Introduction
Severe Acute Respiratory Syndrome Coronavirus-2
(SARS-CoV-2) is a coronavirus-related disease that has been
spreading globally since December 2019. The disease is known as
Coronavirus disease 2019 (COVID-19) and its clinical spectrum
varies significantly from asymptomatic or mild, common cold- or
influenza-like disease to a more severe lower respiratory tract
illness, associated with acute respiratory distress syndrome,
pulmonary failure, septic shock and/or multiple organ dysfunction
(1,2). Various underlying comorbidities are
considered risk factors for progression to severe COVID-19. Thus,
authoritative agencies, such as the US and European Centers for
Disease Control and Prevention have issued warnings for several
factors, which may contribute to severe outcomes in at-risk
populations, e.g. aged ≥65 years old, diabetes type 1 or type 2
diabetes mellitus, obesity (body mass index ≥35 kg/m2),
pregnancy, cancer, primary or acquired
immunodeficiency/immunosuppression, chronic tobacco smokers and
haemoglobinopathies, as well as cardiovascular, pulmonary, chronic
kidney and liver disease. However, there are also patients with no
apparent co-morbidities who eventually develop severe SARS-CoV-2
infection with poor outcomes (1).
Dyslipidemia is an established risk factor for
severe COVID-19 infection, due to several reasons. Primarily,
patients with dyslipidemia may have high levels of low-density
lipoprotein (LDL). The latter interacts with macrophages in
atherosclerotic plaques, leading to increased inflammatory gene
expression (3). Furthermore, excess
LDL accumulation in macrophage cells results in cholesterol crystal
deposition, leading to inflammasome activation and the secretion of
proinflammatory cytokines, such as IL-1β and IL-18. However, the
presence of high levels of pro-inflammatory cytokines has been
linked with severe outcomes via the ‘cytokine release syndrome’,
characterized by systemic inflammation and multiorgan dysfunction
(4). Furthermore, patients with
dyslipidemia may also have low levels of high-density lipoprotein
(HDL). This fraction itself is involved in the regulation of the
innate immune response. HDL down regulates T-cell activation and
inflammatory mediators' expression in macrophages and dendritic
cells, via interaction with ATP-binding cassette protein A1 (ABCA1)
or ABCG1. HDL levels in the acute phase of coronavirus infection
have also been associated with disease activity, as a decrease in
the number of small HDL particles is inversely associated with the
disease activity score and C-reactive protein levels (5). The aforementioned alterations
contribute to the dysregulation of the innate immune response, the
first-line defense against any invading pathogens (6). In patients with dyslipidemia, the
accumulation of LDL and triglycerides may cause additional
endothelial dysfunction (7). The
latter is accentuated during COVID-19 infections, as the SARS-CoV-2
receptor angiotensin 2-converting enzyme (ACE2) is also expressed
by endothelial cells (8). The
combination of these risk factors leads to the development of
cardiovascular complications associated with severe clinical
outcomes. Beyond innate immunity, dyslipidemia is also a critical
regulator of adaptive immunity, as it has an impact on the
differentiation and function of CD4+ T cells, CD8+T cells and
B-cells (9).
The lipid profiles of patients with COVID-19 are
quite variable (10,11). A likely explanation is that the
genetics and epigenetics of dyslipidemia and other pathologic
states may differ among patients with COVID-19. In the context of
the growing need to understand the pathogenic mechanisms of this
aggressive RNA virus and its relation to dyslipidemia and other
cardiovascular traits, the present study aimed to analyze a
comprehensive panel of specific genes involved in cardio-pulmonary,
metabolic and vascular disorders associated with COVID-19.
Materials and methods
Subjects and genetic analysis
In the present study, 60 consecutive cases were
enrolled retrospectively (2019-2021) divided into four groups,
i.e.: i) The control group, volunteers who came for a regular check
up (n=14), with an age range of 28-69 years (43% men and 57% women;
none of the subjects in this group had COVID-19 or any other
underlying pathologies); ii) adult patients (visiting the Lipid
Outpatient Departments through convenience sampling) with a type of
dyslipidemia and predisposition to atherosclerotic disease (n=18),
with an age range of 10-56 years (72% men and 28% women), and none
of the subjects in this group had COVID-19; iii) patients with
COVID-19 and mild or no symptoms (L/NO S) (n=16) with an age range
of 22-67 years (38% men and 62% women); iv) patients with COVID-19,
who were hospitalized with severe COVID-19 symptoms in the
Intensive Care Unit (ICU) of Sotiria Hospital (Athens, Greece)
(n=12) with an age range of 39-91 years (50% men and 50% women).
The collection of samples was performed by the University Research
Institute of Maternal and Child Health and Precision Medicine,
National and Kapodistrian University of Athens, in collaboration
with the ICU of Sotiria Thoracic Diseases Hospital (Athens,
Greece). All patients or their representatives/relatives consented
to their participation in the study.
Demographic, clinical and laboratory data were
recorded, blood was obtained by venipuncture and genetic material
was isolated (NucleoSpin Blood; Macherey Nagel), followed by
simultaneous analysis of the genes low density lipoprotein receptor
(LDLR), apolipoprotein B-100 (APOB-100), proprotein convertase
subtilisin/kexin type 9, lipoprotein (LP)-α, angiopoietin-like 3
gene, APOB, microsomal triglyceride transfer protein, secretion
associated, Ras related GTPase 1B, ATP-binding cassette (ABC)
transporters G5 (ABCG5), angiotensin II type I receptor (AGTR1),
11-β-hydroxysteroid dehydrogenase type 2, epithelial sodium channel
(EnaC), inducible nitric oxide synthases chromosome 17 (NOS2),
APOE, LP lipase, APOA5, APOC3, cholesteryl ester transfer protein,
scavenger receptor class B type 1, phospholipid transfer protein,
NPC intracellular cholesterol transporter 1 (NPC1), NPC2
(NIEMANN-Pick C), sphingomyelin phosphodiesterase 1 (SMPD1), fat
mass and obesity-associated gene (FTO), dual specificity tyrosine
phosphorylation regulated kinase 1B, melanocortin 4 receptor and
chymase 1, using a custom-made next-generation sequencing panel,
designed by the correspondent author and manufactured by SOPHiA
GENETICS. The exon and adjacent intrinsic/exon regions of the
above-mentioned genes were sequenced on the Illumina MiSeq platform
(Illumina, Inc.), followed by bioinformatics analyses of the
sequencing files from the validated platform of the company Sophia
Genetics DDM and the Varaft annotation tool (https://bio.tools/varaft).
Statistical analysis
Values are expressed as the mean and stand ard
deviation or the median and interquartile range. Differences
between independent samples were assessed with Student's t-test or
the Mann-Whitney U-test considering the assumption of normality,
which was checked through kurtosis, skewness and the Shapiro-Wilk
test. Statistical analysis was performed using SPSS version 26.0
(IBM Corp). P≤0.05 was considered to indicate a statistically
significant difference.
