mammalian GPI-anchored proteins. Open Biol.
10(190290)2020.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Coudert E, Gehant S, De Castro E, Pozzato
M, Baratin D, Neto T, Sigrist CJ, Redaschi N and Bridge A: UniProt
Consortium. Annotation of biologically relevant ligands in
UniProtKB using ChEBI. Bioinformatics. 39(btac793)2023.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Wu T, Yin F, Guang S, He F, Yang L and
Peng J: The glycosylphosphatidylinositol biosynthesis pathway in
human diseases. Orphanet J Rare Dis. 15(129)2020.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Komath SS, Fujita M, Hart GW, Ferguson MAJ
and Kinoshita T: Glycosylphosphatidylinositol anchors. In: Varki A,
Cummings RD, Esko JD, Stanley P, Hart GW, Aebi M, Mohnen D,
Kinoshita T, Packer NH, Prestegard JH (eds) et al:
Essentials of Glycobiology [Internet]. 4th edition. Cold Spring
Harbor (NY): Cold Spring Harbor Laboratory Press: Chapter 12, 2022.
Available from: https://www.ncbi.nlm.nih.gov/books/NBK579963/.
|
|
5
|
Liu YS and Fujita M: Mammalian GPI-anchor
modifications and the enzymes involved. Biochem Soc Trans.
48:1129–1138. 2020.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Liu SS, Liu YS, Guo XY, Murakami Y, Yang
G, Gao XD, Kinoshita T and Fujita M: A knockout cell library of GPI
biosynthetic genes for functional studies of GPI-anchored proteins.
Commun Biol. 4(777)2021.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Yadav U and Khan MA: Targeting the GPI
biosynthetic pathway. Pathog Glob Health. 112:115–122.
2018.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Lopez S, Rodriguez-Gallardo S, Sabido-Bozo
S and Muñiz M: Endoplasmic reticulum export of GPI-anchored
proteins. Int J Mol Sci. 20(3506)2019.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Wang Y, Maeda Y, Liu YS, Takada Y,
Ninomiya A, Hirata T, Fujita M, Murakami Y and Kinoshita T:
Cross-talks of glycosylphosphatidylinositol biosynthesis with
glycosphingolipid biosynthesis and ER-associated degradation. Nat
Commun. 11(860)2020.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Hirata T, Mishra SK, Nakamura S, Saito K,
Motooka D, Takada Y, Kanazawa N, Murakami Y, Maeda Y, Fujita M, et
al: Identification of a Golgi GPI-N-acetylgalactosamine transferase
with tandem transmembrane regions in the catalytic domain. Nat
Commun. 9(405)2018.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Lebreton S, Zurzolo C and Paladino S:
Organization of GPI-anchored proteins at the cell surface and its
physiopathological relevance. Crit Rev Biochem Mol Biol.
53:403–419. 2018.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Bellai-Dussault K, Nguyen TTM, Baratang
NV, Jimenez-Cruz DA and Campeau PM: Clinical variability in
inherited glycosylphosphatidylinositol deficiency disorders. Clin
Genet. 95:112–121. 2019.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Paprocka J, Hutny M, Hofman J, Tokarska A,
Kłaniewska M, Szczałuba K, Stembalska A, Jezela-Stanek A and
Śmigiel R: Spectrum of neurological symptoms in
glycosylphosphatidylinositol biosynthesis defects: Systematic
review. Front Neurol. 12(758899)2022.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Nakakido M, Tamura K, Chung S, Ueda K,
Fujii R, Kiyotani K and Nakamura Y: Phosphatidylinositol glycan
anchor biosynthesis, class X containing complex promotes cancer
cell proliferation through suppression of EHD2 and ZIC1, putative
tumor suppressors. Int J Oncol. 49:868–876. 2016.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Cao J, Wang P, Chen J and He X: PIGU
overexpression adds value to TNM staging in the prognostic
stratification of patients with hepatocellular carcinoma. Hum
Pathol. 83:90–99. 2019.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Martinez-Morales P, Morán Cruz I, Roa-de
la Cruz L, Maycotte P, Reyes Salinas JS, Vazquez Zamora VJ,
Gutierrez Quiroz CT, Montiel-Jarquin AJ and Vallejo-Ruiz V:
Hallmarks of glycogene expression and glycosylation pathways in
squamous and adenocarcinoma cervical cancer. PeerJ.
