Introduction
Pancreatic cancer (PC) is a disease with low
incidence and high mortality rate. American Cancer Society
estimates that ~66,440 people (31,910 female and 34,530 male
patients) will be diagnosed with PC and ~51,750 people (24,480
female and 27,270 male patients) will die of the disease in the
United States in 2024. Despite progress in research on PC and
management of the disease, the 5-year survival rate remains ~10%
(1). PC is the 4th leading cause of
cancer death in the USA, after lung, colon and breast cancer and
the 7th foremost cause of worldwide cancer-related death (1). Poor prognosis of PC is due to
non-specific symptoms and silent growth until advanced progression
of the disease; there are few diagnostic methods sufficiently
sensitive and specific to detect the disease early (2). Furthermore, retroperitoneal position
of the pancreas prevents accurate physical examination of the organ
and there has not been consensus on the optimum usage of diagnostic
imaging for early detection of PC. Even after using the most
advanced imaging techniques, lesions <3 cm in size are not
detected (2,3). Among non-invasive biomarkers,
carbohydrate antigen 19-9 (CA 19-9) is the only molecule used in
management of PC (4). However,
there are reports of alteration of CA 19-9 in benign pancreatic
diseases and gastrointestinal inflammation, thereby decreasing its
specificity as a biomarker for pancreatic malignancy (4,5).
There has been some progress in molecular diagnosis.
Technological advancements facilitate detection of circulating
cancer cells, circulating microRNAs (miRNAs) and proteins for early
diagnosis of PC and predict prognosis of the disease (6). Non-coding (nc)RNAs such as long nc
(lnc)RNAs, circular RNAs (circRNAs) and piwi-interacting RNAs
(piRNAs) serve vital roles in the regulation of tumorigenesis,
tumour progression and prognosis in multiple types of cancer
including colon, breast, lung, gastric and liver cancer, PC,
glioblastoma, leukemia (7).
Multiple studies have established involvement of specific lncRNAs
(Homeobox Transcript Antisense Intergenic RNA, Plasmacytoma Variant
Translocation 1, H19-H19 Imprinted Maternally Expressed Transcript,
myocardial Infarction Associated Transcript, GAS5-Growth Arrest
Specific 5 etc.), circRNAs (circPDAC, circFOXK2-Circular RNA
Forkhead Box K2, ciRS-7-Circular RNA Sponge For MiR-7,
hsa_circ_0007534 etc.) and piRNAs (piR-162725, piR-017061) in the
regulation of gene expression and control of several signal
transduction pathways in PC (7,8).
piRNAs are a type of short, single-stranded RNA 21-35 nucleotides
in length. piRNAs interact with PIWI proteins to silence
transposable elements (TEs) and maintain genome stability and
integrity. piRNAs regulate endogenous genes mainly through RNA
degradation (9,10). piRNAs-mouse PIWI (MIWI) protein
interaction may target mRNAs with imperfect base pairing to promote
their degradation by MIWI-dependent cleavage, thereby regulating
gene expression and contributing to disease phenotype (9,10).
piRNAs serve as non-invasive biomarkers since they are also found
in body fluids such as blood, saliva, gastric juice and urine
(11). To the best of our
knowledge, however, there is little information on the role of
piRNAs in pancreatic ductal adenocarcinoma (PDAC). Transcriptome
analysis of pancreatic tumour tissues has identified lncRNAs,
miRNAs and piRNAs that are altered in a tumour-specific manner
(12). Another study reported
candidate piRNAs isolated from plasma of patients with PC (13) and a separate study listed piRNAs
that are differentially expressed (DE) in patients with PC
(14). On the other hand, other
studies have investigated the functional aspects of selected piRNAs
and their interactions in PC (15,16).
The present study performed small RNA sequencing
analysis of piRNAs from both tissues and plasma of patients with PC
and controls. Target genes were subsequently identified and the
pathways involved in disease development were predicted. A similar
analysis using plasma samples from patients with chronic
pancreatitis (CP) was performed to identify piRNAs that may
contribute to chronic inflammation.
Materials and methods
Patients and bio-specimen
collection
A total of 16 healthy individuals and 15 pancreatic
cancer patients were recruited between April 2015 to August 2019
with age range of 20 to 70 years for PC patients and 20 to 55 years
for normal individuals. In both PC and normal individuals the
female to male ratio was about 3:2. Surgical tissue and plasma
samples of patients with confirmed PC (pancreatic ductal
adenocarcinoma) and not undergoing any chemotherapy were obtained
from the Institute of Postgraduate Medical Education & Research
and the Chittaranjan National Cancer Institute (both Kolkata,
India). The study was approved by the Institutional Ethics
Committee (INST/IEC/2015/218 and IPGME&R/IEC/2022/318 for
Institute of Postgraduate Medical Education & Research-Research
Oversight Committee and CNCI-IEC-SG2-2023-69 for Chittaranjan
National Cancer Institute-Institutional Ethics Committee). Written
informed consent was procured from all participants prior to the
study. A total of 5 ml peripheral venous blood was collected in
vacutainer tubes (BD Biosciences) before routine surgery and plasma
samples were processed as previously described (17). Normal plasma samples were collected
from healthy individuals with no history of pancreatic disease and
were processed in the same way. Tumour and adjacent normal
pancreatic tissue (>5 cm from tumour margin) were collected from
patients with PC. The samples were stored at -80˚C until use.
Simultaneously, resected specimens were processed for
histopathological assessment to confirm malignant or benign nature
(Table SI).
RNA isolation and quality control
Total RNA enriched with small RNAs was isolated from
the plasma samples using miRNAeasy Serum/Plasma advanced kit
(Qiagen GmbH). Briefly, 200 µl plasma sample was centrifuged at
high speed of 16,000 x g for 5 min at 4˚C to remove any cellular
debris or particulate matter that may interfere with downstream RNA
isolation. Next, QIAzol lysis reagent was followed by vortexing and
phase separation after adding chloroform as per the manufacturer's
instructions. The aqueous phase was then separated, and ethanol
precipitation of RNA was performed followed by passage through the
column provided with the kit. Column-bound RNA was washed and
eluted using the buffer solution, according to the manufacturer's
instructions. Quantification was performed using a multi-channel
spectrophotometer (ND 8000; Thermo Fisher Scientific, Inc.)
according to the manufacturer's instructions. Additionally, total
RNA was isolated from tissue using QIAzol (Qiagen GmbH) and
PureLink RNA mini kit (Ambion; Thermo Fisher Scientific, Inc.)
according to the manufacturer's instructions. The quality of
isolated total RNA was determined using Agilent RNA 6000 Nano chips
in a 2100 Bioanalyzer (Agilent Technologies, Inc.) and NanoDrop
spectrophotometer (Thermo Fisher Scientific, Inc.). Quantification
was performed using Qubit and the Quant-iT RNA assay kit broad
range (Thermo Fisher Scientific, Inc.). Total RNA samples with RNA
integrity number >7 were selected for small RNA library
preparation and Illumina sequencing.
Small RNA library preparation and
sequencing
The quality of isolated small RNA was checked using
small RNA chips in a 2100 Bioanalyzer (Agilent Technologies, Inc.)
and quantitation was performed using a Qubit Fluorometer. Small RNA
sequencing library preparation was performed using
Illumina® TruSeq® Small RNA Library Prep kit
(Illumina, Inc.; cat. no. RS-200-0012) according to the
manufacturer's instructions. A total of 10 ng isolated small RNA
was used for library preparation. The first step was to ligate
adapters to 3' and 5' ends of the RNA molecule. Subsequently
reverse transcription and amplification were performed to generate
a cDNA library, using the reagents provided in kit following
manufacturer's instructions. Gel purification step that selects
bands 145-160 bp long was performed to prepare the final small RNA
sequencing library for clustering and sequencing. The quality of
small RNA sequencing libraries was checked using high sensitivity
D1000 screen tape in a 2200 TapeStation (Agilent Technologies,
Inc.) and final library quantification was performed using a Qubit
Fluorometer (Thermo Fisher Scientific, Inc.). Single end 1X 50 bp
sequencing of pooled libraries was performed in a Novaseq 6000
(Illumina, Inc.).
Analysis of sequencing data.
Preprocessing and quality control of piRNA sequencing data
Initial quality control and visualization of small
RNA sequencing data were performed using FastQC (version 0.12.0)
(18) and MultiQC (v1.24) (19). Adapter trimming (Illumina TruSeq
small RNA adapters) was performed using the TrimGalore tool
(v0.6.10) (20). Sequence reads of
a length of 24-35 nucleotides were retained in the analysis. Poor
quality reads (Phred score <20) were filtered out.
