Introduction
Stroke, which is the leading cause of death in
China, mainly comprises cerebral hemorrhage and cerebral
infarction, of which cerebral ischemia accounts for nearly 80% of
stroke cases (1,2). According to survey results, stroke has
become the second leading cause of death worldwide, with high rates
of incidence, recurrence and high disability, especially among the
elderly (3,4). Thrombolytic and neuroprotective
therapies are the main clinical treatments of this disease;
however, in cases of cerebral ischemia, thrombolysis and
reperfusion can cause secondary damage to the brain, leading to
death and disability (5-9).
This undoubtedly brings economic pressure and physical and
emotional pain to the families of patients. Therefore, there is an
urgent need to find effective drugs for the treatment of
stroke.
Traditional Chinese medicine has unique advantages
and roles in the treatment of diseases. Panax notoginseng
(Burk.) F.H. Chen is a geo-authentic Chinese medicinal material
from Wenshan, Yunnan, which is used as a medicine with dried roots
and rhizomes. Although there is high annual production of Panax
notoginseng stems and leaves, the utilisation rate of the 226
saponin constituents is extremely low. The most abundant of these
saponins is ginsenoside Rb3 (G-Rb3;
C53H90O22). A large amount of
resource waste could be reduced by full utilization of
G-Rb3. It has been shown to have a variety of biological
activities, including cardiovascular protection and brain
protection (10). Liu et al
(11) found that G-Rb3
inhibits apoptosis and protects against myocardial
ischemic-reperfusion. Ginsenosides protect against the damage
brought about by oxygen-glucose deprivation/reoxygenation (OGD/R)
in hippocampal neuron HT22 cells through an anti-oxidative stress
mechanism (12). These findings
showed that G-Rb3 has great potential in the treatment
of stroke; thus, in the present study it was aimed to define more
clearly the mechanism by which G-Rb3 protects against
cerebral ischemic-reperfusion injury (CIRI). The present study
aimed to use a HT22 cell-based replica of the OGD/R model, combined
with metabolomics and PCR array analyses, to determine whether
G-Rb3 ameliorates OGD/R injury through inhibition of
cell apoptosis.
Liquid chromatography (LC)-mass spectrometry (MS)
has been proved to be a powerful and reliable analytical method
with high sensitivity and structural separation ability (13,14).
The occurrence of CIRI leads to alterations in systemic
metabolites, which cause a series of complex cascade reactions that
ultimately lead to apoptosis (15,16).
Metabolomics analysis can be used to the biological functions and
metabolic pathways of metabolites in vivo (17,18).
Therefore, untargeted metabolomics were also used to conduct an
in-depth exploration of the changes in metabolite levels related to
the actions of G-Rb3 against OGD/R injury.
Materials and methods
Cell culture and drug delivery
HT22 cells derived from mouse hippocampal neuronal
cell line (cat. no. CC-Y2137; Shanghai Enzyme Research
Biotechnology Co., Ltd.) were utilized. Ηigh sugar medium DMEM
(cat. no. 01-043-1A; Biological Industries) with 1%
penicillin-streptomycin liquid (cat. no. 03-031-5B; Biological
Industries) and 10% fetal bovine serum (cat. no. 504090618;
Shanghai Yeasen Biotechnology Co., Ltd.) were used for cell culture
at 37˚C with 5% CO2 in a HF90 incubator (Shanghai Lishen
Scientific Equipment Co., Ltd.). When the cells proliferated to the
logarithmic growth period, they were transferred into 6-well plates
at a concentration of 1x105/ml for the subsequent
experiments. G-Rb3 (cat. no. CCPE900218; Henan Wanjia
Standard Material R&D Center Co., Ltd.; https://cdn.bzwzzx.com/product/search.html?keywords=G-Rb3&pageindex=1;
purity ≥99.86%) was dissolved in phosphate-buffered saline to a
mother liquor concentration of 1 mmol/l. The cells were randomly
divided into 4 groups: Control group, OGD/R group, G-Rb3
high dose group (10 µmol/l) and G-Rb3 low dose group (5
µmol/l). Since relevant cell safety experiments were already
conducted in the previous study, it was found that the effective
concentrations of G-Rb3 to improve OGD/R were 2.5, 5 and
10 µmol/l (12); For the present
study, it was observed that the most effective drug concentrations
were 5 and 10 µmol/l and were therefore selected for the
experiments.
OGD/R model
Na2S2O4 is an
effective oxygen scavenger without harming the cells, thus
Na2S2O4 (cat. no. S817915;
Shanghai Macklin Biochemical Co., Ltd.) was chosen to simulate the
OGD/R model in this experiment. When the cell density increased to
80-90%, except for the control group which was left untreated, the
remaining groups were incubated with 10 mmol/l
Na2S2O4 in sugar-free DMEM (cat.
no. 01-042-1A; Biological Industries) at 37˚C for 2 h and then
replaced with serum-free high-sugar DMEM at 37˚C for 2 h in order
to replicate the in vitro OGD/R model. Meanwhile, in the
G-Rb3 group, the drug started being added 24 h before
modelling, and continued until the end of re-glycation and
reoxygenation.
Trypan blue staining for cell
viability measurement
HT22 cells were used to inoculate 6-well plates at a
uniform inoculum density of 1x105/ml (2 ml cell
suspension per well). When cells proliferated to logarithmic growth
phase, a total of 1 ml EDTA-free trypsin (cat. no. 15050-065;
Thermo Fisher Scientific, Inc.) was added to each well at 37˚C for
1 min, followed by the addition of trypan blue (cat. no. G1019;
Wuhan Servicebio Technology Co., Ltd.), which was mixed with the
cell suspension at a ratio of 1:9 and was then left to stand at
room temperature for 2 min. A 20 µl aliquot of the cell suspension
was aspirated to a hemocytometer for observation under an inverted
microscope (DMI1; Leica Microsystems Ltd.). In total, four large
squares in the field of vision of the hemocytometer were selected
for counting cells. Subsequently, the cell viability was
calculated.