9(e12081)2021.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Baratang NV, Jimenez Cruz DA, Ajeawung NF,
Nguyen TTM, Pacheco-Cuéllar G and Campeau PM: Inherited
glycophosphatidylinositol deficiency variant database and analysis
of pathogenic variants. Mol Genet Genomic Med.
7(e00743)2019.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Cunningham F, Allen JE, Allen J,
Alvarez-Jarreta J, Amode MR, Armean IM, Austine-Orimoloye O, Azov
AG, Barnes I, Bennett R, et al: Ensembl 2022. Nucleic Acids Res. 50
(D1):D988–D995. 2022.PubMed/NCBI View Article : Google Scholar
|
|
19
|
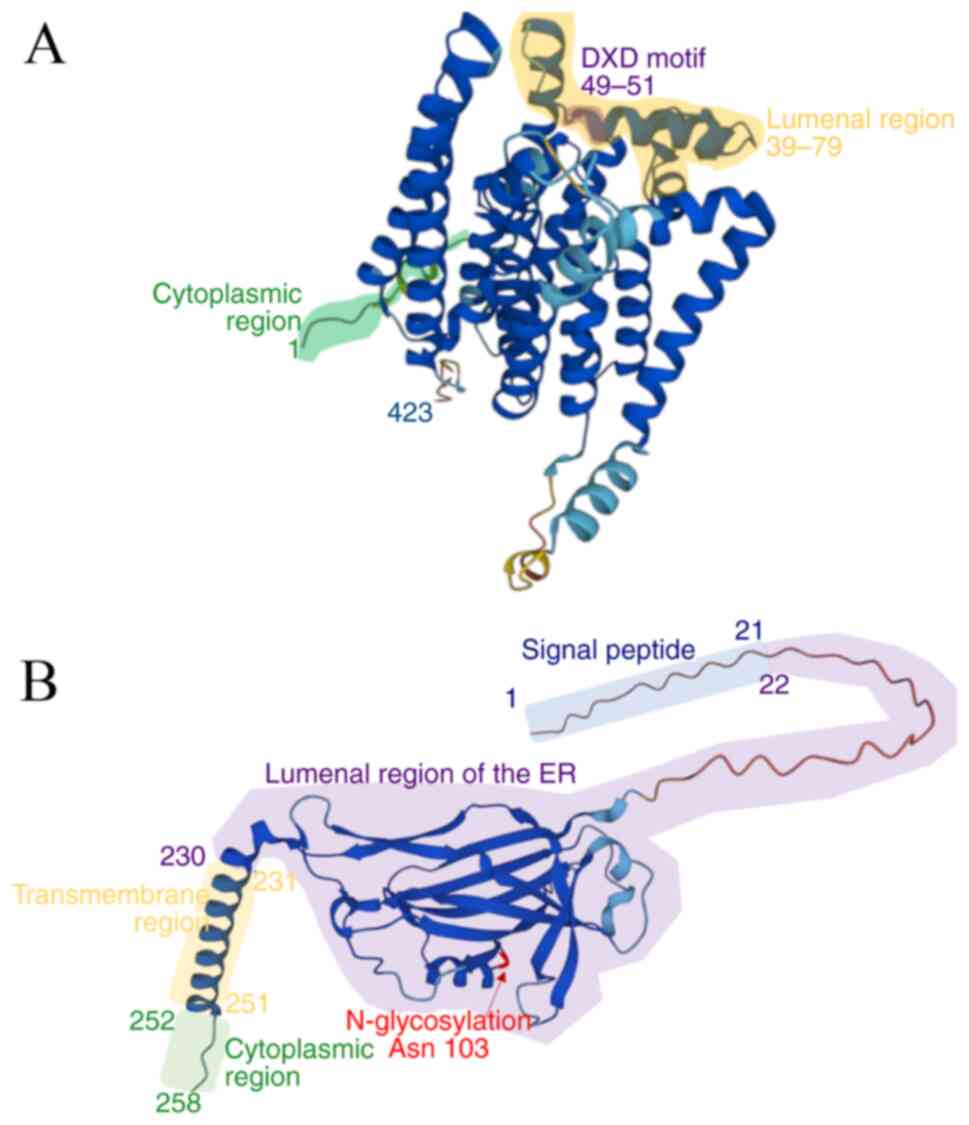
Jumper J, Evans R, Pritzel A, Green T,
Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Žídek A,