Alignment of reads to the reference genome and
quantification of piRNA expression. The filtered sequencing
reads were aligned to the human genome reference (hg19) using
Bowtie2 aligner (Version 2.5.1) (21). Quality-checking of the sequencing
alignment data was performed using SAMtools (v1.21) (22), Sambamba (v0.5.0) (23) and Qualimap (v2.3) (24). Aligned sequencing reads were
overlapped with other small RNA sequencing information from the
DASHR (v2.0) database (in BED file format) (25) to filter out other small ncRNA. Raw
piRNA expression counts were quantified using the Featurecounts
tool (v1.6.0.3) (26), with a
piRNAdb annotation file (version 1.7.5; reference genome, hg19)
(27) in GTF format. For
normalization of the count data, ‘estimateSizeFactors’ was used to
calculate size factors for each sample, using the Median ratios
method and normalized counts were obtained using ‘counts’ of
DESeq2.
Analysis of differential piRNA expression.
The R package DEseq2 (version 1.12.3) (28) was used to identify DE piRNAs. Wald
test was used for assessing statistical significance with adjusted
P-value <0.1 as the threshold and -log2FC >0.58 or <-0.58
for up- and downregulated piRNA, respectively. Visualization of DE
piRNAs was performed through heatmap and volcano plots, using R
packages pheatmap (10.32614/CRAN.package.pheatmap) and dplyr
(10.32614/CRAN.package.dplyr). A detailed schematic of the data
analysis pipeline and quality filtering is shown in Fig. S1.
piRNA target identification
Differentially expressed piRNAs and protein coding
genes were used to predict the target genes for piRNAs using
miRanda target prediction algorithm (29). Briefly, the FASTA sequences of DE
piRNAs and gene coding transcripts were retrieved from piRNAdb
(version 1_7_5; hg19 reference; pirnadb.org/)
and Ensembl
databases(Homo_sapiens.GRCh37.cdna.all.fa;hg19reference; ensembl.org/index.html), respectively, and miRanda was
run using alignment score ≥170 and a binding energy ≤-20 kcal/mol.
Next, the piRNA-target gene pairs were filtered based on degree of
sequence complementarity in the primary (2-11 nucleotides) and
secondary seed site (12-21 nucleotides) with no mismatch and wobble
base pairing within the primary seed site and only one mismatch and
no wobble base pairing within the secondary seed site, as reported
previously with minor modifications (30). Additionally, targets for each piRNA
were manually verified using piRNADb (27).
Functional annotation of
DE-piRNAs
To understand the functional aspects of targets of
DE piRNAs, gene set enrichment analysis (GSEA) was used. Gene
Ontology (GO) (geneontology.org/) and Kyoto Encyclopedia of Genes and
Genomes (KEGG) databases (genome.jp/kegg/) were used to perform the
enrichment analysis. KEGG database integrates genomic information
to explore metabolic pathways, genetic information processing and
cellular processes, whereas GO analysis involves the computational
examination of gene sets to identify and categorize biological
processes (BPs), cellular components (CCs) and molecular functions
(MFs). Enrichr was used for GO and KEGG analysis (31,32).
P<0.05 was considered to indicate significant enrichment. The
Cancer Genome Atlas-Pancreatic Adenocarcinoma (TCGA-PAAD) dataset
was used through Gene Expression Profiling Interactive Analysis;
gepia.cancer-pku.cn/).
Predicting piRNA clusters from
diseased and normal samples
Most piRNAs in the genome originate from
25-35-bp-long discrete loci termed ‘piRNA clusters’, which serve a
key role in the silencing of TEs (33). proTRAC command-line tool (34) was used with default parameters
(sliding window size, 5,000 bp; sliding window increment, 1,000 bp;
minimum fraction of hits with 1T(U) or 10A, 0.75; minimum size of
piRNA cluster, 1,000 bp) to identify piRNA clusters in the tissue
and plasma samples. Overlap of identified clusters with known
repetitive elements in the genome was determined with Repeatmasker
database (reference genome, hg19) annotation (35) and BEDtools package (36).
Statistical analysis
Wald test was used for assessing statistical
significance with adjusted P-value <0.1 as the threshold and
-log2FC >0.58 or <-0.58 in R package DEseq2 was used to
identify DE-piRNAs. Pearson's correlation test. In GO enrichment
analysis, hypergeometric distribution mathematic model was used to
obtain the P-value of the Pathways. In KEGG pathways, multiple
Benjamini and Hochberg testing was performed.
Results
Comparative analysis of DE piRNAs
derived from small RNA sequencing of tissue and plasma
Small RNA sequencing was performed to obtain a
median of ~52.45 million reads (range, 28.32-65.68 million) in
tumor tissues and adjacent normal samples. A median of 15.79
million reads (range, 3.29-92.69 million) was obtained in the case
of the plasma samples (PC and CP cases and respective normal
samples). Detailed metrics and quality control information of small
RNA sequencing in tissue and plasma samples of patients with
pancreatitis and PC are provided in Table SII.
Landscape of DE piRNAs in pancreatic
tumor tissues
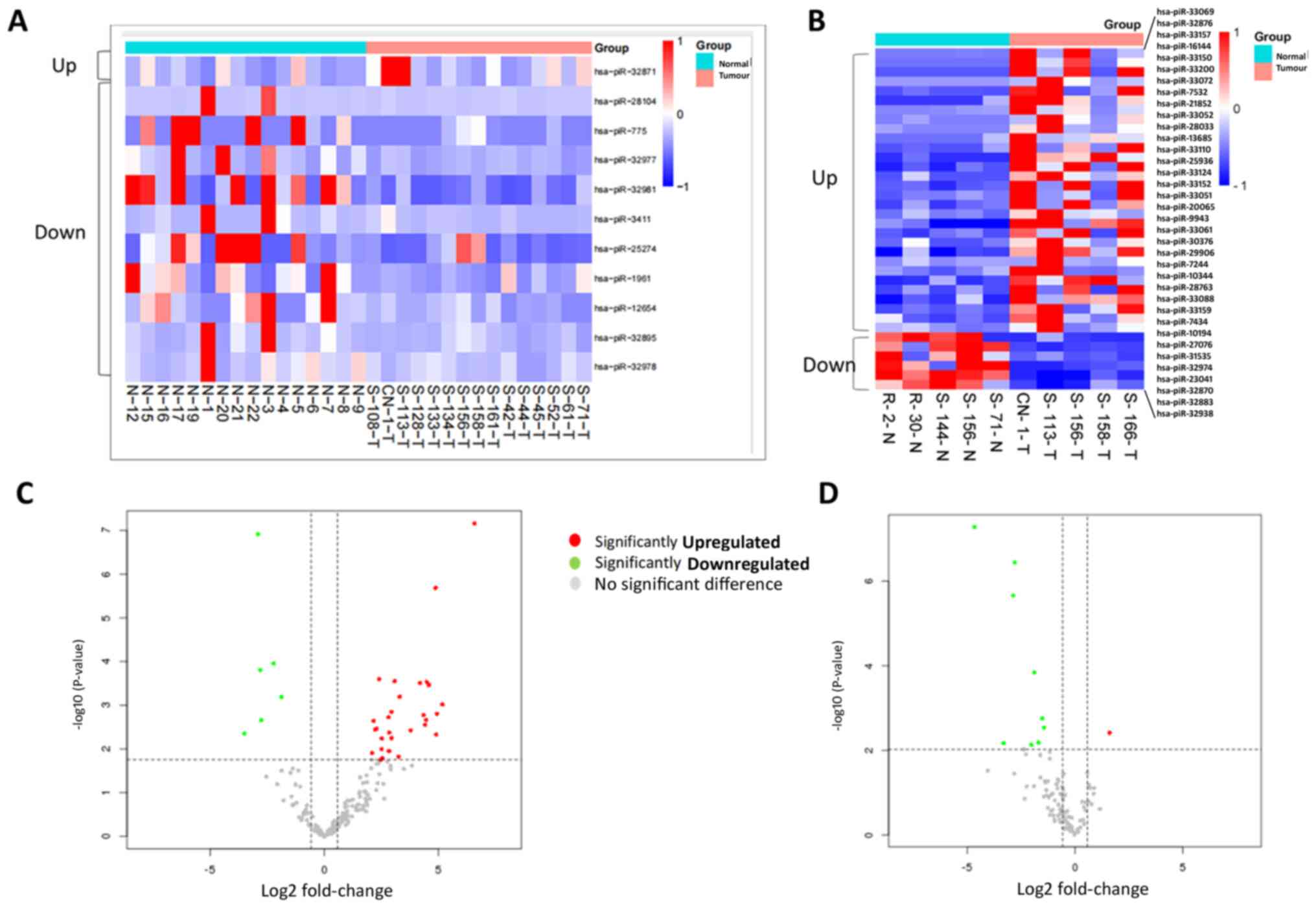
In pancreatic tumor tissues compared with normal
tissue, 30 piRNAs were significantly upregulated and six piRNAs
were significantly downregulated (Table
I; Fig. 1A and C).