Results
Patient data
In the current study, 60 participants were included,
divided into four groups, i.e. i) The non-Covid Control group, ii)
non-Covid dyslipidemic patients, iii) Covid-19 L/NO symptoms group,
and iv) the Covid-19 ICU group. Available demographic data and
biomedical parameters are shown in Tables I and II. The median age was significantly
different among the four groups (non-Covid Controls vs. non-Covid
dyslipid and ICU, non-Covid dyslipid vs. Controls, L/NO and ICU,
L/NO vs. dyslipid and ICU, ICU vs. non-Covid Controls, non-Covid
dyslipid and L/NO; P<0.001), but there were no significant
differences in gender. Glucose levels, serum glutamic-oxaloacetic
transaminase, serum glutamic-pyruvic transaminase and γ-glutamyl
transferase levels were significantly higher in the ICU group
compared to the L/NO S group (P<0.05). Of note, total
cholesterol (TC) and LDL cholesterol levels were lower in the ICU
group compared with the L/NO S group, whereas triglycerides (TGs)
were higher.
 | Table IDemographic data. |
Table I
Demographic data.
| Item | Control (n=14) | Dyslipidemic
(n-18) | Covid-ICU
(n=12) | Covid-L/NO S
(n=16) | P-value |
|---|
| Gender | | | | | 0.189a |
|
Female | 8(57) | 5(28) | 6(50) | 10(62) | |
|
Male | 6(43) | 13(72) | 6(50) | 6(38) | |
| Age, years | | | | |
<0.001b |
|
Range | 26-72 | 8-66 | 39-91 | 21-54 | |
|
Mean | 44 | 22 | 28 | 40 | |
|
Median | 49 | 36 | 65 | 38 | |
| Ethnicity | | | | | 0.485c |
|
Greek
(Caucasians) | 14 | 16(89) | 11(92) | 16(100) | |
|
Other | - | 2(11) | 1(8) | - | |
 | Table IIDifferences in laboratory parameters
between Group 4 (ICU) vs. Group 3 (L/NO S). |
Table II
Differences in laboratory parameters
between Group 4 (ICU) vs. Group 3 (L/NO S).
| Parameter (normal
range) | ICU | L/NO S | P-value |
|---|
| HCT, % (females,
37-47; males, 42-50) | 38.30 (3.80) | 41.00 (3.00) | 0.030 |
| PLTs, K/µl
(152-433) | 190.50
(146.50) | 236.00 (57.00) | 0.235 |
| INR | 1.15±0.19 | 0.95±0.07 | 0.204 |
| PT, sec
(11-13) | 14.20 (1.60) | 19.45 (18.30) | >0.999 |
| CRP, mg/dl (low
risk, <0.1; average risk, 0.1-0.3; high risk, >0.3) | 6.85 (7.06) | 2.70 (1.70) | 0.019 |
| GLU, mg/dl
(70-99) | 197.10±86.27 | 94.31±9.95 | 0.004 |
| CREA, mg/dl
(female: 0.5-1.1; male: 0.70-1.30) | 0.77±0.23 | 0.91±0.15 | 0.091 |
| SGOT, IU/l
(10-48) | 50.00 (45.00) | 18.00 (9.00) | 0.001 |
| SGPT, IU/l
(10-40) | 57.00 (65.00) | 19.00 (7.00) | <0.001 |
| γ-GT, IU/l
(females, 8-40; males, 9-50) | 77.00 (156.00) | 19.00 (9.00) | <0.001 |
| K, mmol/l
(3.5-5.0) | 4.08±0.40 | 4.41±0.34 | 0.044 |
| Na, mmol/l
(136-145) | 138.00 (4.00) | 143.00 (5.00) | 0.042 |
| TBIL, mg/dl
(0.3-1.0) | 0.73±0.34 | 1.19±0.79 | 0.333 |
| Ferr, ng/ml
(females, 24-307; males, 24-336) | 498.00
(889.90) | 48.00 (22.00) | <0.001 |
| TC, mg/dl
(<200) | 129.82±28.33 | 215.38±46.01 | <0.001 |
| LDL, mg/dl
(<100) | 75.45±23.14 | 133.46±36.49 | <0.001 |
| HDL, mg/dl
(females, >50; males, >40 mg/dl) | 33.00 (12.00) | 55.00 (10.00) | 0.001 |
Genetic analysis
None of the subjects in Group 1 (control) had a
mutation or pathogenic variant in the studied genes.
The investigation of the patients in Group 2 of
dyslipidemic patients revealed, as predicted, mutations in the
genes LDLR, MTTP, NOS2, FTO,
APOB and AGTR1, compatible with the underlying
dyslipidemia.
In Group 3 of patients with L/NO S COVID-19, the
variant NM_001038:exon2:c.112C>T:p.P38S was detected in the
sodium channel epithelial 1 subunit α (SCNN1A) gene, which
encodes the α subunit of the ENaC, with a minor allele frequency
(Maf) <0.01, which is characterized as a variant of uncertain
significance according to the National Center for Biotechnology
Information (NCBI; https://www.ncbi.nlm.nih.gov/snp/rs3764873#clinical_significance).
In the COVID-19 Group 4 of patients with severe
symptoms, the variant NM_000336:exon5:c.786G>A:p.T262T was
detected in the SCNN1B gene, which encodes for the β subunit
of ENaC, with a Maf <0.01 (Table
III).
 | Table IIIVariants detected in the four study
groups. |
Table III
Variants detected in the four study
groups.
| A, Group 1:
Control |
|---|
| Gene | AA change | FREQ HET/500 | FREQ HOM/500 | SNP 150 | UMD prediction | Gnomad |
|---|
| PCSK9 ΝΜ
174936 |
Exon5:c.709C>T:p.R237W | 0 | 0 | rs148195424 | US | 0.0007 |
| APOB ΝΜ 000384 |
Exon29:c.12382G>A:p.V4128M | 6 | 0 | rs1801703 | P | 0.0063 |
| |
Exon26:c.10131G>A:p.L3377L | 6 | 0 | rs1799812 | P | 0.0063 |
| |
Exon26:c.9075delA:p.L3025fs | 0 | 0 | - | NA | - |
| |
Exon26:c.4449A>G:p.E1483E | 2 | 0 | rs151018874 | P | 0.0022 |
| |
Exon22:c.3337G>C:p.D1113H | 16 | 1 | rs12713844 | P | 0.0068 |
| B, Group 2:
Dyslipidemic |
| Gene | AA change | FREQ HET/500 | FREQ HOM/500 | Av SNP 150 | UMD prediction | Gnomad
(P-values) |
| AGTR1
NM_009585 |
exon2:c.30T>C:p.G10G | 0 | 0 | rs747843312 | FH | 0 |
| APOB NM_000384 |