Potapenko A, et al: Highly accurate protein structure prediction
with AlphaFold. Nature. 596:583–589. 2021.PubMed/NCBI View Article : Google Scholar
|
|
20
|
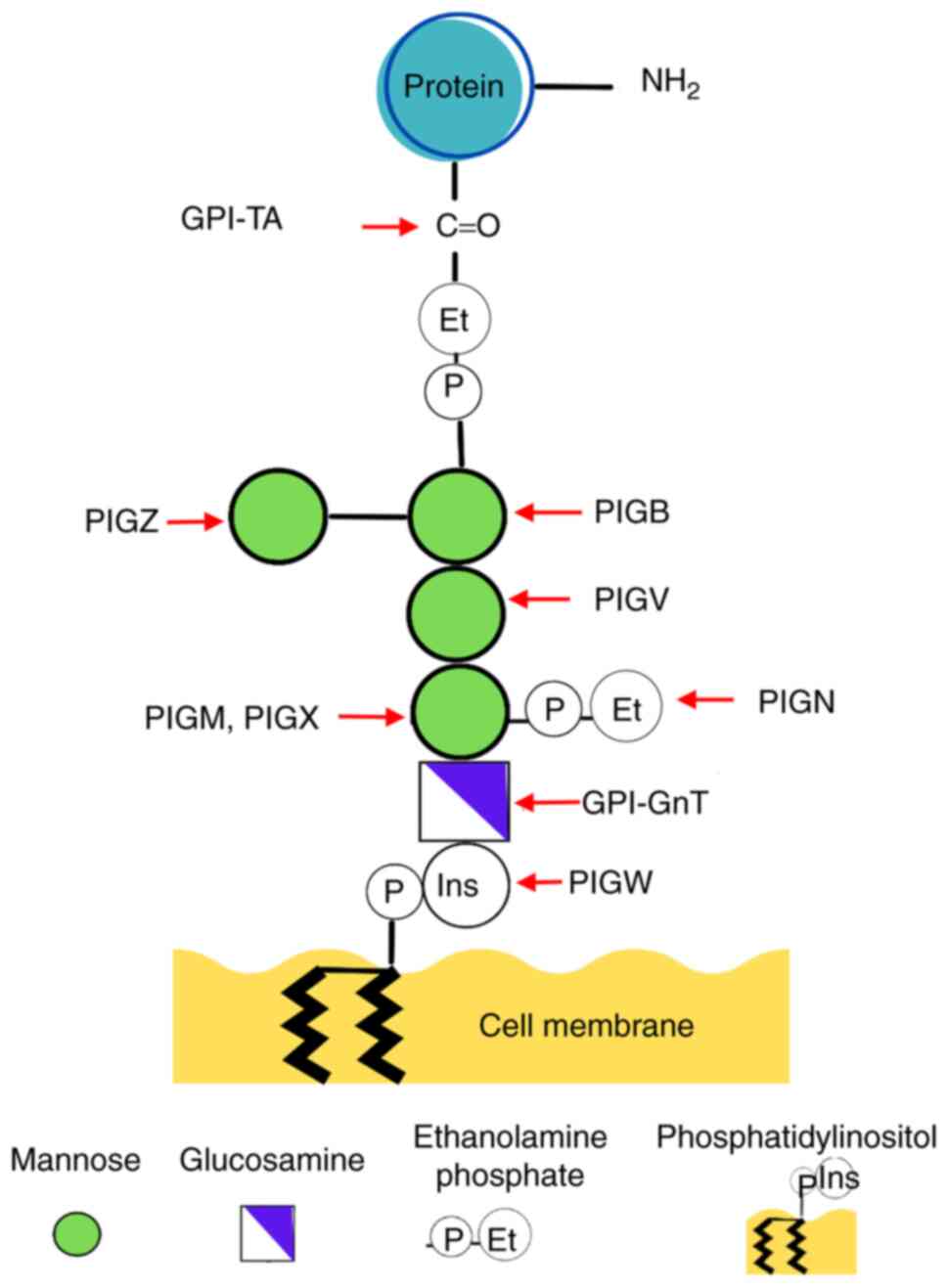
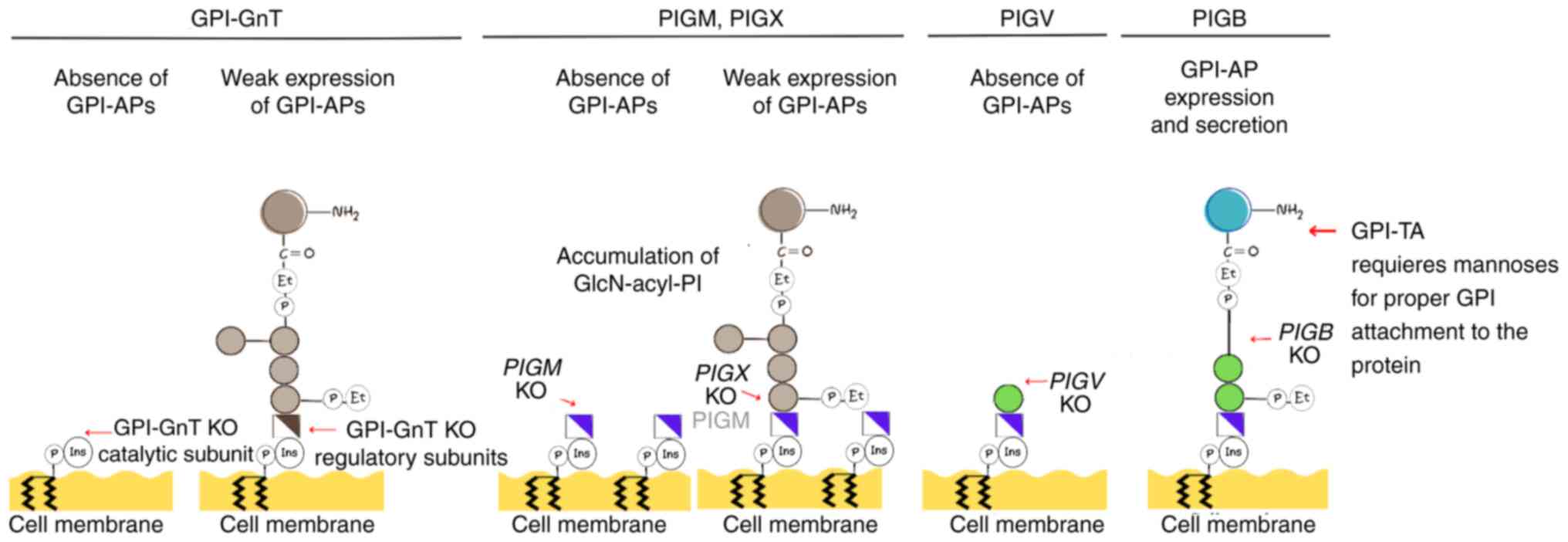
Maeda Y, Watanabe R, Harris CL, Hong Y,
Ohishi K, Kinoshita K and Kinoshita T: PIG-M transfers the first
mannose to glycosylphosphatidylinositol on the lumenal side of the
ER. EMBO J. 20:250–261. 2001.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Sayers EW, Bolton EE, Brister JR, Canese
K, Chan J, Comeau DC, Connor R, Funk K, Kelly C, Kim S, et al:
Database resources of the national center for biotechnology
information. Nucleic Acids Res. 50 UD1):D20–D26. 2022.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Kearse MG and Wilusz JE: Non-AUG
translation: A new start for protein synthesis in eukaryotes. Genes
Dev. 31:1717–1731. 2017.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Oughtred R, Rust J, Chang C, Breitkreutz
BJ, Stark C, Willems A, Boucher L, Leung G, Kolas N, Zhang F, et
al: The BioGRID database: A comprehensive biomedical resource of
curated protein, genetic, and chemical interactions. Protein Sci.
30:187–200. 2021.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Almeida AM, Murakami Y, Layton DM, Hillmen
P, Sellick GS, Maeda Y, Richards S, Patterson S, Kotsianidis I,
Mollica L, et al: Hypomorphic promoter mutation in PIGM causes
inherited glycosylphosphatidylinositol deficiency. Nat Med.