 | Table IDifferentially expressed piRNAs in
normal and pancreatic cancer tissue. |
Table I
Differentially expressed piRNAs in
normal and pancreatic cancer tissue.
| A. Upregulated
piRNAs |
|---|
| piRNA | Sequence | Log2
fold-change | Adjusted
P-value |
|---|
| hsa-piR-33069 |
5'-AGACCTATGAAGAGATTGAAGAAGAAACTGAGGTCC-3' | 6.580811883 | 1.21x10-5 |
| hsa-piR-33150 |
5'-GGCGTGTGATGATTACCTGAGTATTTCTGACG-3' | 4.873765728 | 1.38206x10-4 |
| hsa-piR-33200 |
5'-TTTGCCATGATGAGAATTTATCTGAGG-3' | 4.58814715 | 6.842285x10-3 |
| hsa-piR-33072 |
5'-AGCCCTGAGGATGAAAGAACTATCCCTGAAGGGC-3' | 4.478109188 | 6.842285x10-3 |
| hsa-piR-28033 |
5'-GGCCAGCCTGGTCCACATGGGTCGGAA-3' | 4.200503537 | 6.842285x10-3 |
| hsa-piR-33124 |
5'-CTGTCCTTGATGTTACTGCTGTTCTGAGACAT-3' | 3.085192218 | 6.842285x10-3 |
| hsa-piR-28763 |
5'-GTTTAGACGGGCTCACATCACCCCATAAACA-3' | 2.409542772 | 6.842285x10-3 |
| hsa-piR-33110 |
5'-CGAGAATGATGAACGATGCTTCCAGATTCTGACAC-3' | 3.302763067 | 1.0722637x10-2 |
| hsa-piR-32876 |
5'-ATATCATGATGTTACTTTGATTCTCTGACC-3' | 5.176551309 | 1.4853741x10-2 |
| hsa-piR-33051 |
5'-AAACAATGATGGAGTTGCAAGGGTCTGAGC-3' | 2.954871635 | 2.0095922x10-2 |
| hsa-piR-33157 |
5'-GTGCTGGGATGAACGTTTTAACATCTGAGCAG-3' | 4.929144892 | 2.0802856x10-2 |
| hsa-piR-33052 |
5'-AAACTGATGATGCTTGAATTCCTGTTTACTCTGAAG-3' | 4.352093533 | 2.0979839x10-2 |
| hsa-piR-33061 |
5'-ACATGTGATGAGATCGTTGCTCTGATGG-3' | 2.816116428 | 2.1997222x10-2 |
| hsa-piR-7532 |
5'-TCTCATAATGAAGACATAGCCGATTCTCTGC-3' | 4.464241618 | 2.24885x10-2 |
| hsa-piR-7434 |
5'-TCTCAAAGTGAAAGGACCAGTTCGAAT-' | 2.161793669 | 2.24885x10-2 |
| hsa-piR-21852 |
5'-TGTGCTGACCATGGGCCCTGAGCGTCCT-3' | 4.420598295 | 2.6320863x10-2 |
| hsa-piR-13685 |
5'-TGCAGAGATCATACCCCAGAACCAAAAGGCC-3' | 3.789043899 | 3.0982636x10-2 |
| hsa-piR-33088 |
5'-CACCGTGATGAATAGATACTCTGAAGC-3' | 2.28739202 | 3.0982636x10-2 |
| hsa-piR-33159 |
5'-GTTCCAGGATGAAACCATGCGTATCTGAGC-3' | 2.222917393 | 3.0982636x10-2 |
| hsa-piR-20065 |
5'-TGGTCATTGACAATGGCTCCGGCATGTGC-3' | 2.846357394 | 3.3871133x10-2 |
| hsa-piR-16144 |
5'-TGCTGGGAAACGCAAAGCATCCGGAC-3' | 4.905862403 | 3.4132054x10-2 |
| hsa-piR-33152 |
5'-GGGCTGATGATGACCTCTGCAACTCTGAAGCAA-3' | 2.959235719 | 3.9836546x10-2 |
| hsa-piR-29906 |
5'-TACACCTAAGAAACAAGGAGGACTGGGA-3' | 2.521517367 | 3.9848324x10-2 |
| hsa-piR-7244 |
5'-TCGTTGCGGATGGCCAGCTGGAGGTGA-3' | 2.513102981 | 6.612213x10-2 |
| hsa-piR-9943 |
5'-TGACGGTTCCCTGTCTCTGAAAGACCTT-3' | 2.837158255 | 7.18151x10-2 |
| hsa-piR-10194 |
5'-TGAGAACCAATGGGAAGGAGCCTGAGC-3' | 2.094934269 | 7.6956152x10-2 |
| hsa-piR-25936 |
5'-TTTGAGGGTGATGATGGATTCTGTGT-3' | 3.248962354 | 8.9802476x10-2 |
| hsa-piR-30376 |
5'-TACCTCATGAAGATCCTCACCGAGCGCGGC-3' | 2.548138494 | 9.404138x10-2 |
| hsa-piR-10344 |
5'-TGAGACCAATGAAATCGCCAATGCCAAC-3' | 2.470225538 | 9.7043131x10-2 |
| hsa-piR-27076 |
5'-GCAAGGTGGGTCTCAGAGGTGATCGGCGA-3' | 2.088493166 | 9.7043131x10-2 |
| B. Downregulated
piRNAs |
| hsa-piR-31535 |
5'-TAGGACATTATGACGTGCTTGGGTTC-3' | -3.492343956 | 3.3893191x10-2 |
| hsa-piR-32974 |
5'-GTCCTGCAATTCACATTAATTCTCACAGCT-3' | -2.903453446 | 1.21x10-5 |
| hsa-piR-23041 |
5'-CCCCTGGTGGTCTAGTGGTTAGGATTCGGC-3' | -2.806147201 | 6.265475x10-3 |
| hsa-piR-32870 |
5'-AGGGTGGTTCAGTGGTAGAATTCTCG-3' | -2.772172955 | 2.24885x10-2 |
| hsa-piR-32883 |
5'-CAAGAATTCTACCACTGAACAACCAATGC-3' | -2.218575542 | 5.546742x10-3 |
| hsa-piR-32938 |
5'-GCATTGGTGGTTCAGTAGTAGAATTCTCG-3' | -1.866421057 | 1.0722637x10-2 |
Exploratory analysis of circulating
piRNAs in plasma samples
There were 16 normal samples and 15 samples from
patients with PC. One piRNA was up- and 10 were downregulated
(Table SIII; Fig. 1B and D).
Correlation between plasma and tissue
samples of matched patients
Matched plasma and tissue specimens were evaluated
for piRNA expression profiles in four PDAC plasma samples.
Normalized expression levels of 20 piRNAs were positively
correlated in plasma and tissues of patients with PDAC (Pearson's
correlation coefficient >0.3; Table
SIV). This suggests a common trend of piRNA deregulation
between biospecimens and increases the chances of tumour tissue
piRNA alteration being reflected in plasma. A total of three
piRNAs, hsa-piR-23246, hsa-piR-32858 and hsa-piR-9137, were highly
correlated between tumour tissue and plasma of patients with PDAC
and may have an important role in disease development.
piRNA target prediction
piRNAs have been found to operate in a manner
similar to miRNA, as evident from studies, regulating gene
expression through complementary base pairing and exhibiting an
inverse correlation with target mRNA expression (9,10,37).
By using Miranda algorithm and piRNAdb database to predict the
targets of DE piRNAs, 413 mRNA targets for six downregulated DE
piRNAs and 1,984 targets for 30 upregulated DE piRNAs were
identified (Table SV).