Flow cytometry to detect
apoptosis
HT22 cells were used to inoculate 6-well plates at a
uniform inoculum density of 1x105/ml (2 ml cell
suspension per well). The cells were digested with 1 ml of
EDTA-free trypsin at 37˚C for 1 min and were subsequently
collected. Annexin V-AbFluor™ 488/PI apoptosis detection
kit (cat. no. KTA0002; Abbkine Scientific Co., Ltd.) was utilized.
The cells were first resuspended in 100 µl AnnexinV Binding Buffer
diluted in deionized water to which, 5 µl of
AnnexinV-AbFluorTM488 was added, and then incubated on
ice for 15 min. A total of 2 µl propidium iodide dye was pre-added.
Detected was performed by flow cytometry (FACSCelesta; BD
Biosciences) within 30 min. BD FACSDiva™ software
(v8.0.1.1;) was used to analyze data.
Western blotting (WB) detection of
protein expression
The HT22 cells of each group in the logarithmic
growth phase were collected and cultured in 6-well plates at a
density of 6.5x105/ml for 24 h. The cells were lysed on
the ice with RIPA lysis buffer (cat. no. P0013C; Beyotime Institute
of Biotechnology). The supernatant was centrifuged at 12,000 x g at
4˚C for 5 min. Determination of protein concentration by BCA method
(cat. no. P0010; Beyotime Institute of Biotechnology). According to
the molecular weight, 10% separation gel and 5% compression gel
were prepared, 20 µg protein samples were added to each lane, and
separated by SDS-PAGE electrophoresis for 30 min; the
electrophoresis was terminated when the desired target band reached
the appropriate position. The cut PVDF membrane (0.45 µm; cat. no.
IPFL85R; MilliporeSigma) was soaked in methanol for 1 min. After
the transfer, the membrane was blocked with 5% skim milk (cat. no.
P0216; Beyotime Institute of Biotechnology) powder for 1 h at room
temperature. Subsequently, the membranes were incubated at 4˚C
overnight with the following primary antibodies: Bax (1:2,000; cat.
no. 50599-2-Ig; Proteintech Group, Inc.), Bcl-2 (1:1,000; cat. no.
sc-7382; Santa Cruz Biotechnology, Inc.), caspase-3 (1:1,000; cat.
no. 9662; Cell Signaling Technology, Inc.) and β-actin (1:1,000;
cat. no. ab8226; Abcam). Then, the membranes were incubated for 1 h
at room temperature with the following secondary antibodies: Goat
anti-rabbit IgG (1:10,000; cat. no. ab6721; Abcam) and rabbit
anti-mouse IgG (1:10,000; cat. no. ab6728; Abcam). The
aforementioned membrane washing procedure was repeated. Next, the
luminescent reagent liquid A and liquid B were mixed in equal
volume and were applied in the membrane. After 5 min, the protein
bands were detected with Tanon-6600 luminescence imaging
workstation (Shanghai Tianneng Life Science Co., Ltd.). Protein
expression was analyzed by using the ImageJ v2 software (National
Institutes of Health) to analyze the optical density values.
Metabolomics sample collection
HT22 cells were inoculated into 6-well plates at a
concentration of 1x105/ml. The cells were divided into
an OGD/R group and a G-Rb3 group, using one sample per
two wells, with a total of six samples collected for analysis.
Glass beads (cat. no. G8772; Shanghai Lianshuo Biotechnology Co.,
Ltd.) and 1,000 µl acetonitrile (cat. no. AS1121; Shanghai Yaokan
Chemical Industry Co., Ltd.) were added, and the mixture was
vortexed for 30 sec and then ground for 2 min at 60 Hz in a tissue
grinder. The mixture was transferred to a centrifuge tube, spun at
14,000 x g for 10 min at 4˚C, the supernatant was isolated, and 300
µl of a 2-chlorophenylalanine (4 ppm) solution were accurately
prepared using acetonitrile with 0.1% formic acid (cat. no. 28905;
Thermo Fisher Scientific, Inc.) (1:9, v/v). The solution was used
to re-dissolve the samples. The supernatant was filtered through a
0.22 µm membrane and was subsequently added to the detection vials
for LC-MS analysis.
LC/MS conditions
Chromatographic conditions: Waters ACQUITY (Waters
Corporation) and ACQUITY UPLC® HSS T3 (2.1x150 mm, 1.8
µm) (Waters Corporation) column were used with a flow rate of 0.25
ml/min at 40˚C. The injection volume was 2 µl. The mobile phases
were: 0.1% formic acid in acetonitrile (B1) and 0.1% formic acid in
water (A1) in positive ion mode, and the gradient elution program
was as follows: 0-1 min, 2% B1; 1-9 min, 2-50% B1; 9-12 min, 50-98%
B1; 12-13.5 min, 98% B1; 13.5-14 min, 98-2% B1; 14-12 min, 98-2%
B1; 14-20 min, 2% B1. In negative ion mode, the mobile phases were
acetonitrile (B2) and ammonium formate water (A2), and the gradient
elution procedures were as follows: 0-1 min, 2% B2; 1-9 min, 2-50%
B2; 9-12 min, 50-98% B2; 12-13.5 min, 98% B2; 13.5-14 min, 98-2%
B2; 14-17 min, 2% B2.
MS conditions
A Thermo Q Exactive MS detector (Thermo Fisher
Scientific, Inc.) with an electrospray ioniation source was used to
collect data separately in positive and negative ion modes, the
settings were as following: Positive ion spray voltage, 3.50 kV;
negative ion spray voltage, -2.50 kV; sheath gas, 30 arbs;
auxiliary gas, 10 arbs and capillary temperature, 325˚C. The
primary full scan was performed at a resolution of 70,000 m/z, with
a primary ion scanning range of 100-1,000 m/z. Higher-energy
collisional dissociation was used for the secondary cleavage, with
a collision energy of 30 eV and a secondary resolution of 17,500
m/z. The first 10 ions of the acquired signal were fragmented, with
dynamic exclusion used to remove unnecessary tandem MS
information.