12:846–851. 2006.PubMed/NCBI View
Article : Google Scholar
|
|
25
|
GTEx Consortium: The genotype-tissue
expression (GTEx) project. Nat Genet. 45:580–585. 2013.PubMed/NCBI View Article : Google Scholar
|
|
26
|
GTEx Portal: Available in: gtexportal.org/home/gene/PIGX. Reviewed on
05/07/2023.
|
|
27
|
Stelzer G, Rosen R, Plaschkes I, Zimmerman
S, Twik M, Fishilevich S, Stein TI, Nudel R, Lieder I, Mazor Y, et
al: The GeneCards suite: From gene data mining to disease genome
sequence analyses. Curr Protoc Bioinformatics. 54:1.30.1–1.30.33.
2016.PubMed/NCBI View
Article : Google Scholar
|
|
28
|
Samaras P, Schmidt T, Frejno M, Gessulat
S, Reinecke M, Jarzab A, Zecha J, Mergner J, Giansanti P, Ehrlich
HC, et al: ProteomicsDB: A multi-omics and multi-organism resource
for life science research. Nucleic Acids Res. 48 (D1):D1153–D1163.
2020.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Maeda Y, Ashida H and Kinoshita T: CHO
glycosylation mutants: GPI anchor. Methods Enzymol. 416:182–205.
2006.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Hyman R: Somatic genetic analysis of the
expression of cell surface molecules. Trends Genet. 4:5–8.
1988.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Kang JY, Hong Y, Ashida H, Shishioh N,
Murakami Y, Morita YS, Maeda Y and Kinoshita T: PIG-V involved in
transferring the second mannose in glycosylphosphatidylinositol. J
Biol Chem. 280:9489–9497. 2005.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Ashida H, Hong Y, Murakami Y, Shishioh N,
Sugimoto N, Kim YU, Maeda Y and Kinoshita T: Mammalian PIG-X and
yeast Pbn1p are the essential components of
glycosylphosphatidylinositol-mannosyltransferase I. Mol Biol Cell.
16:1439–1448. 2005.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Bektas M, Copley-Merriman C, Khan S, Sarda
SP and Shammo JM: Paroxysmal nocturnal hemoglobinuria: Role of the
complement system, pathogenesis, and pathophysiology. J Manag Care
Spec Pharm. 26 (12-b Suppl):S3–S8. 2020.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Jeong D, Park HS, Kim SM, Im K, Yun J, Lee
YE, Ryu S, Ahn YO, Yoon SS and Lee DS: Ultradeep sequencing
analysis of paroxysmal nocturnal hemoglobinuria clones detected by
flow cytometry: PIG mutation in small PNH clones. Am J Clin Pathol.
156:72–85. 2021.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Pode-Shakked B, Heimer G, Vilboux T,
Marek-Yagel D, Ben-Zeev B, Davids M, Ferreira CR, Philosoph AM,
Veber A, Pode-Shakked N, et al: Cerebral and portal vein
thrombosis, macrocephaly and atypical absence seizures in
glycosylphosphatidyl inositol deficiency due to a PIGM promoter
mutation. Mol Genet Metab. 128:151–161. 2019.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Costa JR, Caputo VS, Makarona K, Layton
DM, Roberts IAG, Almeida AM and Karadimitris A: Cell-type-specific
transcriptional regulation of PIGM underpins the divergent
hematologic phenotype in inherited GPl deficiency. Blood.
124:3151–3154. 2014.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Moehler TM, Seckinger A, Hose D, Andrulis
M, Moreaux J, Hielscher T, Willhauck-Fleckenstein M, Merling A,
Bertsch U, Jauch A, et al: The glycome of normal and malignant
plasma cells. PLoS One. 8(e83719)2013.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Sayeeram D, Katte TV, Bhatia S, Jai Kumar
A, Kumar A, Jayashree G, Rachana DS, Nalla Reddy HV, Arvind
Rasalkar A, Malempati RL and Reddy S DN: Identification of
potential biomarkers for lung adenocarcinoma. Heliyon.