exon26:c.11354C>T:p.T3785I | 2 | 0 | rs143710616 | - | 0 |
| |
exon26:c.5652C>T:p.N1884N | 0 | 0 | rs766106302 | - | 0 |
| |
exon22:c.3426G>A:p.S1142S | 1 | 0 | rs142448733 | FH/HYP | 0.0005 |
| |
exon9:c.1088T>C:p.V363A | 1 | 0 | rs751259935 | - | 0 |
| |
exon16:c.2412C>T:p.R804R | 0 | 0 | rs755974543 | - | 0 |
| FTO
NM_001363897 |
exon3:c.778G>T:p.A260S | 0 | 0 | - | - | - |
| LMF1
NM_001352018 |
exon4:c.115+1G>A | 3 | 0 | rs72759474 | - | 0.0011 |
| LDLR
NM_001195799 |
exon3:c.394T>C:p.C132R | 0 | 0 | rs879254558 | FH | - |
| |
exon4:c.354C>A:p.S118R | 1 | 0 | rs140241383 | FH | 0 |
| | exon7:r.spl | 3 | - | - | - | - |
| |
exon9:c.1142G>A:p.G381D | 0 | 0 | rs28941776 | FH | 0 |
| |
exon12:c.1706-10G>A | 15 | 0 | rs17248882 | FH | 0.0002 |
| |
exon12:c.1550C>T:p.P517L | 0 | 0 | rs28942084 | FH | 0.00616 |
| LMF1
NM_001352019 |
exon9:c.964G>A:p.A322T | 0 | 0 | rs186247027 | - | 0 |
| |
exon8:c.758T>G:p.V253G | 0 | 0 | rs368408082 | - | 0 |
| LPA NM_005577 |
exon16:c.2490T>C:p.N830N | 0 | 0 | - | - | 0 |
| |
exon8:c.1161T>A:p.T387T | 27 | 1 | rs4709450 | - | 0.0222 |
| |
exon8:c.1122T>C:p.N374N | 0 | 0 | - | - | 0 |
| MTTP
NM_001300785 |
exon17:c.2514G>C:p.L838F | 0 | 0 | rs144590904 | ABETA | 0.0004 |
| |
exon26:c.3173A>G:p.D1058G | 0 | 0 | rs371929273 | - | 0 |
| NOS2 NM_000625 |
exon3:c.135G>A:p.Q45Q | 2 | 0 | rs201239372 | - | 0 |
| PLTP NM_000625 |
exon7:c.447G>A:p.R149R | 6 | 0 | rs141035863 | - | 0.0002 |
| SAR1b
NM_016103 |
NM_016103:exon6:c.480+3A>- | 1 | 0 | - | - | - |
| SCARB1
NM_001367987 |
exon10:c.1243G>A:p.G415R | 6 | 0 | rs144985120 | - | 0 |
| SCNN1A
NM_001159576 |
exon5:c.1250A>G:p.E417G | 0 | 0 | rs569195112 | PHA | 0 |
| |
exon1:c.111G>A:p.P37P | 2 | 0 | rs573341191 | - | 0 |
| SCNN1B
NM_000336 |
exon2:c.245C>G:p.S82C | 6 | 6 | rs35731153 | BR | 0.0012 |
| |
exon13:c.1782G>A:p.T594T | 2 | 0 | rs13306628 | PYA | 0.0002 |
| SCNN1D
NM_001130413 |
exon3:c.129G>T:p.L43L | 0 | 0 | rs778315388 | - | 0 |
| |
exon5:c.450G>C:p.G150G | 3 | 0 | rs199854533 | - | 0 |
| |
exon12:c.1621G>C:p.V541L | 3 | 0 | rs202246275 | - | 0.001 |
| SMPD1
NM_000543 |
exon2:c.729C>T:p.A243A | 0 | 0 | rs149476159 | SCL | 0 |
| C, Group 3:
COVID-19 (L/NO S) |
| Gene | AA change | FREQ HET/500 | FREQ HOM/500 | Av SNP 150 | UMD prediction | Gnomad
(P-values) |
| APOB NM_000384 |
exon29:c.12382G>A:p.V4128M | 6 | 0 | rs1801703 | P | 0.0063 |
| |
exon26:c.10131G>A:p.L3377L | 6 | 0 | rs1799812 | P | 0.0063 |
| |
exon26:c.9321C>T:p.N3107N | 0 | 0 | rs72653101 | P | 0.0001 |
| |
exon26:c.4825T>C:p.L1609L | 1 | 0 | rs72653083 | P | 0.0019 |
| |
exon22:c.3383G>A:p.R1128H | 0 | 0 | rs12713843 | P | 0.0041 |
| |
exon22:c.3337G>C:p.D1113H | 16 | 1 | rs12713844 | P | 0.0068 |
| |
exon16:c.2258G>A:p.G753E | 2 | 0 | rs148502464 | P | 0.0002 |
| SCNN1A
NM_001038 | exon2: c.112C>
T: p.P38S | 0 | 0 | - | - | - |
| D, Group 4:
COVID-19 (ICU) |
| Gene | AA change | FREQ HET/500 | FREQ HOM/500 | Av SNP 150 | UMD prediction | Gnomad
(P-values) |
| APOB NM_000384 |
exon26:c.9736T>C:p.F3246L | 0 | 0 | - | PP | - |
| |
exon26:c.9294C>T:p.Y3098Y | 1 | 0 | rs145777339 | P | 0.0016 |
| |
exon26:c.6093T>C:p.S2031S | 0 | 0 | - | P | - |
| |
exon22:c.3337G>C:p.D1113H | 16 | 1 | rs12713844 | P | 0.0067 |
| |
exon16:c.2258G>A:p.G753E | 2 | 0 | | PP | 0.0002 |
| SCNN1B
NM_000336 |
exon5:c.786G>A:p.T262T | 0 | 0 | rs150781093 | P | 0.000028 |
| LDLR | exon7:r.spl | 3 | 0 | - | NA | - |
| NM_000527 |
exon10:c.1332C>T:p.A444A, | 0 | 0 | rs143872778 | P | 0.0002 |
| |
exon14:c.2140+5G>A | 13 | 0 | rs72658867 | NA | 0.0074 |
The P38S and T262T variants were further evaluated
for their allele frequency in the in-house Exome Sequencing
Database of the Laboratory of Medical Genetics of the St. Sophia's
Children's Hospital, NKUA (Athens, Greece). Exome Sequencing (ES)
data from 500 unrelated individuals, originally referred due to a
clinically suspected genetic condition, were also used. The cohort
included a wide range of age groups, but 80% were children and
adolescents up to the age of 18 years (12). Specifically, the allele frequency of
these variants was evaluated in 500 randomly selected samples using
the variant annotation and filter tool VarAFT (13,14).
The structure of the ENaC protein was retrieved by
the Research Collaboratory for Structural Bioinformatics Protein
Data Bank (ID no. 6WTH) at a resolution of 3.06 Å (15). An initial structural study was
performed using the Molecular Operating Environment (MOE; version
2013.08; 2016; Chemical Computing Group Inc.; https://www.chemcomp.com/en/Products.htm) platform for
the optimization of the three-dimensional protein structure and
energy minimizations and dynamic simulations under the CHARMM27
force field (16).
Amongst all retrieved sequences, the threonine
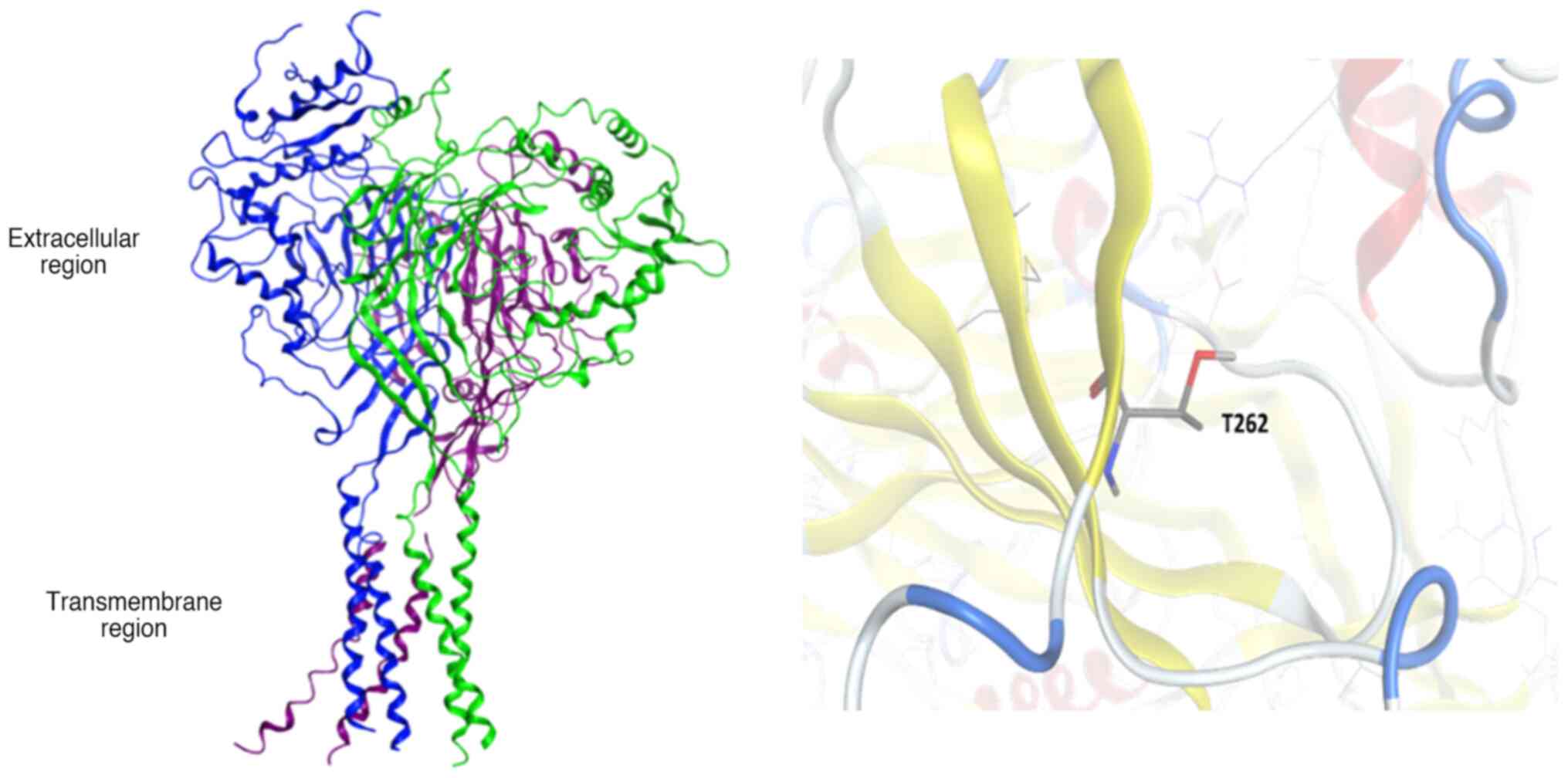
residue in position 262 was 96.95% conserved (Fig. 1).