Metabolic data processing and
analysis
The raw files were converted to mzXML file format
using MSConvert in the ProteoWizard package (v3.0.8789), with
parameters set to ‘bw=2’, ‘ppm=15’, ‘peakwidth=c’ (parameters,
5,30), ‘mzwid=0.015’, ‘mzdiff=0.01’ and ‘method=centWave’ (19-21).
After obtaining the quantitative list of substances, the following
databases: HMDB (22), massbank
(23), LipidMaps (24), mzcloud (25) and Kyoto Encyclopedia of Genes and
Genomes (KEGG) (26) were used to
identify the substances, followed by data rectification. The
quality control samples with relative standard deviation >30% of
the substances were removed. The R language Ropls package (v1.15.0;
Autodesx) was used to perform principal component analysis (PCA),
partial least-square discriminant analysis (PLS-DA). The sample
data were analyzed by orthogonal partial least-square discriminant
analysis (OPLS-DA) to reduce dimensions. The final screening was
performed via calculating the P-value and variable projection
importance (VIP) value, and metabolite molecules were considered
statistically significant when the P-value was <0.05 and the VIP
value was >1. Pathway analysis was mainly carried out by
MetaboAnalyst software (v6.0) developed by Xia-lab of McGill
University in Canada, which is used for enrichment analysis of
differential metabolites obtained from screening, and for browsing
differential metabolites by using pathway maps in KEGG Mapper tool
(27).
Apoptosis PCR array
Firstly, according to the RNA extraction kit (cat.
no. DP430; Tiangen Biotech Co., Ltd.), the RNA of OGD/R group and
G-Rb3 group was extracted, its concentration and purity
were detected by a spectrophotometer and it was subsequently
reversed into cDNA. The substance was diluted and mixed to a total
volume of 100 µl, and was then placed on ice. According to the PCR
array kit for detection of gene expression. (cat. no.
wc-mRNA0263-M; Shanghai WcGene Technology Co., Ltd.; www.wcgene.cn) plates were centrifuged at 100 x g for
20 sec before use and the sealing membrane was carefully torn off
at the end of centrifugation. A total of 920 µl of cDNA were
prepared and mixed the components are revealed in Table I, then add the prepared mixed sample
into a 96-well plate (9 µl per well), the plate was then sealed
with a transparent sealing membrane, centrifuged at 100 x g for 20
sec and was finally assessed on the PCR instrument
(Veriti™ 96-Well Fast Thermal Cycler; Thermo Fisher
Scientific, Inc.). The reaction system was set at 10 µl, and the
conditions are shown in Table II.
The real time PCR reaction was started on the machine, and the
results were exported for data analysis. The PCR primer sequences
were not provided by the company, although the primer sequences of
the differential genes are included in Table SI.
 | Table IFormulation of cDNA and
components. |
Table I
Formulation of cDNA and
components.
| Components | Volume (9 µl per
well in 96-well plates) |
|---|
| WCGENE®
mRNA qPCR mix (2x) | 510 µl |
| cDNA sample | 100 µl |
| RNase-free
ddH2O | 310 µl |
| Total volume | 920 µl |
 | Table IIPCR reaction conditions. |
Table II
PCR reaction conditions.
| Cycle | | Temperature | Time | Remarks |
|---|
|
Pre-denaturation | 1 | 95˚C | 30 sec | - |
| Denaturation | 40 | 95˚C | 5 sec | - |
| Annealing
extension | 40 | 60˚C | 30 sec | Open fluorescence
acquisition channel |
Statistical analysis
Data are expressed as the mean ± standard deviation
(SD) (n=6). Data were statistically analyzed using GraphPad Prism
8.0 software (Dotmatics) and were normally distributed. If the
variances were equal, Bonferroni's multiple comparison test was
performed using one-way analysis of variance (ANOVA). If the
conflicts were not equal, Dunnett's multiple comparison test was
used in Welch's ANOVA test. P<0.05 was considered to indicate a
statistically significant difference.
Results
Trypan blue staining and flow
cytometry
The cell morphology after G-Rb3
intervention indicated that G-Rb3 effectively improved
OGD/R injury (Fig. 1A). The results
of trypan blue staining showed that cell viability was
significantly higher in the drug treatment group than the model
group (P<0.001), at a drug concentration of 10 µmol/l,
indicating that G-Rb3 effectively improved the survival
rate of HT22 cells in the OGD/R model (Fig. 1B and E). Similarly, the apoptosis rate showed
that G-Rb3 effectively inhibited apoptosis (Fig. 1C and F). When the dose of G-Rb3
reached 10 µmol/l, there was significant difference compared with
the model group (P<0.0001). The chemical structural formula of
G-Rb3 is illustrated in Fig.
1D.
 | Figure 1Effect of G-Rb3 on
OGD/R-induced HT22 cell morphology and apoptosis. (A) Morphology of
HT22 cells in each group. (B) Trypan blue staining. Blue color
represents apoptotic cells as indicated by red arrows in the
figure, and unstained cells are normal cells. (C) Detection of
apoptosis by flow cytometry. Each point in the figure represents a
cell, the X-axis and Y-axis represent the fluorescence intensity of
the cells in the two channels of FITC and PI, the upper left
quadrant represents the cell debris, which are the mechanically
damaged cells, the lower left quadrant represents the normal cells,
the upper right quadrant represents the late apoptotic and necrotic
cells, and the lower right quadrant represents the cells with early
apoptosis. The sum of the early apoptotic and late apoptotic cells
is the total number of each group of HT22 cells. The sum of early
apoptotic cells and late apoptotic cells is the number of apoptotic
cells in each group. (D) Chemical structure of G-Rb3.