6(e05452)2020.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Pisapia DJ, Salvatore S, Pauli C, Hissong
E, Eng K, Prandi D, Sailer VW, Robinson BD, Park K, Cyrta J, et al:
Next-generation rapid autopsies enable tumor evolution tracking and
generation of preclinical models. JCO Precis Oncol.
2017(PO.16.00038)2017.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Fu F, Tao X, Jiang Z, Gao Z, Zhao Y, Li Y,
Hu H, Shen L, Sun Y and Zhang Y: Identification of germline
mutations in east-asian young never-smokers with lung
adenocarcinoma by whole-exome sequencing. Phenomics. 3:182–189.
2022.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Murakami Y, Kanazawa N, Saito K, Krawitz
PM, Mundlos S, Robinson PN, Karadimitris A, Maeda Y and Kinoshita
T: Mechanism for release of alkaline phosphatase caused by
glycosylphosphatidylinositol deficiency in patients with
hyperphosphatasia mental retardation syndrome. J Biol Chem.
287:6318–6325. 2012.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Horn D, Krawitz P, Mannhardt A, Korenke GC
and Meinecke P: Hyperphosphatasia-mental retardation syndrome due
to PIGV mutations: Expanded clinical spectrum. Am J Med Genet A.
155A:1917–1922. 2011.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Horn D, Wieczorek D, Metcalfe K, Barić I,
Paležac L, Cuk M, Petković Ramadža D, Krüger U, Demuth S, Heinritz
W, et al: Delineation of PIGV mutation spectrum and associated
phenotypes in hyperphosphatasia with mental retardation syndrome.
Eur J Hum Genet. 22:762–767. 2014.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Fu L, Liu Y, Chen Y, Yuan Y and Wei W:
Mutations in the PIGW gene associated with hyperphosphatasia and
mental retardation syndrome: A case report. BMC Pediatrics.
19(68)2019.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Aguilera-Romero A and Muñiz M: GPI
anchors: Regulated as needed. J Cell Biol.
222(e202303097)2023.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Liu YS, Wang Y, Zhou X, Zhang L, Yang G,
Gao XD, Murakami Y, Fujita M and Kinoshita T: Accumulated
precursors of specific GPI-anchored proteins upregulate GPI
biosynthesis with ARV1. J Cell Biol. 222(e202208159)2023.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Wang J, Huo K, Ma L, Tang L, Li D, Huang
X, Yuan Y, Li C, Wang W, Guan W, et al: Toward an understanding of
the protein interaction network of the human liver. Mol Syst Biol.
13(965)2017.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Huttlin EL, Ting L, Bruckner RJ, Gebreab
F, Gygi MP, Szpyt J, Tam S, Zarraga G, Colby G, Baltier K, et al:
The BioPlex network: A systematic exploration of the human
interactome. Cell. 162:425–440. 2015.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Huttlin EL, Bruckner RJ, Paulo JA, Cannon
JR, Ting L, Baltier K, Colby G, Gebreab F, Gygi MP, Parzen H, et
al: Architecture of the human interactome defines protein
communities and disease networks. Nature. 545:505–509.
2017.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Brown DA and Jacobson K: Microdomains,
lipid rafts and caveolae (San Feliu de Guixols, Spain, 19-24 May
2001). Traffic. 2:668–672. 2001.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Galbiati F, Ranani B and Lisanti MP:
Emerging themes in lipid rafts and caveolae. Cell. 106:403–411.
2001.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Suzuki KG, Fujiwara TK, Sanematsu F, Iino
R, Edidin M and Kusumi A: GPI-anchored receptor clusters
transiently recruit Lyn and G alpha for temporary cluster
immobilization and Lyn activation: Single-molecule tracking study
1. J Cell Biol. 177:717–730. 2007.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Schneider WM, Luna JM, Hoffmann HH,
Sánchez-Rivera FJ, Leal AA, Ashbrook AW, Le Pen J, Ricardo-Lax I,
Michailidis E, Peace A, et al: Genome-scale identification of
SARS-CoV-2 and pan-coronavirus host factor networks. Cell.
184:120–132.e14. 2021.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Sievers F, Wilm A, Dineen DG, Gibson TJ,
Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, et al:
Fast, scalable generation of high-quality protein multiple sequence
alignments using Clustal Omega. Mol Syst Biol.
7(539)2011.PubMed/NCBI View Article : Google Scholar
|