Pathway analysis of DE piRNA
targets
To determine the role of deregulated piRNAs in
patients with PDAC, pathway analysis using predicted genes of DE
piRNAs was performed. The top 10 GO classifications of BP, CC and
MF were used (Fig. 2A and B). Using targets of upregulated piRNAs,
top GO processes included ‘monoatomic cation transport’ and
‘maturation of SSU-rRNA’ in the BP subgroup, ‘pre-ribosome, small
subunit precursor’ and ‘90S pre-ribosome’ in the CC and ‘mRNA 5'
UTR binding’ and ‘histone demethylase activity’ in the MF subgroup
(Table SVI, Table SVII, Table SVIII, Table SIX, Table SX and Table SXI). Similarly, using
downregulated piRNA and targets, top GO processes were identified
as cAMP-mediated signaling regulation, regulation of transcription
and regulation of arginine-histamine methylation in BP group. Rough
endoplasmic reticulum membrane and RISC complex were the top CC
subgroup, while histone demethylase activity and promoter-specific
chromatin binding were the top GO processes in the MF subgroup.
Additionally, 154 KEGG pathways were identified among dysregulated
piRNAs and their targets (Tables
SXII and SXIII). Pathway
enrichment analysis showed that several key pathways such as
pathways of glycolysis and gluconeogenesis, pathways of bile
secretion, pathways in cancer, glucagon signaling pathway, ‘insulin
signaling pathway’ and MAPK signaling pathway were enriched
(Fig. 3A and B) (Tables
SXII and SXIII). These
results indicated alteration of multiple malignancy-specific
pathways in PC.
Predicting piRNA clusters
proTRAC was used to predict genomic location
enriched with piRNA clusters in PC and normal samples. A total of
262 clusters were identified in plasma samples (Table SXIV) containing 40 piRNAs. Among
these, 18 were exclusive to PC patients, 15 were common to both
patient and normal samples and seven were unique to normal samples.
A total of 34 clusters was identified in the patient genome while
analyzing the tissue samples. Out of these, 17 clusters showed high
normalized count reads. From these high-density clusters, 10
clusters demonstrated the presence of 25 enriched piRNAs (Table SXV). Similarly, within normal
tissue samples, 24 clusters were identified alongside 12 highly
concentrated clusters. One cluster was the origin of six piRNAs, of
which hsa-piR-32859, hsa-piR-22269, hsa-piR-20792, hsa-piR-32002
and hsa-piR-15181 were DE.
Although there was an asymmetric distribution
throughout the chromosomes, clusters of piRNAs were more prevalent
in chromosomes 9, 10, 18, 20 and Y (Table SXV; Fig. 4A). There were no clusters detected
on chromosomes 1, 4, 5, 6, 7, 8, 12, 14, 19, 22 and X. Furthermore,
the nucleotide preference among the piRNAs was also investigated.
Specifically, piRNAs encoded from the plus strand exhibited the
strongest preference for 1U compared with those from the minus
strand (9,10). However, a higher bias for 10A was
observed in the minus compared with the plus strand (Fig. 4B). This indicated the importance of
orientation in acquiring or determining the type of biogenesis
pathway of piRNA production. Based on this genomic feature,
orientation is key in determining the type of pathway by which
piRNA is produced. Fig. S2 shows a
representative image of piRNA cluster visualization.
Functional annotation of the origin of
piRNA clusters
TEs or jumping genes occupy 45% of the entire human
genome. TEs such as long Interspersed Nuclear Elements and SINE
(Short Interspersed Nuclear Elements) are small repetitive
sequences and easily enter or jump into any position of genome
(38). This type of insertion leads
to mutations and contributes to cancer development (38). To understand how piRNAs silence TEs,
their origin was analyzed from the genome cluster and targeted
coordinates in those regions were identified (Table SXV; Fig. 5) (39-41).
There was a higher density of piRNA origin found in
tumour samples compared with normal samples. Additionally, density
of piRNA clusters present in tumour samples was higher in SINE
repeats. Long Terminal Repeats regions that harbour piRNAs were
identified only in tumour samples. In normal samples, piRNAs were
mapped to the DNA transposons like Tc1/mariner, DNA/TcMar-Tigger,
DNA/hAT-Charlie and DNA/hAT-Tip100.
DE piRNAs in plasma of patients with
CP and healthy individuals
Pre-operative plasma samples from eight patients
with CP were used and circulating piRNA expression pattern was
compared with that of healthy control individuals (n=16). A total
of four upregulated piRNAs and 15 downregulated piRNAs were found
in the plasma of patients with CP (Table SXVI; Fig. S3). piRNAdb was used to identify
potential targets of those piRNA (Table SXVII). Most of the target genes
contribute to chronic inflammation in multiple organs (Table II). The results of the present
study suggested similar changes of piRNAs in pancreatic tissues of
patients with CP that cause deregulation of target genes
contributing to chronic inflammation. Additionally, the involvement
of these target genes in PC was investigated using TCGA-PAAD.
Almost all the genes were upregulated in pancreatic tumour tissue,
indicating that the inflammatory condition in CP may also be
present in pancreatic tumour tissue and promote carcinogenesis.
 | Table IIPro-inflammatory target genes for
differentially expressed piwi-interacting RNAs in plasma of
patients with chronic pancreatitis. |
Table II
Pro-inflammatory target genes for
differentially expressed piwi-interacting RNAs in plasma of
patients with chronic pancreatitis.
| Target gene | Role in modulating
inflammatory pathways | GEPIA PAAD
fold-change (TCGA data) | (Refs.) |
|---|
| HES7 | Significantly
increased expression facilitates development of severe/very severe
COPD | 1.8 | (66) |
| TRPM8 | Induces
ophthalmological neuroinflammatory disease | 5.8 | (74,89) |
| INTS4 | Increases cell
proliferation and inflammation signaling during development of
glioma | 1.7 | (71,78) |
| SCAMP4 | Promotes systemic
inflammation and contributes to development of SLE | 2.4 | (67,85) |
| API5 | Facilitates
TLR4-dependent activation of antigen-presenting cells | 2.3 | (68,82) |
| IFT88 | Promotes
inflammatory responses in non-ciliated macrophage | 2.0 | (72,73,81) |
| PDE3A | Promotes
proinflammatory functions in platelets | 1.6 | (65,83) |
| TM9SF2 | Oncogene in colon
cancer and promotes inflammation | 2.7 | (64,84) |
| EFCAB11 | Upregulated in
inflammatory conditions resulting in asthma | 7.1 | (62,88) |
| SYNRG | Upregulated in
sepsis associated lung inflammation | 2.3 | (61) |
| SLCO5A1 | Upregulated in
oesophageal epithelial cells upon induction of inflammation by
acidic bile salt | 2.9 | (71) |
| EED | Upregulated in
neuroinflammation | 2.1 | (76,87) |
| G3BP2 | Promotes
oscillatory shear stress-induced inflammation in endothelial
cells | 4.3 | (70,77) |
| RYR2 | Promotes
inflammation in spinal cord and diabetic cardiomyopathy; induces
oxidative stress | 0.22 | (75,79) |
| WWP2 | E3 ubiquitin ligase
that regulates pro-fibrogenic monocyte infiltration and activity in
heart fibrosis | 1.9 | (63,80) |
| MACF1 | Alteration
associated with metabolic syndrome and inflammation | 3.2 | (69,86) |
Discussion
Circulating ncRNAs are being studied in greater
detail for their role as potential disease biomarkers (6,7). It is
hypothesized that the changes seen in various body fluids are the
reflection of the changes in corresponding diseased tissue and may
additionally indicate altered regulation of gene expression.