(E) Statistics of cell survival rate by trypan blue staining.
###P=0.0003 and Q=0.0006 vs. control group;
**P=0.0016 and Q=0.0016 vs. OGD/R group. (F) Statistical
graph of total apoptosis rate in each group.
####P<0.0001 vs. control group; **P=0.0086
and Q=0.0086, ****P<0.0001 vs. OGD/R group.
Statistical results were all analyzed by one-way ordinary ANOVA
(n=6). G-Rb3, ginsenoside Rb3OGD/R,
oxygen-glucose deprivation/reoxygenation. |
WB assay results
Protein expression levels of Bax, Bcl-2, caspase-3
and cleaved caspase-3 are shown in Fig.
2A. There was no significant difference in the expression of
caspase-3 protein compared with either the control or model groups
(Fig. 2D). The OGD/R group showed
increased levels of Bax and cleaved caspase-3 protein expression
(Fig. 2B and E), and decreased levels of Bcl-2 protein
expression (Fig. 2C), compared with
the control group. The levels of Bax and cleaved caspase-3 protein
expression were decreased (Fig. 2B
and E) and the levels of Bcl-2
protein expression were increased, compared with the
G-Rb3 group (Fig. 2C).
The original blots of Fig. 2 are
shown in Fig. S1.
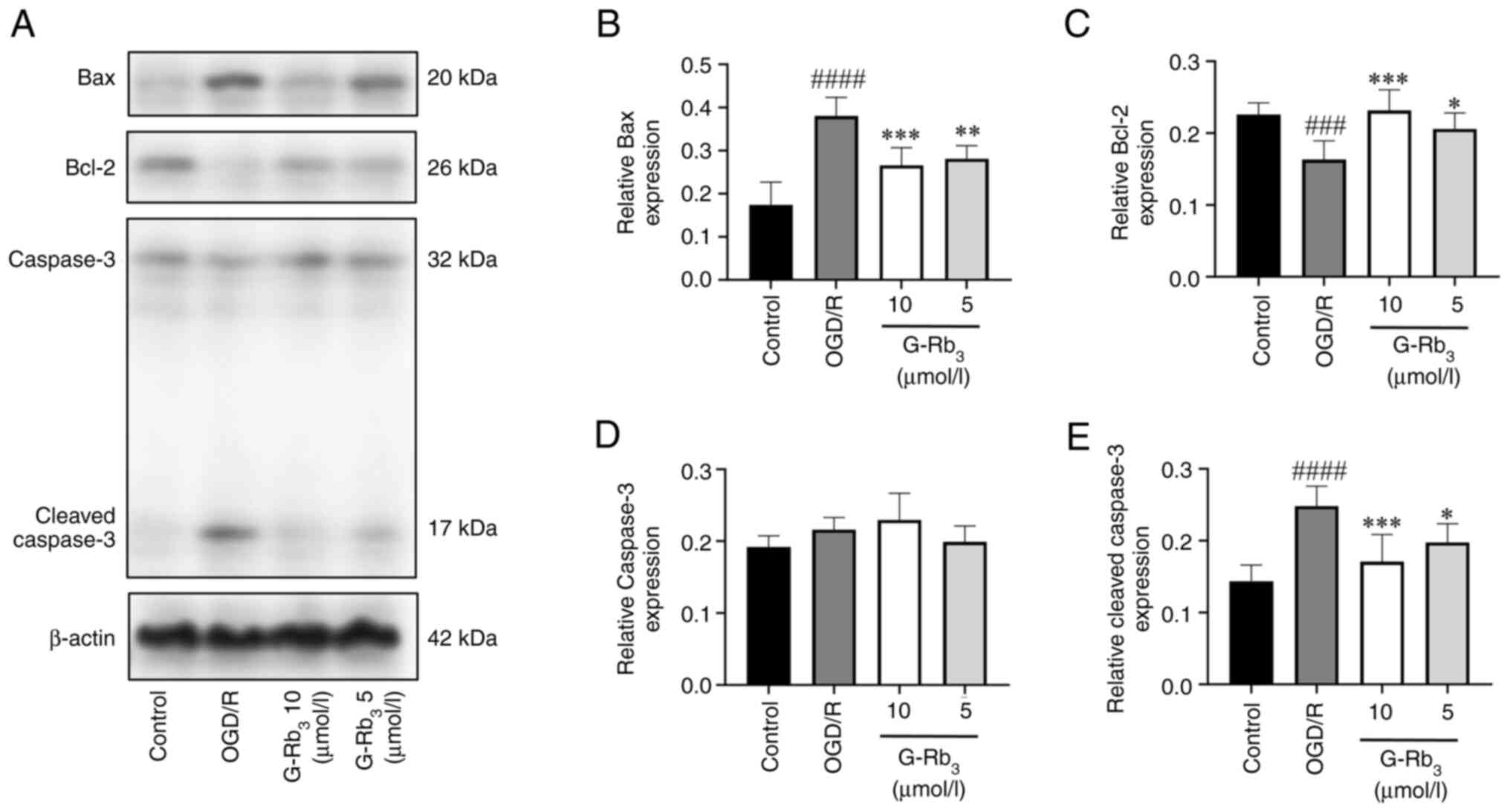 | Figure 2Western blot analysis of related
protein expression. (A) Bax, Bcl-2, caspase-3 and cleaved caspase-3
protein expression are illustrated. (B) Bax protein expression
statistic graph. Compared with the control group, the OGD/R group
had a significant difference (####P<0.0001) with
upregulated expression, compared with the OGD/R group, and
downregulated expression in the G-Rb3 group
(***P=0.0004 and Q=0.0008; **P=0.0019 and
Q=0.0019). (C) Statistical graph of Bcl-2 protein expression, which
was downregulated in the OGD/R group compared with the control
group (###P=0.0005 and Q=0.0015) and upregulated in the
G-Rb3 group compared with the OGD/R group with
significant differences (***P=0.0002 and Q=0.0003;
*P=0.0144 and Q=0.0144). (D) Caspase-3 protein
expression statistics were not significantly different between the
model group and the G-Rb3 group. (E) Cleaved caspase-3
protein expression statistics had significant difference and
upregulated expression in the OGD/R group compared with the control
group (####P<0.0001), downregulated expression in the
G-Rb3 group compared with the OGD/R group, and
significant difference (***P=0.0005 and Q=0.001;
*P=0.0185 and Q=0.185). G-Rb3, ginsenoside
Rb3; OGD/R, oxygen-glucose deprivation/reoxygenation;
kDa, kilodalton. |
Feasibility evaluation of metabolomics
experimental data
The screening methods for differential metabolites
mainly include PCA, PLS-DA and OPLS-DA. PCA is an unsupervised
discriminant analysis method, which reveals clustering of the
samples within the groups and dispersion of the samples between the
groups. The results were reliable (Fig.