The human ENaC protein is comprised of the three
subunits, α, β and γ. Each subunit consists of two transmembrane
helices of 25-30 amino-acid length and a short intracellular region
at the N- and C-terminus, whereas the large extracellular region of
the protein encompasses multiple domains. Residue T262-SCNN1B is
exposed on the extracellular space and no known mutations have been
reported so far (Fig. 2).
The structural analysis of the SCNN1B protein in the
present study revealed an overall rigid conformation. The
crystallographic structure of the ENaC β subunit was fixed to
remove geometric restraints using the ‘Structure Preparation’
module of the MOE platform. Energy minimization and molecular
dynamic simulations resulted in a stable conformation with T262
located on the surface of the extracellular domain. In addition,
computational mutation analysis in position 262 did not reveal any
conformational changes in the overall structure.
Genetic networks and codon bias
usage
The recovered genes were further analyzed for their
ranking and function via GeneMania network (http://www.genemania.org). The constructed genetic
network for SCNN1A and SCNN1B resulted in multiple interactions
(Fig. 3). A total of 20 genes,
including ENaC subunits α β and γ, with-no-lysine kinases and serum
and glucocorticoid-induced protein kinase, were revealed due to
their physical interactions, i.e., the overall tertiary structure
of ENaC, as well as the ENaC-specific kinases.
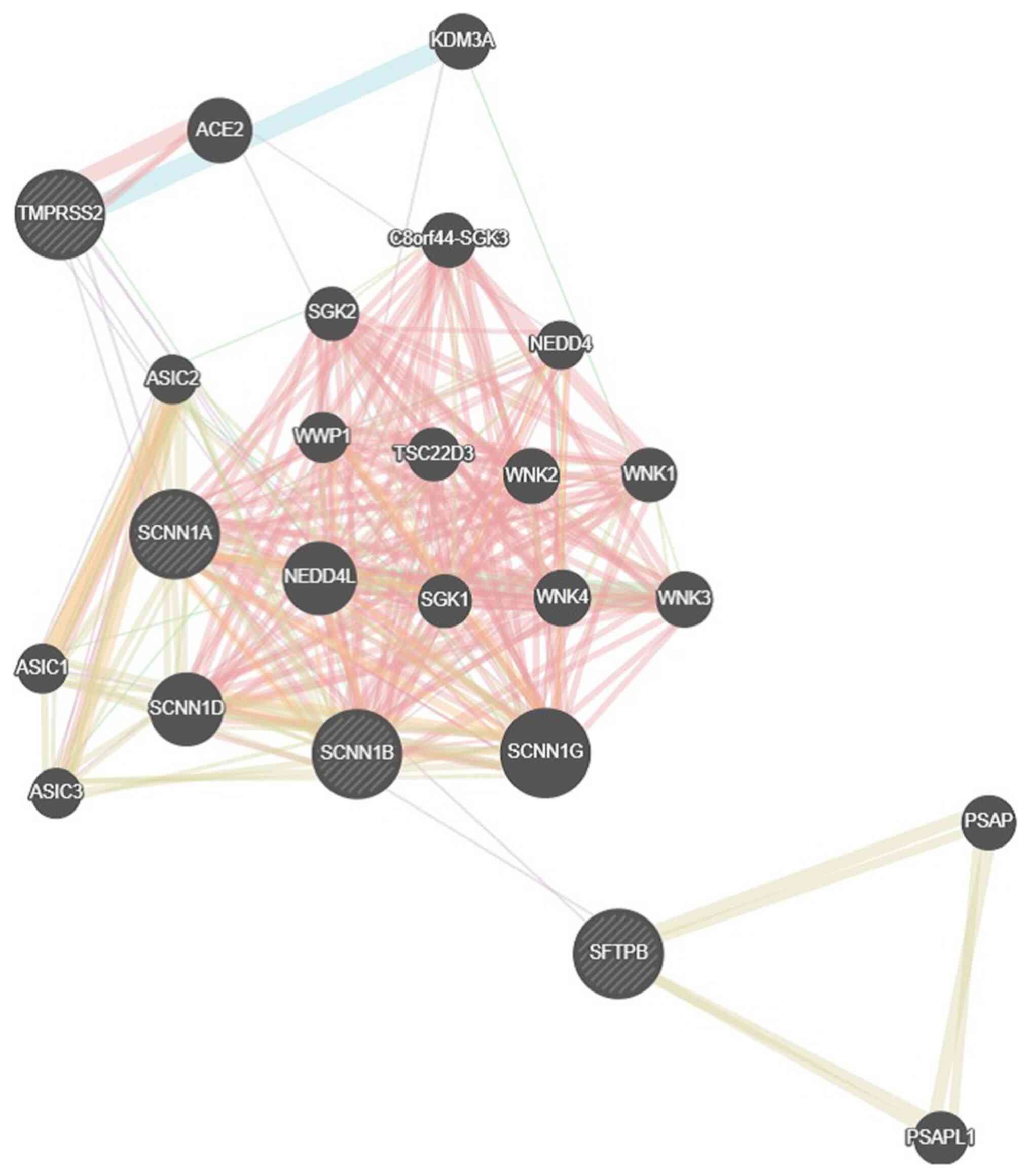 | Figure 3GeneMANIA genetic network for the
SCNN1A-SCNN1B genes. SCNN1A, sodium channel epithelial 1 subunit α
(related to the genes SCNN1G, SFTPB, ACE2, KDM3, TMPRSS2, ASIC1,
ASIC2, ASIC3, SGK1, SGK2, C8ORF44, WWP1, NEDD4, NEDD4L, TSC22D3,
WNK2, WNK1, WNK4, WNK3, PSAP and PSAPL1). |
To explore codon usage bias in the identified SCNN1B
variant, a statistical analysis was performed based on the
retrieved homologous sequences of SCNN1B. Multiple sequence
alignments result in a 99.6% identity of threonine residues in
position 262, and thus, a codon bias usage analysis was performed
to calculate the frequency of threonine codon ACA against codon
ACG. The analysis of the corresponding data for T262 codon usage of
all the retrieved sequences revealed a high frequency of ~72,3% for
the ACA codon and 12,1% for the ACG codon. The frequency of
occurrence for the human ENaC-b subunit of the ACA and ACG codon
for threonine was also calculated and was found to be 39 vs. 7%,
respectively.