Although cell-free and exosomal miRNAs have been studied, to the
best of our knowledge, there are few studies on piRNAs (6,7). PC is
very aggressive in nature and investigations on circulating miRNAs
are quite a few (42,43). However, there is also not much
information on circulating piRNAs in PC. Hence, it is necessary to
identify non-invasive biomarkers for timely diagnosis. One study
has reported identification of piR-168112 and piR-162725 in both PC
cells and patient plasma. Expression level of piR-162725 was
measured in patients along with CA19-9 level. Combined analysis of
both the values of piR-162725 and CA19-9 in all the patients
increased the sensitivity to 89.7%, which is about 15% more than
CA19-9 sensitivity alone (13).
piRNA expression patterns were investigated in
tissue and plasma sample of the same patients. A positive
correlation of expression of certain piRNAs was shown between
tissue and plasma. Additional piRNAs were shown to be deregulated
in plasma samples as well as in tissue samples (hsa-piR-23246,
hsa-piR-32858 and hsa-piR-32858). hsa-piR-23041 is downregulated in
PDAC tissue sample as per the piRDB database. To the best of our
knowledge, there is little information on disease association of
piRNAs compared with other ncRNAs. piRNAs have evolved as a
countermeasure to suppress TEs. piRNA clusters are sites throughout
the genome from where most piRNAs are synthesized. These clusters
generally overlap with a large number of TEs. Hence, piRNA
sequences derived from each cluster are homologous to TEs in the
same cluster and to similar TEs residing in other parts of the
genome (44,45). Therefore, it is key to determine
expression data of piRNAs from the clusters, while considering
suppression of TEs by piRNAs in both cis- and trans- context. From
the present expression and cluster analysis, hsa-piR-15181,
hsa-piR-22269, hsa-piR-32859, hsa-piR-32002 and hsa-piR-20792 were
overexpressed in tumour samples and were identified in relevant
piRNA clusters through proTRAC analysis. There are two types of
piRNA biogenesis pathways. The primary maturation pathway produces
piRNAs and the secondary maturation pathway amplifies those piRNAs.
The primary maturation pathway shows a bias for U at position 5'
(46) and the secondary
amplification pathway shows a bias for A at position 10. Here, it
was shown that bias differed between groups. The number of piRNAs
with a bias for A at position 10 was comparatively lower than the
bias for ‘U’. This bias for ‘A’ is indicative of the fact that
there is more piRNA formation through primary maturation pathway.
It is necessary to conduct additional research to determine
biogenesis of piRNAs. The presence of TEs in tumor samples suggests
piRNAs are generated more frequently to silencing TEs. piRNAs are
hypothesized to modulate other cellular functions by targeting
specific mRNAs and hence, identification of their targets may
identify the pathways they modulate to mediate disease development
or progression. Metabolic reprogramming has been proposed as a key
hallmark of malignancy. The uptake and catabolism of amino acids
are aberrantly altered and in general, amino acids promote the
survival and proliferation of cancer cells under cell stress and
provide growth advantage to the tumour (47,48).
Significant downregulation of multiple amino acid catabolism
pathways was revealed in the present study, as well as fatty acid
degradation pathways, suggesting potential modulation of metabolic
reprogramming by piRNAs. Glutaryl-CoA dehydrogenase (GCDH) is a key
enzyme involved in the degradative pathway of L-lysine,
L-hydroxylysine and L-tryptophan metabolism (49). This gene was a direct target of
upregulated piR-7244. GCDH gene has been previously reported as a
tumour suppressor gene in hepatocellular carcinoma (50) and may function in the same manner in
PDAC. Similarly, diacylglycerol Kinase Gamma (DGKG) gene is a
member of the type I subfamily of diacylglycerol kinases, which are
involved in lipid metabolism (51).
DGKG is a target of upregulated piR-10194 identified in our
results. The present study found alteration of lipid catabolizing
pathways from our pathway analysis. Therefore, DGKG gene expression
might contribute to observed suppression of lipid catabolizing
pathways.
Nucleotide-binding oligomerization domain receptor-2
(NOD2) exerts oncogenic effects via activation of the NF-κB and ERK
signaling pathways. Activation of NOD2 signaling through
upregulation of either NF-kB or ERK signaling pathways revealed
that gasdermin D is involved in this pathway (52). To the best of our knowledge, there
are no studies of gasdermin D in PC, however other gasdermin family
proteins (gasdermin E) have been shown to promote chemo-resistance
in PC (53). Ras-related nuclear
protein-guanine nucleotide release factor) has also been observed
as an upregulated target of the present downregulated piRNAs and is
an important component of the microtubule nucleation process.
Microtubule dynamics is an important player in cancer (54) and nucleation is the most important
regulatory step. Unfolded protein response (UPR) is constitutively
active in PDAC, likely contributing to disease progression and
acquisition of therapeutic resistance (55). Disabled Homolog 2 Interacting
Protein) and DAXX (Death Domain Associated Protein), which serve as
regulators of UPR (unfolded protein response) (56), were targets of downregulated piRNAs
hsa-piR-28096 and hsa-piR-23041, respectively, indicating the
potential role of these altered piRNAs in modulating UPR-driven
signaling in pancreatic cancer. Among upregulated pathways, the
insulin and the AGE-RAGE signaling pathways are implicated in PC
(17). Notable genes such as SMAD3
(Sma- And Mad-Related Protein 3), TGFBR2 (Transforming Growth
Factor beta Receptor 2) and PPP1R3B (Protein Phosphatase 1
Regulatory Subunit 3B) were upregulated (targets of downregulated
piRNAs) may play an important role in piRNA-mediated development of
PC. Another upregulated pathway, focal adhesion and associated
focal adhesion kinase, has also been reported in the metastasis of
PC and the integrin signaling pathway is instrumental in this
process (57). Caveolins have also
been found to modulate integrin function (58) and 3D collagen architecture is also
reported to regulate cell adhesion (59). The identification of upregulated
target genes regulating the focal adhesion pathway (integrin
Subunit Beta 6, CAV3-Caveolin 3, COL4A6-Collagen Type IV Alpha 6
Chain) provides insight to the possible mechanism. All the
aforementioned findings indicate the potential roles of altered
piRNAs as well as their altered targets in carcinogenesis.
According to the results of the present study, hsa-piR-23246,
hsa-piR-32858 and hsa-piR-9137 may serve as plasma biomarkers.
DE piRNAs were also identified in the plasma of
patients with CP. To the best of our knowledge, the present study
is the first on the alteration of piRNAs in patients with CP. CP is
a progressive fibro-inflammatory disorder and is considered a
pre-malignant condition for PC (60). Therefore, it is key to identify
characteristic changes in the serum or tissue piRNAs in these
patients to identify inflammation and malignancy specific
alterations. After identification of the target genes, their
biological functions were investigated; >50% of the target genes
were proinflammatory and were reported to promote inflammation in
other organs (61-76).
Among these genes, transient Receptor Potential Cation Channel
Subfamily M Member 8), SCAMP4 (Secretory Carrier Membrane Protein
4), TM9SF2 (Transmembrane 9 Superfamily Member 2) and G3BP2 [GTPase
Activating Protein (SH3 Domain) Binding Protein 2] expression is
also increased in patients with PDAC (77-89).
CP increases the risk of PC, and overexpression of these genes not
only promotes CP, but also maintains the inflammatory milieu in
pancreatic tumour tissue.
The present results suggested that piRNAs
hsa-piR-23246, hsa-piR-32858 and hsa-piR-9137 may be used as
potential biomarkers to distinguish pancreatic malignancy.
Additionally, alteration of specific piRNAs in pancreatic tumour
tissues could drive the process of tumorigenesis. However, the
present study did not assess the expression status of these three
piRNAs in other gastrointestinal disease, which would determine the
specificity and sensitivity of the signature. Validation of the
piRNAs in a different cohort of patient samples and healthy
individuals was not performed. Functional validation of the altered
piRNAs and their target genes should be performed in future.
Supplementary Material
Schematic overview of the data
analysis methodology. pi, piwi-interacting; nt, nucleotide; KEGG,
Kyoto Encyclopedia of Genes and Genomes.
piRNA cluster visualization.
Representative clusters obtained from (A) normal and (B) tumour
plasma samples. pi, piwi-interacting RNA.
Expression of plasma-specific piRNAs
in patients with CP and healthy individuals. (A) Heatmap showing
expression patterns of piRNAs across all adjacent normal tissues
(n=5) and tumour tissues (n=5). Higher expression is shown in blue
and the lower expression is shown in red. (B) Volcano plot showing
differentially expressed piRNAs in tissue from tumour and adjacent
normal samples. pi, piwi-interacting; CP, chronic
pancreatitis.
Patient sample information.
Small RNA sequencing in normal and
patient samples.
Differentially expressed piRNAs in
normal and pancreatic cancer plasma samples
piRNA correlation between tissue and
plasma samples.
Target list of piwi-interacting
RNAs.
GO annotation analysis of biological
process for targets of downregulated piwi-interacting RNAs.
GO annotation analysis of cellular
components for targets of downregulated piwi-interacting RNAs.
GO annotation analysis of molecular
function for targets of downregulated piwi-interacting RNAs.
GO annotation analysis of biological
process for targets of upregulated piwi-interacting RNAs.
GO annotation analysis of cellular
components for targets of upregulated piwi-interacting RNAs.