3A and B). The advantage of
PLS-DA over PCA is that it is a supervised discriminant analysis
method, in which the samples have to be specified and grouped, and
the separation of groups was improved compared with the PCA
analysis (Fig. 3C and D). The OPLS-DA is based on PLS-DA, with
orthogonal transformation correction, which improves the model's
resolving ability and validity. The samples were further resolved
with OPLS-DA analysis to characterize the true differences between
groups and identify biomarkers from them. In both positive and
negative ion modes, intra-group sample clustering and inter-group
sample dispersion could be observed in the OGD/R group and the
G-Rb3 group (Fig. 3E and
F), suggesting that the results can
be used for further analysis.
 | Figure 3Detection of metabolomics data and
differential metabolite screening. (A and B) PCA scores plots.
Arranged in order to positive and negative ions. (C and D) PLS-DA
score plots. The samples were specified and grouped in positive and
negative ion order using PLS-DA to eliminate random errors
unrelated to the purpose of the study. (E and F) OPLS-DA scores
plots. The more clustered the samples within the group and the more
dispersed the samples between the groups, the more reliable the
results. (G) Clustering heat map of 31 differential metabolites.
Red color means higher expression, blue color means lower
expression, the top clustering line is the clustering line of OGD/R
group and G-Rb3 group, the left side is the clustering
line of metabolites, and the expression of metabolites can be
clearly observed on the right side. (H) Differential metabolite
volcano plot. Red dots represent upregulated differential
metabolites, blue dots represent downregulated differential
metabolites. (I) Differential metabolite enrichment analysis score
plot. Horizontal coordinate differential abundance score is the
total number of upregulated metabolites-total number of
downregulated metabolites/total number of metabolites. Vertical
coordinate is the pathway, and the size of the dot represents the
number of enriched differential metabolites in the pathway. M,
Model group; R, G-Rb3 group; PCA, principal component
analysis; PLS-DA, partial least-square discriminant analysis;
OPLS-DA, PLS-DA. |
Screening for differential
metabolites
In this method, P-value <0.05 and VIP value >1
were used to screen for differential metabolites between groups,
and a total of 31 metabolites showed significant differences
(Table III). In total, 12
metabolites were upregulated: 3-indoleacetonitrile, 4-pyridoxic
acid, enalaprilat, (R) 2,3-dihydroxy-3-methylvalerate, sorbitol,
yohimbine, 4-fumarylacetoacetate; pimelic acid, oleamide,
dihydrouracil, linoleic acid and cis-4-hydroxy-D-proline. In
addition, 19 metabolites were downregulated: geranyl diphosphate,
cis-aconitate acid, D-arabitol, D-lyxose, D-galactose;
alpha-D-glucose, adipic acid, isocitric acid, 6-phosphogluconic
acid, 3-hydroxymethylglutaric acid, linoleic acid, thiamine,
aminoethylphosphate, L-glutamate-gamma-semialdehyde, geranyl
diphosphate, procollagen 5-hydroxy-L-lysine, phosphoglycolic acid,
guanosine and suberic acid. The screening results are presented in
Table SII.
 | Table IIIDifferential metabolite
statistics. |
Table III
Differential metabolite
statistics.
| Group | Total number of
metabolites | Upregulated | Downregulated | Total number of
differential metabolites |
|---|
| RVSM | 291 | 12 | 19 | 31 |
Differential metabolite analysis
Heatmaps provide relative quantitative hierarchical
clustering of differential metabolites, where clustered metabolites
have similar expression patterns and may have similar functions or
participate in the same metabolic processes or cellular pathways.
Red color indicates the higher expression level and blue colour
indicates lower expression level (Fig.
3G). The distribution of differential metabolites between the
two groups of samples can be visualized using a volcano plot, where
the horizontal coordinates represent the multiplicity of
differences and the vertical coordinates represent the
significance. Red represents metabolites with significant
upregulation, blue represents metabolites with significant
downregulation and gray represents metabolites with no significant
differences (Fig. 3H). To observe
the overall changes in metabolism, this assessment captured the
average and overall changes in all metabolites in the pathway based
on differential abundance scores. The differential metabolites
between the G-Rb3 and OGD/R groups mainly interacted
through ABC transporters, galactose metabolism, glyoxylate and
dicarboxylate metabolism, citrate cycle, linoleic acid metabolism
and other different pathways affect nerves, energy metabolism and
other systems together. These data illustrated that the higher the
contribution of a detected metabolite under the ABC transporters
and galactose metabolism pathways, the more significant is the
effect of the differential metabolite on these pathways (Fig. 3I).
Apoptosis PCR array
A total of 87 target genes in the apoptosis PCR
array (Fig. 4A) were investigated,
according to the ‘log2FoldChange’ value as a reference.