Discussion
Patients with SARS-CoV-2 infection may experience a
wide range of clinical manifestations, from being asymptomatic to
critical illness and even death. Several studies have suggested
that variability in the genotype distribution of diverse gene
polymorphisms may explain the variability in disease prevalence,
morbidity and mortality of patients with COVID-19 among different
regions of the world (17). In the
current study, which aimed to identify a possible association of
the genetic profile predisposing to cardiovascular diseases in
patients with COVID-19, several findings have emerged: i) There was
a positive genetic confirmation of inherited dyslipidemia in
patients without COVID-19; ii) the ICU-COVID-19 participants
exhibited significantly lower cholesterol levels; and iii) among
all the observed variants in the present study, the rare variant
P38S of the ENaC-α subunit in the group of patients with
L/NO S COVID-19 and the rare variant T262T in the ENaC-β
subunit were identified only in the ICU patients.
Lipid disorders may increase the risk of a severe
course of COVID-19, but also the infection itself may alter the
patient's metabolic profile, mainly by impairing the function of
HDLs (18). However, our
genetically confirmed dyslipidemic patients appeared to not be
vulnerable to severe or mild COVID-19 disease. Data from a study
support the same impact of dyslipidemia in 5,279 patients. It was
demonstrated that its occurrence was not associated with an
increased risk of hospitalization (P=0.51) or mortality in patients
with COVID-19 (P=0.79) (19).
Similarly, another retrospective study of 211 patients failed to
reveal any association of dyslipidemia with an increased risk of
progression to severe COVID-19 disease (P=0.940) (20).
Another important observation was the low TC, LDL
and HDL levels between patients in the ICU-COVID-19 and L/NO S
groups (129.82±28.33 vs. 215.38±46.01, P<0.001; 75.45±23.14 vs.
133.46±36.49, P<0.001; and 33.00±12.00 vs. 55.00±10.00,
P=0.001). A recent prospective study of 108 patients with
SARS-CoV-2, which evaluated their lipid profiles in a long-term
follow-up, showed significantly lower TCs (140 vs. 175 mg/dl;
P<0.001) and LDL cholesterol levels (71.3 vs. 98 mg/dl; P=0.002)
(21).
Furthermore, in another observational
cross-sectional study, which included 1,411 hospitalized patients
with COVID-19, the usefulness of serum TC, LDL, non-HDL, HDL
cholesterol and TGs in the prognosis was assessed. Similar to the
present results, they observed that low HDL and high TGs before or
during hospitalization were strong predictors of severe COVID-19.
The researchers emphasized the notion that the lipid profile should
be considered a sensitive marker of inflammation in patients with
COVID-19. A possible explanation for the aforementioned outcomes is
that patients with acute infections experience a hypercatabolic
status combined with malnutrition; however, the contradiction of
increased TG levels remains an issue (22).
The detected ENaC gene variants in the patients with
COVID-19 were in two of the three homologous subunits (α, β and γ)
of the heterotrimeric functional channels, which are selectively
permeable to ions of sodium (Na+) (23,24).
These channels are constitutively active, allowing sodium
reabsorption from the lumen into the apical cell membrane across
epithelial cells, thus regulating the volume of the extracellular
fluid and influencing arterial blood pressure. Aldosterone
regulates their activity in the renal tubules and the distal colon,
while atrial natriuretic peptide negatively modulates their
function, leading to natriuresis and diuresis (25-30).
Of note, ENaC channels are also expressed in the lingual epithelium
and taste receptors, implicated in salt-taste perception and in
non-epithelial cells, such as endothelial cells and vascular smooth
muscle cells, where they act as mechanosensors (24,27,31).
Channels lacking the α subunit are completely
nonfunctional, whereas channels lacking the β or γ subunits are
hypofunctional (32). Human airways
express a lesser-studied ENaC δ subunit, which is phylogenetically
close to the ENaC-α subunit (33).
Inactivating the α-ENaC subunit in mice leads to defective lung
liquid clearance and premature death (34). Inactivating the β- and γ-subunits of
ENaC also leads to early death in newborn mice due to fluid and
electrolyte imbalances, suggesting that ENaC expression is critical
for fetal lung fluid absorption.
Each of the ENaC subunits have a similar structure:
A cytoplasmic N-terminus, an extracellular loop, two short
hydrophobic segments (transmembrane domains 1 and 2) and a
cytoplasmic C-terminus. The N- and C-termini are turned to the
cytoplasmic surface, whereas the extracellular loop is turned to
the extracellular space. The C-terminus of all ENaC subunits has a
highly conserved sequence - the proline tyrosine motif (29). Cleavage of the extracellular domains
renders ENaC constitutively active, whereas intracellular
conditions and signaling involving the N- and C-termini of the ENac
subunits modulate the ‘open’ vs. ‘closed’ probability
(Po) of active channels (35). Point mutations at a highly conserved
glycine residue in the N termini of any of the three subunits
markedly decrease the Po via alterations in
channel open and closed times (36).