GO annotation analysis of molecular
function for targets of upregulated piwi-interacting RNAs.
Kyoto Encyclopedia of Genes and
Genomes pathway corresponding to target genes of downregulated
piwi-interacting RNAs.
Kyoto Enyclopedia of Genes and Genomes
pathway corresponding to target genes of upregulated
piwi-interacting RNAs.
Cluster analysis of piwi-interacting
RNA plasma samples.
Cluster analysis of piwi-interacting
RNA tissue samples.
Differentially expressed piRNAs found
in normal and chronic pancreatitis plasma samples.
Targets of differentially expressed
piRNAs discovered in normal and chronic pancreatitis plasma
samples.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by Department of
Science and Technology and Biotechnology, Government of West Bengal
[grant no. 16(Sanc)/BT/P/Estt/RD-16/2017], Intramural Funding from
Biotechnology Research and Innovation Council-National Institute of
Biomedical Genomics (No. 60105), Council for Scientific and
Industrial Research [grant nos. 09/1033(0007)/2018-EMR-I) and
BRIC-NIBMG(RCB/NIBMG-Ph.D./2019/1001] and Department of
Biotechnology, Government of India (DBT/2019/NIBMG/1225) and
BRIC-NIBMG(RCB/NIBMG-PhD/2019/1011).
Availability of data and materials
The data generated in the present study may be found
in the Indian Biological Data Centre under accession number
INCARP000298 or at the following URL: (inda.rcb.ac.in/indasecure/userstudydetails).
Authors' contributions
BS performed experiments, analyzed data and wrote
the manuscript. SC and BM performed sequence analysis and wrote the
manuscript. SR, HS, IG and KD designed the study and wrote the
manuscript. NKB designed and supervised the study. SG
conceptualized, designed and supervised the study. All authors have
read and approved the final manuscript. SG and BS confirm the
authenticity of the raw data.
Ethics approval and consent to
participate
The present study was approved by Institutional
Ethics Committee of National Institute of Biomedical Genomics
(Kalyani, India; approval no. CERTIFICATE-SG1-MARCH 05 2014),
Institute of Post Graduate Medical Education & Research
(Kolkata, India; approval nos. INST/IEC/2015/218 and
IPGME&R/IEC/2022/318) and Chittaranjan National Cancer
Institute (Kolkata, India; approval no. CNCI-IEC-SG2-2023-69).
Written informed consent was obtained from the study
participants.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bengtsson A, Andersson R and Ansari D: The
actual 5-year survivors of pancreatic ductal adenocarcinoma based
on real-world data. Sci Rep. 10(16425)2020.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Zhang L, Sanagapalli S and Stoita A:
Challenges in diagnosis of pancreatic cancer. World J
Gastroenterol. 24:2047–2060. 2018.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Lee ES and Lee JM: Imaging diagnosis of
pancreatic cancer: A state-of-the-art review. World J
Gastroenterol. 20:7864–7877. 2014.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Poruk KE, Gay DZ, Brown K, Mulvihill JD,
Boucher KM, Scaife CL, Firpo MA and Mulvihill SJ: The clinical
utility of CA 19-9 in pancreatic adenocarcinoma: Diagnostic and
prognostic updates. Curr Mol Med. 13:340–351. 2013.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Kim S, Park BK, Seo JH, Choi J, Choi JW,
Lee CK, Chung JB, Park Y and Kim DW: Carbohydrate antigen 19-9
elevation without evidence of malignant or pancreatobiliary
diseases. Sci Rep. 10(8820)2020.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Wang K, Wang X, Pan Q and Zhao B: Liquid
biopsy techniques and pancreatic cancer: Diagnosis, monitoring, and
evaluation. Mol Cancer. 22(167)2023.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Yan H and Bu P: Non-coding RNA in cancer.
Essays Biochem. 65:625–639. 2021.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Li Y, Al Hallak MN, Philip PA, Azmi AS and
Mohammad RM: Non-coding RNAs in pancreatic cancer diagnostics and
therapy: Focus on lncRNAs, circRNAs, and piRNAs. Cancers (Basel).
13(4161)2021.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Liu Y, Dou M, Song X, Dong Y, Liu S, Liu
H, Tao J, Li W, Yin X and Xu W: The emerging role of the piRNA/piwi
complex in cancer. Mol Cancer. 18(123)2019.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Wu Z, Yu X, Zhang S, He Y and Guo W: Novel
roles of PIWI proteins and PIWI-interacting RNAs in human health
and diseases. Cell Commun Signal. 21(343)2023.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Limanówka P, Ochman B and Świętochowska E:
PiRNA obtained through liquid biopsy as a possible cancer
biomarker. Diagnostics (Basel). 13(1895)2023.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Müller S, Raulefs S, Bruns P, Afonso-Grunz
F, Plötner A, Thermann R, Jäger C, Schlitter AM, Kong B, Regel I,
et al: Next-generation sequencing reveals novel differentially
regulated mRNAs, lncRNAs, miRNAs, sdRNAs and a piRNA in pancreatic
cancer. Mol Cancer. 14(94)2015.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Li W, Gonzalez-Gonzalez M, Sanz-Criado L,
Garcia-Carbonero N, Celdran A, Villarejo-Campos P, Minguez P,
Pazo-Cid R, Garcia-Jimenez C, Orta-Ruiz A, et al: A novel PiRNA
enhances CA19-9 sensitivity for pancreatic cancer identification by
liquid biopsy. J Clin Med. 11(7310)2022.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Kumar SR, Kimchi ET, Manjunath Y,
Gajagowni S, Stuckel AJ and Kaifi JT: RNA cargos in extracellular
vesicles derived from blood serum in pancreas associated
conditions. Sci Rep. 10(2800)2020.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Xie J, Xing S, Shen BY, Chen HT, Sun B,
Wang ZT, Wang JW and Lu XX: PIWIL1 interacting RNA piR-017061
inhibits pancreatic cancer growth via regulating EFNA5. Hum Cell.
34:550–563. 2021.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Zhong Y, Tian Y, Wang Y, Bai J, Long Q,
Yan L, Gong Z, Gao W and Tang Q: Small extracellular vesicle
piR-hsa-30937 derived from pancreatic neuroendocrine neoplasms
upregulates CD276 in macrophages to promote immune evasion. Cancer
Immunol Res. 12:840–853. 2024.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Chhatriya B, Mukherjee M, Ray S, Sarkar P,
Chatterjee S, Nath D, Das K and Goswami S: Comparison of tumour and
serum specific microRNA changes dissecting their role in pancreatic
ductal adenocarcinoma: A meta-analysis. BMC Cancer.
19(1175)2019.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Andrews S: FastQC: A quality control tool
for high throughput sequence data, 2010.
|
|
19
|
Ewels P, Magnusson M, Lundin S and Käller
M: MultiQC: Summarize analysis results for multiple tools and
samples in a single report. Bioinformatics. 32:3047–3048.
2016.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Krueger F: Trim Galore: A wrapper tool
around Cutadapt and FastQC to consistently apply quality and
adapter trimming to FastQ files, with some extra functionality for
MspI-digested RRBS-type (reduced representation bisufite-seq)
libraries, 2012.
|
|
21
|
Langmead B and Salzberg SL: Fast
gapped-read alignment with Bowtie 2. Nat Methods. 9:357–359.
2012.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Li H, Handsaker B, Wysoker A, Fennell T,
Ruan J, Homer N, Marth G, Abecasis G and Durbin R: 1000 Genome
Project Data Processing Subgroup. The sequence alignment/map format
and SAMtools. Bioinformatics. 25:2078–2079. 2009.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Tarasov A, Vilella AJ, Cuppen E, Nijman IJ
and Prins P: Sambamba: Fast processing of NGS alignment formats.
Bioinformatics. 31:2032–2034. 2015.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Garcia-Alcalde F, Okonechnikov K,
Carbonell J, Cruz LM, Götz S, Tarazona S, Dopazo J, Meyer TF and
Conesa A: Qualimap: Evaluating next-generation sequencing alignment
data. Bioinformatics. 28:2678–2679. 2012.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Leung YY, Kuksa PP, Amlie-Wolf A,
Valladares O, Ungar LH, Kannan S, Gregory BD and Wang LS: DASHR:
Database of small human noncoding RNAs. Nucleic Acids Res.