In total, the results revealed that 30 target genes were
upregulated and 57 target genes were downregulated (Table SIII). The differential genes
screened according to |log2FoldChange |≥1 were the
following: Trp63, Trp73, Dapk1, Casp14
and Cd70, all of which were downregulated in expression by
the action of G-Rb3 (Fig.
4B).
Discussion
In the present study, hippocampal neuron HT22 cells
were used to replicate the OGD/R model for simulation of CIRI, and
to explore the mechanism of G-Rb3 against OGD/R injury
in apoptosis at the metabolite level. Under normal conditions,
Bcl-2 family member Bax exists in the cytoplasm as a monomer.
Apoptosis is caused by the interaction between pro- and
anti-apoptotic members of the Bcl-2 family, which activates the
release of the hydrolase caspase-3 into the cytoplasm. This in turn
activates the form of caspase-3 which can damage the cytoskeleton
and organelles, and degrade DNA and other proteins (28,29).
Following intervention with G-Rb3, the expression
between Bax and Bcl-2 is balanced and inhibited, thereby protecting
HT22 cells from damage caused by CIRI. Apoptosis is the most
important determinant of stroke, and inhibiting apoptosis is a key
treatment factor (30,31). Although preliminary studies by the
group have shown that G-Rb3 may reduce the damage caused
by OGD/R through antioxidant effect, the mechanism of action at the
metabolite level remains unclear. Therefore, based on the authors'
research group, cell viability was detected by trypan blue staining
and cell apoptosis was assessed with flow cytometry. The Bax and
Bcl-2 proteins play a key regulatory role in the process of
apoptosis. Specifically, the ischemic stroke stimulates Bax
translocation, triggering apoptosis, which Bcl-2 acts to prevent.
Therefore, the ratio of Bax to Bcl-2 has an important role in
apoptosis (32,33). Caspase-3 is an enzyme that promotes
apoptotic proteins, playing a key regulatory role in a variety of
apoptotic pathways and leading to apoptosis due to cleavage when
apoptotic signals are received (34,35).
In the present study, WB assays were conducted to show that
G-Rb3 increased the expression of anti-apoptotic protein
Bcl-2, and decreased that of pro-apoptotic protein Bax and
caspase-3 protein. Taken together, these results demonstrated that
G-Rb3 alleviated OGD/R injury by inhibiting
apoptosis.
Starting at the metabolite level in the exploration
of the mechanism of action of G-Rb3, it was revealed
that its protective effect on OGD/R-induced HT22 cells was mainly
mediated through changes in metabolites such as nitrogen-containing
organic compounds and lipid compounds. A total of 31 metabolites
were analyzed for between-group differences between the
G-Rb3 and OGD/R groups. A total of 12 metabolites were
upregulated after G-Rb3 intervention, such as
3-indolacetonitrile, enalaprilat, (R)
2,3-dihydroxy-3-methylvalerate, sorbitol, 4-pyridoxic acid,
4-fumarylacetoacetate and pimelic acid. On the other hand, a total
of 19 metabolites were downregulated, such as D-arabitol, D-lyxose,
cis-aconitate, 6-phosphogluconic acid, isocitric acid, adipic acid
and guanosine. One of these metabolites, guanosine, belongs to the
group of endogenous guanine nucleosides that have been shown to
protect neurons from damage and induced cell death; it as a trophic
factor to promote neuroprotection during oxygen-glucose
deprivation, and to exert anti-inflammatory effects (36-38).
D-galactose is a naturally occurring aldose in the body, which is
usually metabolized into glucose by galactokinase and uridine
transferase, and stored as glycogen in liver, muscle and adipose
tissue. When administered at high doses, exogenous D-galactose
induces senescent effects in several organs by increasing oxidative
stress, apoptosis and inflammation. This in turn leads to cognitive
decline; however, ameliorating oxidative stress in hippocampal
neurons can alleviate such damage. Ιn vitro assays revealed
that D-galactose decreases the viability of HT22 cells and causes
apoptosis (39-41).
Yohimbine is a selective α2-adrenergic blocking agent
with neuroprotective effect (42).
Sorbitol is an osmotic dehydrating diuretic drug that acts, by
increasing plasma osmolality, drawing water out of the eye, brain
and othertissues into the blood vessels, reducing tissue edema,
protecting brain tissue and indirectly protecting HT22 cells
(43). Previous studies have shown
a close relationship between stroke and high blood pressure
(44-46).
Hypertension causes >1.5 million strokes each year and the
second highest number of deaths worldwide. Antihypertensive therapy
helps to prevent most ischemic strokes. Enalaprilat belongs to the
angiotensin-converting enzyme inhibitors drug class, which
significantly reduces systolic and diastolic blood pressure. The
drugs normalize central and cerebral hemodynamic parameters and
reduce the degree of strain on the renin-angiotensin-aldosterone
regulatory system, thus protecting stroke patients and indirectly
protecting HT22 cell damage (44-46).
Linoleic acid protects OGD/R damage by inhibiting the increase of
Ca2+ induced by OGD, causing an increase in reactive
oxygen species levels and reducing apoptosis (47). The aforementioned metabolites that
were altered after G-Rb3 intervention are closely
related to apoptosis. Therefore, it was hypothesized that increases
or decreases in the levels of these metabolites might be related to
G-Rb3 mediated protection against OGD/R induced HT22
cell damage through inhibition of apoptosis. Subsequent KEGG
enrichment analysis demonstrated that G-Rb3 might act by
regulating metabolic pathways involving ABC transporters, galactose
metabolism and citrate cycle. This led to the hypothesis that
guanosine may have potential as a biomarker for the diagnosis of
CIRI attenuated by G-Rb3.