Also, ENaC-mediated Na+ conductance is
controlled by internalization and proteasomal degradation following
ubiquitination of the intracellular N-termini of the ENaC subunits
(37). The latter process regulates
the accessibility of cleavage sites in the extracellular domain to
channel-activating proteases through conformational changes. Knight
et al (38) demonstrated
that intracellular sodium regulates the proteolytic activation of
ENaC possibly by altering the accessibility of protease cleavage
sites. Although these observations indicate that intracellular
signaling or conditions can significantly influence extracellular
cleavage and activation of ENaC, the molecular mechanism of such
transmembrane allosteric regulation of ENaC remains elusive. The
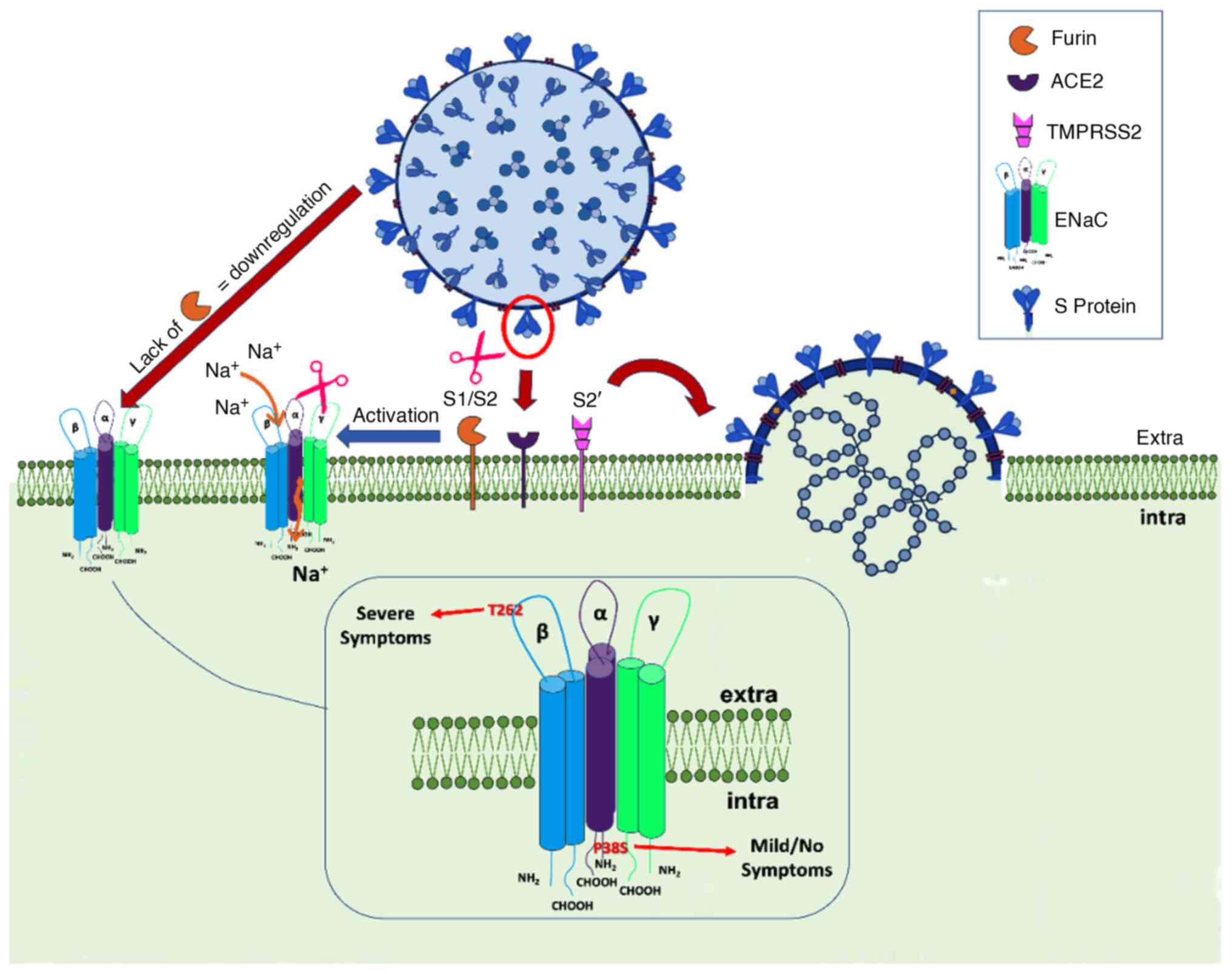
present observation of the N-terminal P38S may have a similar
impact in decreasing the Po affecting ENaC
channel activity, whereas the extracellular T262T may have a
regulatory role, given that extracellular domains of ENaC act as
receptors for regulators controlling the activity of the channel
(Fig. 4).
The expression and activity of ENaC are regulated by
the RAAS member aldosterone and furin (37). SARS-CoV-2 spike protein harbors a
furin cleavage site, which is similar to the ENaC furin-cleavable
peptide. More specifically, the SARS-CoV-2 Spike (S) protein
contains a putative furin recognition motif (680SPRRAR↓SV687) on
the S1/S2 site, which is similar to the PRSVRSV motif of Middle
Eastern respiratory syndrome coronavirus and serves as a protease
recognition site. Similar sequence patterns have been identified in
certain members of Alphacoronavirus, Betacoronavirus and
Gammacoronavirus, whereas they are absent in Coronaviruses of
zoonotic origin (Pangolin-CoV and Bat-CoV RaTG13) (39). This motif may represent an
evolutionary advantage of SARS-CoV-2, facilitating its entry into
host cells. Of note, when examining >10 million peptides of
~20,000 human proteins from UniProtKB, peptide PRRARSV is present
solely in the human ENaC-α subunit. Proteolytic activation by the
protease furin is a prerequisite for ENaC-α activation. However,
proteolytic activation of S protein by cleavage at S1/S2 is also
important for efficient viral entry into host target cells and
plays a role in host species selectivity and infectivity (39,40).
These findings suggest that SARS-CoV-2 has developed a mimicry
mechanism of a human protease substrate of furin, thus hijacking
protease pathways of ENaC-α for its activation in
SARS-CoV-2-infected cells, compromising at the same time ENaC-a
activation (41).
In addition, the present results showed that the
ENaC-α gene is co-expressed with the transmembrane protease serine
2 (TMPRSS2) gene. ENaC-α exerts its function by binding to
ACE2 and is recognized by TMPRSS2. The site at which TMPRSS2 cuts
ENaC-α is identical to a small part of the SARS-CoV-2 S-protein.
Given the high structural similarity between the S-protein and
ENaC-α, neither ACE2 nor TMPRSS2 can discriminate between the virus
and these molecules, allowing viral particles to enter host cells
(41,42).
In the present study, it was also observed that
ENaC-β is co-expressed and interacts genetically with NEDD4 like E3
ubiquitin protein ligase (NEDD4L). Nedd4L regulates the trafficking
of membrane receptors, transporters and ion channels, such as the
ENaC and as a member of HECT domain E3 ubiquitin protein ligase,
has been implicated in the cell egress phase of certain RNA
viruses, possibly high jacking the endosomal sorting complexes
required for the transport known as ESCRT-0, ESCRT-I, ESCRT-II, and
ESCRT-III. Together with a number of accessory proteins, these
ESCRT complexes enable a unique mode of membrane remodeling that
results in membranes bending/budding away from the cytoplasm.
Novelli et al (43)
identified the HECT family members of E3 ligases as likely novel
biomarkers for COVID-19.
In addition, SCNN1B is co-expressed with the
gene NEDD4L, which is involved in the regulation of insulin
and insulin-like growth factor (IGF-1) signaling by regulating the
amount of insulin receptor and IGF-1 receptor on the cell surface.
The deletion of NEDD4 in mice leads to a reduced number of
effector T-cells and a slower T-cell response to antigens,
suggesting that NEDD4 may be implicated in the conversion of native
T-cells into activated T-cells. Of note, both genes are
co-expressed with the SFTPB gene, which encodes the
pulmonary-associated surfactant B protein, an amphipathic
surfactant protein essential for lung function and homeostasis. The
latter genes encode the apolipoproteins that form ~8% of the
surfactant fluid (consisting of surfactant protein A (SP-A; 5.5%,
comprising of SP-A1 and SP-A2), SP-B (1%), SP-C (1%) and SP-D
(0.5%) (44). Pulmonary Surfactant
Metabolism Dysfunction comprises a genetically heterogeneous group
of disorders that result in severe respiratory insufficiency or
failure in full-term neonates or infants. These disorders are
associated with various pathologic entities, including pulmonary
alveolar proteinosis, desquamative interstitial pneumonitis or
cellular nonspecific interstitial pneumonitis. Thus, the
co-expression of the EAaC-α and ENaC-β genes with
SFTPB may reveal the same transcriptional regulatory
program, a functional relation and a common biological process
(43,44).
The surface of SARS-CoV-2 viral bodies is covered by
numerous glycosylated S proteins. These proteins bind to the
membrane-bound ACE2 as a first step in the entry of viral particles
into the host cell. Their entry into the cell depends on the
cleavage of protein S (in Arg-667/Ser-668) by a serine protease.
Anand et al (41) showed
that this cleavage site has a sequence pattern that is homologous
to the furin cleavage site in the ENaC channel. Gentzsch and
Rossier (45) reported that the
virus compromises the function of almost all organs by infecting
the endothelium of blood vessels, where ENaC also plays an
important role, causing inflammation and the release of cytokines
(46).