44:D216–D222. 2016.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Liao Y, Smyth GK and Shi W: featureCounts:
An efficient general purpose program for assigning sequence reads
to genomic features. Bioinformatics. 30:923–930. 2014.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Piuco R and Galante PAF: piRNAdb: A
piwi-interacting RNA database. bioRxiv, 2021.
|
|
28
|
Love MI, Huber W and Anders S: Moderated
estimation of fold change and dispersion for RNA-seq data with
DESeq2. Genome Biol. 15(550)2014.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Riffo-Campos AL, Riquelme I and
Brebi-Mieville P: Tools for sequence-based miRNA target prediction:
What to choose? Int J Mol Sci. 17(1987)2016.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Das B, Jain N and Mallick B: piR-39980
mediates doxorubicin resistance in fibrosarcoma by regulating drug
accumulation and DNA repair. Commun Biol. 4(1312)2021.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Chen EY, Tan CM, Kou Y, Duan Q, Wang Z,
Meirelles GV, Clark NR and Ma'ayan A: Enrichr: Interactive and
collaborative HTML5 gene list enrichment analysis tool. BMC
Bioinformatics. 14(128)2013.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Kuleshov MV, Jones MR, Rouillard AD,
Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM,
Lachmann A, et al: Enrichr: A comprehensive gene set enrichment
analysis web server 2016 update. Nucleic Acids Res. 44:W90–W97.
2016.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Yamanaka S, Siomi MC and Siomi H: piRNA
clusters and open chromatin structure. Mob DNA.
5(22)2014.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Rosenkranz D and Zischler H: proTRAC-a
software for probabilistic piRNA cluster detection, visualization
and analysis. BMC Bioinformatics. 13(5)2012.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Chen N: Using RepeatMasker to identify
repetitive elements in genomic sequences. Curr Protoc
Bioinformatics Chapter 4: Unit 4.10, 2004.
|
|
36
|
Quinlan AR and Hall IM: BEDTools: A
flexible suite of utilities for comparing genomic features.
Bioinformatics. 26:841–842. 2010.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Zuo Y, Liang Y, Zhang J, Hao Y, Li M, Wen
Z and Zhao Y: Transcriptome analysis identifies piwi-interacting
RNAs as prognostic markers for recurrence of prostate cancer. Front
Genet. 10(1018)2019.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Lander ES, Linton LM, Birren B, Nusbaum C,
Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al:
Initial sequencing and analysis of the human genome. Nature.
409:860–921. 2001.PubMed/NCBI View Article : Google Scholar
|
|
39
|
El-Sawy M, Kale SP, Dugan C, Nguyen TQ,
Belancio V, Bruch H, Roy-Engel AM and Deininger PL: Nickel
stimulates L1 retrotransposition by a post-transcriptional
mechanism. J Mol Biol. 354:246–257. 2005.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Giorgi G, Marcantonio P and Del Re B:
LINE-1 retrotransposition in human neuroblastoma cells is affected
by oxidative stress. Cell Tissue Res. 346:383–391. 2011.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Stribinskis V and Ramos KS: Activation of
human long interspersed nuclear element 1 retrotransposition by
benzo(a)pyrene, an ubiquitous environmental carcinogen. Cancer Res.
66:2616–2620. 2006.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Prinz C, Fehring L and Frese R: MicroRNAs
as indicators of malignancy in pancreatic ductal adenocarcinoma
(PDAC) and cystic pancreatic lesions. Cells.
11(2374)2022.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Mok ETY, Chitty JL and Cox TR: miRNAs in
pancreatic cancer progression and metastasis. Clin Exp Metastasis.
41:163–186. 2024.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Ernst C, Odom DT and Kutter C: The
emergence of piRNAs against transposon invasion to preserve
mammalian genome integrity. Nat Commun. 8(1411)2017.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Ho S, Theurkauf W and Rice N: piRNA-guided
transposon silencing and response to stress in drosophila germline.
Viruses. 16(714)2024.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Siomi MC, Sato K, Pezic D and Aravin AA:
PIWI-interacting small RNAs: The vanguard of genome defence. Nat
Rev Mol Cell Biol. 12:246–258. 2011.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Nong S, Han X, Xiang Y, Qian Y, Wei Y,
Zhang T, Tian K, Shen K, Yang J and Ma X: Metabolic reprogramming
in cancer: Mechanisms and therapeutics. MedComm (2020).
4(e218)2023.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Wei Z, Liu X, Cheng C, Yu W and Yi P:
Metabolism of amino acids in cancer. Front Cell Dev Biol.
8(603837)2021.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Sauer SW: Biochemistry and bioenergetics
of glutaryl-CoA dehydrogenase deficiency. J Inherit Metab Dis.
30:673–680. 2007.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Lao Y, Cui X, Xu Z, Yan H, Zhang Z, Zhang
Z, Geng L, Li B, Lu Y, Guan Q, et al: Glutaryl-CoA dehydrogenase
suppresses tumor progression and shapes an anti-tumor
microenvironment in hepatocellular carcinoma. J Hepatol:
S0168-8278(24)00369-6, 2024 (Epub ahead of print).
|
|
51
|
Jiang LQ, de Castro Barbosa T, Massart J,
Deshmukh AS, Löfgren L, Duque-Guimaraes DE, Ozilgen A, Osler ME,
Chibalin AV and Zierath JR: Diacylglycerol kinase-δ regulates AMPK
signaling, lipid metabolism, and skeletal muscle energetics. Am J
Physiol Endocrinol Metab. 310:E51–E60. 2016.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Ren Y, Liu SF, Nie L, Cai SY and Chen J:
Involvement of ayu NOD2 in NF-κB and MAPK signaling pathways:
Insights into functional conservation of NOD2 in antibacterial
innate immunity. Zool Res. 40:77–88. 2019.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Lv J, Liu Y, Mo S, Zhou Y, Chen F, Cheng
F, Li C, Saimi D, Liu M, Zhang H, et al: Gasdermin E mediates
resistance of pancreatic adenocarcinoma to enzymatic digestion
through a YBX1-mucin pathway. Nat Cell Biol. 24:364–372.
2022.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Ritter A and Kreis NN: Microtubule
dynamics and cancer. Cancers (Basel). 14(4368)2022.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Robinson CM, Talty A, Logue SE, Mnich K,
Gorman AM and Samali A: An emerging role for the unfolded protein
response in pancreatic cancer. Cancers (Basel).
13(261)2021.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Yang X, Khosravi-Far R, Chang HY and
Baltimore D: Daxx, a novel Fas-binding protein that activates JNK
and apoptosis. Cell. 89:1067–1076. 1997.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Kanchanawong P and Calderwood DA:
Organization, dynamics and mechanoregulation of integrin-mediated
cell-ECM adhesions. Nat Rev Mol Cell Biol. 24:142–161.
2023.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Israeli-Rosenberg S, Chen C, Li R, Deussen
DN, Niesman IR, Okada H, Patel HH, Roth DM and Ross RS: Caveolin
modulates integrin function and mechanical activation in the
cardiomyocyte. FASEB J. 29:374–384. 2015.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Velez DO, Ranamukhaarachchi SK, Kumar A,
Modi RN, Lim EW, Engler AJ, Metallo CM and Fraley SI: 3D collagen
architecture regulates cell adhesion through degradability, thereby
controlling metabolic and oxidative stress. Integr Biol (Camb).
11:221–234. 2019.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Le Cosquer G, Maulat C, Bournet B,
Cordelier P, Buscail E and Buscail L: Pancreatic cancer in chronic
pancreatitis: Pathogenesis and diagnostic approach. Cancers
(Basel). 15(761)2023.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Ahmad S, Ahmed MM, Hasan PMZ, Sharma A,
Bilgrami AL, Manda K, Ishrat R and Syed MA: Identification and
validation of potential miRNAs, as biomarkers for sepsis and
associated lung injury: A network-based approach. Genes (Basel).
11(1327)2020.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Alrashoudi RH, Crane IJ, Wilson HM,
Al-Alwan M and Alajez NM: Gene expression data analysis identifies
multiple deregulated pathways in patients with asthma. Biosci Rep.
38(BSR20180548)2018.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Chen H, Chew G, Devapragash N, Loh JZ,
Huang KY, Guo J, Liu S, Tan ELS, Chen S, Tee NGZ, et al: The E3
ubiquitin ligase WWP2 regulates pro-fibrogenic monocyte
infiltration and activity in heart fibrosis. Nat Commun.