Based on the metabolomics-based analysis, the
expression of apoptotic genes using PCR array with
|log2FC|≥1 was verified as the screening condition. This
identified the following five differential genes, all of which are
pro-apoptotic: i) Trp63 and Trp73, the main members
of the Trp53 family, which induce apoptosis when they are
overexpressed (48,49); ii) Dapk1, the key gene in the
process of ischemic neuronal death (50); iii) Casp14, a member of the
caspase family and a central player in the execution phase of
apoptosis (51); and iv)
Cd70, which belongs to the tumour necrosis factor family and
induces apoptosis by regulating T-cell activity (52-54).
The results of the present study showed that G-Rb3
significantly downregulated these five pro-apoptotic genes, with
consistent results in PCR array and WB assays. Combined with the
flow cytometry and trypan blue staining results, the data
consistently support a possible role for G-Rb3 in the
inhibition apoptosis.
Owing to the relative complexity of the pathogenesis
of stroke, there are certain limitations to the present study, such
as the lack of verification of metabolites and metabolic pathways
and the research angle being relatively simple. In addition, it was
not verified whether other active components of Panax
notoginseng have a protective effect on cerebral ischemia and
reperfusion. The lack of in-depth analysis affects the
comprehensiveness of the research results. Validation of
metabolomics results by animal experiments will be considered.
In conclusion, it was found in the present study
that G-Rb3 protects against OGD/R injury through a
mechanism involving altered guanosine and regulation of the ABC
transporters metabolic pathway to inhibit apoptosis (Fig. 5). It can be hypothesized that
G-Rb3 can improve CIRI. While there remain numerous
other complex pathological factors of stroke to be examined in the
future, the present study provides a reference for the clinical
application of G-Rb3 in the treatment of stroke.
Supplementary Material
High-resolution scan of the original
blots from Fig. 2. (A) The original
blots of Bax protein. (B) The original blots of Bcl-2 protein. (C)
The original blots of caspase-3 protein. (D) The original blots of
β-actin.
Primer sequences.
Screening results for differential
metabolites in the model andtreatment groups.
PCR array.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by the Regularity and
Mechanism of Prescriptions Containing Panax notoginseng based on
Data Mining and Network Pharmacology (grant no. 2022J0953).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author. The metabolomics data that
have been generated in the present study may be found in the
EMBL-EBI MetaboLights database under accession number MTBLS10570 or
at the following URL: https://www.ebi.ac.uk/metabolights/MTBLS10570.
Authors' contributions
FL was the main contributor to the present study to
formulate the experimental scheme. JT and XD designed the
experiments and performed data analysis. MZ and JT conducted the
statistical analysis and confirm the authenticity of all the raw
data. XY and TX examined the relevant indicators of the experiment
and contributed to statistical analysis of the data. CW interpreted
the research results and modified the manuscript. All authors read
and approved the final version of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Markus HS: Stroke genetics. Hum Mol Genet.
20:R124–R131. 2011.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Abbas M, Malicke DT and Schramski JT:
Stroke anticoagulation. In: StatPearls. StatPearls Publishing,
Treasure Island, FL, 2024.
|
|
3
|
Campbell BCV and Khatri P: Stroke. Lancet.
396:129–142. 2020.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Campbell BCV, De Silva DA, Macleod MR,
Coutts SB, Schwamm LH, Davis SM and Donnan GA: Ischaemic stroke.
Nat Rev Dis Primers. 5(70)2019.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Xu C, Wang W, Wang B, Zhang T, Cui X, Pu Y
and Li N: Analytical methods and biological activities of Panax
notoginseng saponins: Recent trends. J Ethnopharmacol.
236:443–465. 2019.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Liang Z, Liu K, Li R, Ma B, Zheng W, Yang
S, Zhang G, Zhao Y, Chen J and Zhao M: An instant beverage rich in
nutrients and secondary metabolites manufactured from stems and
leaves of Panax notoginseng. Front Nutr.
9(1058639)2022.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Chen XM, Chen HS, Xu MJ and Shen JG:
Targeting reactive nitrogen species: a promising therapeutic
strategy for cerebral ischemia-reperfusion injury. Acta
pharmacologica Sin. 34:67–77. 2013.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Thorén M, Dixit A, Escudero-Martínez I,
Gdovinová Z, Klecka L, Rand VM, Toni D, Vilionskis A, Wahlgren N
and Ahmed N: Effect of recanalization on cerebral edema in ischemic
stroke treated with thrombolysis and/or endovascular therapy.
Stroke. 51:216–223. 2020.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Takahashi H, Yamamoto T and Tsuboi A:
Molecular mechanisms underlying activity-dependent ischemic
tolerance in the brain. Neurosci Res. 186:3–9. 2023.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Zheng MM, Zhang F and Zhang Q: Research
progress in biological activity of ginsenoside Rb3. Central South
Pharm. 9:1249–1252. 2017.(In Chinese).
|
|
11
|
Liu X, Jiang Y, Yu X, Fu W, Zhang H and
Sui D: Ginsenoside-Rb3 protects the myocardium from
ischemia-reperfusion injury via the inhibition of apoptosis in
rats. Exp Ther Med. 8:1751–1756. 2014.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Kim DH, Kim DW, Jung BH, Lee JH, Lee H,
Hwang GS, Kang KS and Lee JW: Ginsenoside Rb2 suppresses the
glutamate-mediated oxidative stress and neuronal cell death in HT22
cells. J Ginseng Res. 43:326–334. 2019.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Qian T, Cai Z, Wong RNS, Mak NK and Jiang
ZH: In vivo rat metabolism and pharmacokinetic studies of
ginsenoside Rg3. J Chromatogr B Analyt Technol Biomed Life Sci.