As seen by the multiple sequence analysis, T262, as
well as other amino acid residues in its proximity, are highly
conserved (Fig. 1). In an effort to
reveal a specific mechanism that may result in the association of
the SCNN1B variant and the severe pathological phenotype in
patients with SARS-CoV-2, a statistical analysis of the codon usage
bias of this synonymous mutation was performed in the present
study. The codons that correspond to the detected variant are ACA
for ‘wild-type’ threonine and ACG for the ‘mutated’ threonine. The
analysis of the corresponding data for T262 codon usage of all the
retrieved sequences reveals a high frequency of ~72.3% for the ACA
codon and 12.1% for the ACG codon. The frequency of occurrence for
the human ENaC-β subunit of the ACA and ACG codon for threonine was
also calculated and was found to be 39 vs. 7%, respectively. At an
intra-species level, codon usage for threonine in Homo
sapiens corresponds to 15.1% (ACA) against 6.1% (ACG), also
revealing the preference for ACA usage (47). Codon usage bias is well established
and plays a crucial role in regulating gene expression. Not only
synonymous codons and their corresponding tRNA availability are a
way of fine-tuning the expression of genes; it has also been shown
that synonymous codons cluster in the coding sequence, resulting in
co-occurrence bias that mediates high expression levels.
The analysis of the ENaC structure and the SCNN1B
T262T variant revealed a stable conformation of the extracellular
domain and the neighboring region of T262 that is highly conserved.
No structural feature was identified that could indicate a
mechanism linked to SARS-CoV-2 infection, particularly for position
262. However, the codon usage bias for this synonymous mutation
could point to a regulatory mechanism in terms of gene expression.
The detected NM_000336:exon5:c.786G>A variant is most likely to
result in a lack of tRNA availability for the alternative codon,
leading to deficient SCNN1B expression.
In conclusion, a dysfunctional lipid profile due to
the genetic phenotype or underlying diseases may be considered a
prediction tool for COVID-19 severity. In addition, the
identification of the two rare ENaC variants in ICU and L/NO S
patients in the coding region may be predictive of whether the ENaC
channel is involved in ENaC-mediated SARS-CoV-2 entry. Therefore,
the effect of SARS-CoV-2 infection on ENaC function in different
cells of the upper and lower respiratory tract and at different
stages of the disease should be studied in a larger population to
reinforce this hypothesis and further clarify its possible
pathophysiologic role in COVID-19 severity and progression.
Physicians should also be engaged in close
monitoring of dyslipidemia patients with suspected COVID-19, for
detecting signs of disease progression in a timely fashion.
Finally, the presence of dyslipidemia may be an important factor in
future risk stratification models for COVID-19.
Acknowledgements
Not applicable.
Funding
Funding: The authors gratefully acknowledge the financial
support of Synenosis, Greek Shipowners' Social Welfare Company and
especially the Angelakos Evangelos family.
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author (raw data are available at
http://www.ncbi.nlm.nih.gov/bioproject/1136239).
Authors' contributions
EK, KH, AN, KG, DV, EP, NM, NR, SM and GPC conceived
the study design and were involved in data interpretation. PB, AA,
AtK and AnK, VE and JTS collected and analysed the data. GPC made
critical revisions to the manuscript. EK and GPC checked and
confirmed the authenticity of the raw data. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
The study was approved by each Ethical Committee of
the University Research Institute of Maternal and Child Health and
Precision Medicine and UNESCO Chair on Adolescent Health Care and
the National and Kapodistrian University of Athens and the ICU,
First Department of Pulmonary Medicine, National and Kapodistrian
University of Athens and Sotiria Hospital (Athens, Greece). All
patients provided written informed consent to participate in this
study according to the General Data Protection Regulation.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Huang C, Wang Y, Li X, Ren L, Zhao J, Hu
Y, Zhang L, Fan G, Xu J, Gu X, et al: Clinical features of patients
infected with 2019 novel coronavirus in Wuhan, China. Lancet.
395:497–506. 2020.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Wang D, Hu B, Hu C, Zhu F, Liu X, Zhang J,
Wang B, Xiang H, Cheng Z, Xiong Y, et al: Clinical characteristics
of 138 hospitalized patients with 2019 novel coronavirus-infected
pneumonia in Wuhan, China. JAMA. 323:1061–1069. 2020.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Tall AR and Yvan-Charvet L: Cholesterol,
inflammation and innate immunity. Nat Rev Immunol. 15:104–116.
2015.PubMed/NCBI View
Article : Google Scholar
|
|
4
|
Soy M, Keser G, Atagündüz P, Tabak F,
Atagündüz I and Kayhan S: Cytokine storm in COVID-19: Pathogenesis
and overview of anti-inflammatory agents used in treatment. Clin
Rheumatol. 39:2085–2094. 2020.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Kaji H: High-density lipoproteins and the
immune system. J Lipids. 2013(684903)2013.PubMed/NCBI View Article : Google Scholar
|
|
6
|
McKechnie JL and Blish CA: The innate
immune system: Fighting on the front lines or fanning the flames of
COVID-19? Cell Host Microbe. 27:863–869. 2020.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Kim JA, Montagnani M, Chandrasekran S and
Quon MJ: Role of lipotoxicity in endothelial dysfunction. Heart
Fail Clin. 8:589–607. 2012.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Froldi G and Dorigo P: Endothelial
dysfunction in Coronavirus disease 2019 (COVID-19): Gender and age
influences. Med Hypotheses. 144(110015)2020.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Kim D, Chung H, Lee JE, Kim J, Hwang J and
Chung Y: Immunologic aspects of dyslipidemia: A critical regulator
of adaptive immunity and immune disorders. J Lipid Atheroscler.
10:184–201. 2021.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Lei P, Zhang L, Han P, Zheng C, Tong Q,
Shang H, Yang F, Hu Y, Li X and Song Y: Liver injury in patients
with COVID-19: Clinical profiles, CT findings, the correlation of
the severity with liver injury. Hepatol Int. 14:733–742.
2020.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Malik J, Laique T, Ishaq U, Ashraf A,
Malik A, Ali M, Zaidi SMJ, Javaid M and Mehmood A: Effect of
COVID-19 on lipid profile and its correlation with acute phase
reactants. medRxiv: doi: https://doi.org/10.1101.
|
|
12
|
Marinakis NM, Svingou M, Veltra D, Kekou
K, Sofocleous C, Tilemis FN, Kosma K, Tsoutsou E, Fryssira H and
Traeger-Synodinos J: Phenotype-driven variant filtration strategy
in exome sequencing toward a high diagnostic yield and
identification of 85 novel variants in 400 patients with rare
Mendelian disorders. Am J Med Genet A. 185:2561–2571.
2021.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Tilemis FN, Marinakis NM, Veltra D,
Svingou M, Kekou K, Mitrakos A, Tzetis M, Kosma K, Makrythanasis P,
Traeger-Synodinos J and Sofocleous C: Germline CNV detection
through whole-exome sequencing (WES) data analysis enhances
resolution of rare genetic diseases. Genes (Basel).
14(1490)2023.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Desvignes JP, Bartoli M, Delague V, Krahn
M, Miltgen M, Béroud C and Salgado D: VarAFT: A variant annotation
and filtration system for human next generation sequencing data.
Nucleic Acids Res. 46 (W1):W545–W553. 2018.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Noreng S, Posert R, Bharadwaj A, Houser A
and Baconguis I: Molecular principles of assembly, activation, and
inhibition in epithelial sodium channel. Elife.