13(7375)2022.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Clark CR, Maile M, Blaney P, Hellweg SR,
Strauss A, Durose W, Priya S, Habicht J, Burns MB, Blekhman R, et
al: Transposon mutagenesis screen in mice identifies TM9SF2 as a
novel colorectal cancer oncogene. Sci Rep. 8(15327)2018.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Coenen DM, Heinzmann ACA, Oggero S, Albers
HJ, Nagy M, Hagué P, Kuijpers MJE, Vanderwinden JM, van der Meer
AD, Perretti M, et al: Inhibition of phosphodiesterase 3A by
cilostazol dampens proinflammatory platelet functions. Cells.
10(1998)2021.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Di Stefano A, Gnemmi I, Rosani U,
Maniscalco M, D'Anna SE, Brun P, Carriero V, Bertolini F, Balbi B
and Ricciardolo FLM: Upregulation of notch signaling and
cell-differentiation inhibitory transcription factors in stable
chronic obstructive pulmonary disease patients. Int J Mol Sci.
25(3287)2024.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Ghanem MH, Shih AJ, Vashistha H, Coke LN,
Li W, Kim SJ, Simpfendorfer KR and Gregersen PK: Investigations
into SCAMP5, a candidate lupus risk gene expressed in plasmacytoid
dendritic cells. Lupus Sci Med. 8(e000567)2021.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Kim YS, Park HJ, Park JH, Hong EJ, Jang
GY, Jung ID, Han HD, Lee SH, Vo MC, Lee JJ, et al: A novel function
of API5 (apoptosis inhibitor 5), TLR4-dependent activation of
antigen presenting cells. Oncoimmunology.
7(e1472187)2018.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Kraja AT, Chasman DI, North KE, Reiner AP,
Yanek LR, Kilpeläinen TO, Smith JA, Dehghan A, Dupuis J, Johnson
AD, et al: Pleiotropic genes for metabolic syndrome and
inflammation. Mol Genet Metab. 112:317–338. 2014.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Li T, Qiu J, Jia T, Liang Y, Zhang K, Yan
W, Hou Z, Yang S, Liu L, Xiong W, et al: G3BP2 regulates
oscillatory shear stress-induced endothelial dysfunction. Genes
Dis. 9:1701–1715. 2022.PubMed/NCBI View Article : Google Scholar
|
|
71
|
Lin YC, Chang PC, Hueng DY, Huang SM and
Li YF: Decoding the prognostic significance of integrator complex
subunit 9 (INTS9) in glioma: links to TP53 mutations, E2F
signaling, and inflammatory microenvironments. Cancer Cell Int.
23(154)2023.PubMed/NCBI View Article : Google Scholar
|
|
72
|
Mc Fie M, Koneva L, Collins I, Coveney CR,
Clube AM, Chanalaris A, Vincent TL, Bezbradica JS, Sansom SN and
Wann AKT: Ciliary proteins specify the cell inflammatory response
by tuning NFκB signalling, independently of primary cilia. J Cell
Sci. 133(jcs239871)2020.PubMed/NCBI View Article : Google Scholar
|
|
73
|
Patankar M, Li M, Khalatbari A, Castle JD,
Hu L, Zhang C and Shaker A: Inflammatory and proliferative pathway
activation in human esophageal myofibroblasts treated with acidic
bile salts. Int J Mol Sci. 23(10371)2022.PubMed/NCBI View Article : Google Scholar
|
|
74
|
Ramachandran R, Hyun E, Zhao L, Lapointe
TK, Chapman K, Hirota CL, Ghosh S, McKemy DD, Vergnolle N, Beck PL,
et al: TRPM8 activation attenuates inflammatory responses in mouse
models of colitis. Proc Natl Acad Sci USA. 110:7476–7481.
2013.PubMed/NCBI View Article : Google Scholar
|
|
75
|
Tian CJ, Zhang JH, Liu J, Ma Z and Zhen Z:
Ryanodine receptor and immune-related molecules in diabetic
cardiomyopathy. ESC Heart Fail. 8:2637–2646. 2021.PubMed/NCBI View Article : Google Scholar
|
|
76
|
Weng HR, Taing K, Chen L and Penney A:
EZH2 methyltransferase regulates neuroinflammation and neuropathic
pain. Cells. 12(1058)2023.PubMed/NCBI View Article : Google Scholar
|
|
77
|
Asadi MR, Rahmanpour D, Moslehian MS,
Sabaie H, Hassani M, Ghafouri-Fard S, Taheri M and Rezazadeh M:
Stress granules involved in formation, progression and metastasis
of cancer: A scoping review. Front Cell Dev Biol.
9(745394)2021.PubMed/NCBI View Article : Google Scholar
|
|
78
|
Federico A, Rienzo M, Abbondanza C, Costa
V, Ciccodicola A and Casamassimi A: Pan-cancer mutational and
transcriptional analysis of the integrator complex. Int J Mol Sci.
18(936)2017.PubMed/NCBI View Article : Google Scholar
|
|
79
|
Jiang H, Wang G, Gu J, Xiao Y, Wang P,
Huang X, Sha H, Wang Z and Ma Q: Resveratrol inhibits the
expression of RYR2 and is a potential treatment for pancreatic
cancer. Naunyn Schmiedebergs Arch Pharmacol. 395:315–324.
2022.PubMed/NCBI View Article : Google Scholar
|
|
80
|
Jo EH, Kim MY, Lee HJ and Park HS:
Ubiquitin E3 ligases in cancer: somatic mutation and amplification.
BMB Rep. 56:265–274. 2023.PubMed/NCBI View Article : Google Scholar
|
|
81
|
Khan NA, Garg AD, Agostinis P and Swinnen
JV: Drug-induced ciliogenesis in pancreatic cancer cells is
facilitated by the secreted ATP-purinergic receptor signaling
pathway. Oncotarget. 9:3507–3518. 2018.PubMed/NCBI View Article : Google Scholar
|
|
82
|
Koci L, Chlebova K, Hyzdalova M, Hofmanova
J, Jira M, Kysela P, Kozubik A, Kala Z and Krejci P: Apoptosis
inhibitor 5 (API-5; AAC-11; FIF) is upregulated in human carcinomas
in vivo. Oncol Lett. 3:913–916. 2012.PubMed/NCBI View Article : Google Scholar
|
|
83
|
Kumazoe M, Takai M, Hiroi S, Takeuchi C,
Yamanouchi M, Nojiri T, Onda H, Bae J, Huang Y, Takamatsu K, et al:
PDE3 inhibitor and EGCG combination treatment suppress cancer stem
cell properties in pancreatic ductal adenocarcinoma. Sci Rep.
7(1917)2017.PubMed/NCBI View Article : Google Scholar
|
|
84
|
Li Q, Lei C, Lu C, Wang J, Gao M and Gao
W: LINC01232 exerts oncogenic activities in pancreatic
adenocarcinoma via regulation of TM9SF2. Cell Death Dis.
10(698)2019.PubMed/NCBI View Article : Google Scholar
|
|
85
|
Mao F, Duan H, Allamyradov A, Xin Z, Du Y,
Wang X, Xu P, Li Z, Qian J and Yao J: Expression and prognostic
analyses of SCAMPs in pancreatic adenocarcinoma. Aging (Albany NY).
13:4096–4114. 2021.PubMed/NCBI View Article : Google Scholar
|
|
86
|
Miao Z, Ali A, Hu L, Zhao F, Yin C, Chen
C, Yang T and Qian A: Microtubule actin cross-linking factor 1, a
novel potential target in cancer. Cancer Sci. 108:1953–1958.
2017.PubMed/NCBI View Article : Google Scholar
|
|
87
|
Paradise BD, Barham W and Fernandez-Zapico
ME: Targeting epigenetic aberrations in pancreatic cancer, a new
path to improve patient outcomes? Cancers (Basel).
10(128)2018.PubMed/NCBI View Article : Google Scholar
|
|
88
|
Witkiewicz AK, Balaji U, Eslinger C,
McMillan E, Conway W, Posner B, Mills GB, O'Reilly EM and Knudsen
ES: Integrated patient-derived models delineate individualized
therapeutic vulnerabilities of pancreatic cancer. Cell Rep.
16:2017–2031. 2016.PubMed/NCBI View Article : Google Scholar
|
|
89
|
Yee NS, Brown RD, Lee MS, Zhou W, Jensen
C, Gerke H and Yee RK: TRPM8 ion channel is aberrantly expressed
and required for preventing replicative senescence in pancreatic
adenocarcinoma: Potential role of TRPM8 as a biomarker and target.
Cancer Biol Ther. 13:592–599. 2012.PubMed/NCBI View Article : Google Scholar
|