9(e59038)2020.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Foloppe N and MacKerell AD Jr: All-Atom
empirical force field for nucleic Acids: 2) parameter optimization
based on small molecule and condensed phase macromolecular target
data. J Comput Chem. 21:86–104. 2000.
|
|
17
|
Yamamoto T, Uchiumi C, Suzuki N, Yoshimoto
J and Murillo-Rodriguez E: The psychological impact of ‘mild
lockdown’ in Japan during the COVID-19 pandemic: A nationwide
survey under a declared state of emergency. Int J Environ Res
Public Health. 7(9382)2020.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Li H, Xiang X, Ren H, Xu L, Zhao L, Chen
X, Long H, Wang Q and Wu Q: Serum amyloid A is a biomarker of
severe coronavirus disease and poor prognosis. J Infect.
80:646–655. 2020.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Petrilli CM, Jones SA, Yang J, Rajagopalan
H, O'Donnell L, Chernyak Y, Tobin KA, Cerfolio RJ, Francois F and
Horwitz LI: Factors associated with hospital admission and critical
illness among 5279 people with coronavirus disease 2019 in New York
City: Prospective cohort study. BMJ. 369(m1966)2020.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Chang MC, Park YK, Kim BO and Park D: Risk
factors for disease progression in COVID-19 patients. BMC Infect
Dis. 20(445)2020.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Aparisi A, Martín-Fernández M,
Ybarra-Falcón C, Gil J, Carrasco-Moraleja M, Martinez-Paz P,
Cusacovich I, Gonzal-Benito H, Fuertes R, Marcos-Mangas M, et al:
Dyslipidemia and Inflammation as Hallmarks of oxidative stress in
COVID-19: A follow up study. Int J Mol Sci.
23(15350)2022.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Masana L, Correig E, Ibarretxe D, Anoro E,
Arroyo JA, Jericó C, Guerrero C, Miret M, Näf S, Pardo A, et al:
Low HDL and high triglycerides predict COVID-19 severity. Sci Rep.
11(7217)2021.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Mano I and Driscoll M: DEG/ENaC channels:
A touchy superfamily that watches its salt. Bioessays. 21:568–578.
1999.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Drummond HA, Grifoni SC and Jernigan NL: A
New Trick for an Old Dogma: ENaC proteins as mechanotransducers in
vascular smooth muscle. Physiology (Bethesda). 23:23–31.
2008.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Govindan R, Banerjee P, Dhania NK and
Senapati S: FTIR based approach to study EnaC mechanosensory
functions. Prog Biophys Mol Biol. 167:79–86. 2021.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Kashlan OB and Kleyman TR: ENaC structure
and function in the wake of a resolved structure of a family
member. Am J Physiol Renal Physiol. 301:F684–F696. 2011.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Baldin JP, Barth D and Fronius M:
Epithelial Na+ channel (ENaC) formed by one or two subunits forms
functional channels that respond to shear force. Front Physiol.
11(141)2020.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Champigny G, Voilley N, Lingueglia E,
Friend V, Barbry P and Lazdunski M: Regulation of expression of the
lung amiloride-sensitive Na+ channel by steroid hormones. EMBO J.
13:2177–2181. 1994.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Hanukoglu I and Hanukoglu A: Epithelial
sodium channel (ENaC) family: Phylogeny, structure-function, tissue
distribution, and associated inherited diseases. Gene. 579:95–132.
2016.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Kellenberger S and Schild L: International
Union of Basic and Clinical Pharmacology. XCI. structure, function,
and pharmacology of acid-sensing ion channels and the epithelial
Na+ Channel. Pharmacol Rev. 67:1–35. 2015.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Golestaneh N, Klein C, Valamanesh F,
Suarez G, Agarwal MK and Mirshahi M: Mineralocorticoid
receptor-mediated signaling regulates the ion gated sodium channel
in vascular endothelial cells and requires an intact cytoskeleton.
Biochem Biophys Res Commun. 280:1300–1306. 2001.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Canessa CM, Schild L, Buell G, Thorens B,
Gautschi I, Horisberger JD and Rossier BC: Amiloride-sensitive
epithelial Na+ channel is made of three homologous subunits.
Nature. 367:463–467. 1994.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Waldmann R, Champigny G, Bassilana F,
Voilley N and Lazdunski M: Molecular cloning and functional
expression of a novel amiloride-sensitive Na+ channel. J Biol Chem.
270:27411–27414. 1995.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Hummler E, Barker P, Gatzy J, Beermann F,
Verdumo C, Schmidt A, Boucher R and Rossier BC: Early death due to
defective neonatal lung liquid clearance in alpha-ENaC-deficient
mice. Nat Genet. 12:325–328. 1996.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Tong Q, Gamper N, Medina JL, Shapiro MS
and Stockand JD: Direct activation of the epithelial Na(+) channel
by phosphatidylinositol 3,4,5-trisphosphate and
phosphatidylinositol 3,4-bisphosphate produced by phosphoinositide
3-OH kinase. J Biol Chem. 279:22654–22663. 2004.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Gründer S, Firsov D, Chang SS, Jaeger NF,
Gautschi I, Schild L, Lifton RP and Rossier BC: A mutation causing
pseudohypoaldosteronism type 1 identifies a conserved glycine that
is involved in the gating of the epithelial sodium channel. EMBO J.
16:899–907. 1997.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Ruffieux-Daidié D, Poirot O, Boulkroun S,
Verrey F, Kellenberger S and Staub O: Deubiquitylation regulates
activation and proteolytic cleavage of ENaC. J Am Soc Mephrol.
19:2170–2180. 2008.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Knight KK, Wentzlaff DM and Snyder PM:
Intracellular sodium regulates proteolytic activation of the
epithelial sodium channel. J Biol Chem. 283:27477–27482.
2008.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Örd M, Faustova I and Loog M: The sequence
at Spike S1/S2 site enables cleavage by furin and
phospho-regulation in SARS-CoV2 but not in SARS-CoV1 or MERS-CoV.
Sci Rep. 10(16944)2020.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Bonny O and Hummler E: Dysfunction of
epithelial sodium transport: From human to mouse. Kidney Int.
57:1313–1318. 2000.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Anand P, Puranik A, Aravamudan M,
Venkatakrishnan AJ and Soundararajan V: SARS-CoV-2 strategically
mimics proteolytic activation of human ENaC. Elife.
9(e58603)2020.PubMed/NCBI View Article : Google Scholar
|
|
42
|
V'kovski P, Kratzel A, Steiner S, Stalder
H and Thiel V: Coronavirus biology and replication: Implications
for SARS-CoV-2. Nat Rev Microbiol. 19:155–170. 2021.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Novelli G, Biancolella M, Mehrian-Shai R,
Colona VL, Brito AF, Grubaugh ND, Vasiliou V, Luzzatto L and
Reichardt JKV: COVID-19 one year into the pandemic: From genetics
and genomics to therapy, vaccination, and policy. Hum Genomics.
15(27)2021.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Gandhi CK, Chen C, Wu R, Yang L, Thorenoor
N, Thomas NJ, DiAngelo SL, Spear D, Keim G, Yehya N and Floros J:
Association of SNP-SNP interactions of surfactant protein genes
with pediatric acute respiratory failure. J Clin Med.
9(1183)2020.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Gentzsch M and Rossier BC: A
pathophysiological model for COVID-19: Critical importance of
transepithelial sodium transport upon airway infection. Function
(Oxf). 1(zqaa024)2020.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Ji HL, Song W, Gao Z, Su XF, Nie HG, Jiang
Y, Peng JB, He YX, Liao Y, Zhou YJ, et al: SARS-CoV proteins
decrease levels and activity of human ENaC via activation of
distinct PKC isoforms. Am J Physiol Lung Cell Mol Physiol.
296:L372–L383. 2009.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Quax TE, Claassens NJ, Söll D and van der
Oost J: Codon bias as a means to fine-tune gene expression. Mol
Cell. 59:149–161. 2015.PubMed/NCBI View Article : Google Scholar
|