816:223–232. 2005.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Qian T, Cai Z, Wong RNS and Jiang ZH:
Liquid chromatography/mass spectrometric analysis of rat samples
for in vivo metabolism and pharmacokinetic studies of ginsenoside
Rh2. Rapid Commun Mass Spectrom. 19:3549–3554. 2005.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Zhou S, Gao X, Chen C, Zhang J, Zhang Y,
Zhang L and Yan X: Porcine cardiac blood-Salvia miltiorrhiza root
alleviates cerebral ischemia reperfusion injury by inhibiting
oxidative stress induced apoptosis through PI3K/AKT/Bcl-2/Bax
signaling pathway. J Ethnopharmacol. 316(116698)2023.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Yuan Y, Tian Y, Jiang H, Cai LY, Song J,
Peng R and Zhang XM: Mechanism of PGC-1α-mediated mitochondrial
biogenesis in cerebral ischemia-reperfusion injury. Front Mol
Neurosci. 16(1224964)2023.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Schrimpe-Rutledge AC, Codreanu SG, Sherrod
SD and McLean JA: Untargeted metabolomics strategies-challenges and
emerging directions. J Am Soc Mass Spectrom. 27:1897–1905.
2016.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Wishart DS: Metabolomics for investigating
physiological and pathophysiological processes. Physiol Rev.
99:1819–1875. 2019.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Smith CA, Want EJ, O'Maille G, Abagyan R
and Siuzdak G: XCMS: Processing mass spectrometry data for
metabolite profiling using nonlinear peak alignment, matching, and
identification. Anal Chem. 78:779–787. 2006.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Navarro-Reig M, Jaumot J, García-Reiriz A
and Tauler R: Evaluation of changes induced in rice metabolome by
Cd and Cu exposure using LC-MS with XCMS and MCR-ALS data analysis
strategies. Anal Bioanal Chem. 407:8835–8847. 2015.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Cheng J, Li G, Yang L, Chen P and Duan X:
Alcohol extract of Rubia yunnanensis: Metabolic alterations and
preventive effects against OGD/R-induced oxidative damage in HT22
cells. Biomed Rep. 20(75)2024.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Wishart DS, Tzur D, Knox C, Eisner R, Guo
AC, Young N, Cheng D, Jewell K, Arndt D, Sawhney S, et al: HMDB:
The human metabolome database. Nucleic Acids Res. 35 (Database
Issue):D521–D526. 2007.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Horai H, Arita M, Kanaya S, Nihei Y, Ikeda
T, Suwa K, Ojima Y, Tanaka K, Tanaka S, Aoshima K, et al: MassBank:
A public repository for sharing mass spectral data for life
sciences. J Mass Spectrom. 45:703–714. 2010.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Sud M, Fahy E, Cotter D, Brown A, Dennis
EA, Glass CK, Merrill AH Jr, Murphy RC, Raetz CR, Russell DW and
Subramaniam S: LMSD: LIPID MAPS structure database. Nucleic Acids
Res. 35 (Database Issue):D527–D532. 2007.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Abdelrazig S, Safo L, Rance GA, Fay MW,
Theodosiou E, Topham PD, Kim DH and Fernández-Castané A: Metabolic
characterisation of Magnetospirillum gryphiswaldense MSR-1 using
LC-MS-based metabolite profiling. RSC Adv. 10:32548–32560.
2020.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Xia J and Wishart DS: Web-based inference
of biological patterns, functions and pathways from metabolomic
data using MetaboAnalyst. Nat Protoc. 6:743–760. 2011.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Barman J, Kumar R, Saha G, Tiwari K and
Dubey VK: Apoptosis: Mediator molecules, interplay with other cell
death processes and therapeutic potentials. Curr Pharm Biotechnol.
19:644–663. 2018.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Flores-Romero H, Ros U and Garcia-Saez AJ:
Pore formation in regulated cell death. EMBO J.
39(e105753)2020.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Yuan J, Zeng L, Sun Y, Wang N, Sun Q,
Cheng Z and Wang Y: SH2B1 protects against OGD/R-induced apoptosis
in PC12 cells via activation of the JAK2/STAT3 signaling pathway.
Mol Med Rep. 18:2613–2620. 2018.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Peña-Blanco A and García-Sáez AJ: Bax, Bak
and beyond-mitochondrial performance in apoptosis. FEBS J.
285:416–431. 2018.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Brady HJ and Gil-Gómez G: Bax. The
pro-apoptotic Bcl-2 family member, Bax. Int J Biochem Cell Biol.
30:647–650. 1998.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Edlich F: BCL-2 proteins and apoptosis:
Recent insights and unknowns. Biochem Biophys Res Commun.
500:26–34. 2018.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Trubiani O, Guarnieri S, Paganelli R and
Di Primio R: Involvement of caspace-3 in the cleavage of terminal
transferase. Int J Immunopathol Pharmacol. 15:201–208.
2002.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Dal-Cim T, Ludka FK, Martins WC, Reginato
C, Parada E, Egea J, López MG and Tasca CI: Guanosine controls
inflammatory pathways to afford neuroprotection of hippocampal
slices under oxygen and glucose deprivation conditions. J
Neurochem. 126:437–450. 2013.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Rathbone M, Pilutti L, Caciagli F and
Jiang S: Neurotrophic effects of extracellular guanosine.
Nucleosides Nucleotides Nucleic Acids. 27:666–672. 2008.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Oliveira KA, Dal-Cim TA, Lopes FG, Nedel
CB and Tasca CI: Guanosine promotes cytotoxicity via adenosine
receptors and induces apoptosis in temozolomide-treated A172 glioma
cells. Purinergic Signal. 13:305–318. 2017.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Schneider EH, Hofmeister O, Kälble S and
Seifert R: Apoptotic and anti-proliferative effect of guanosine and
guanosine derivatives in HuT-78 T lymphoma cells. Naunyn
Schmiedebergs Arch Pharmacol. 393:1251–1267. 2020.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Shwe T, Pratchayasakul W, Chattipakorn N
and Chattipakorn SC: Role of D-galactose-induced brain aging and
its potential used for therapeutic interventions. Exp Gerontol.
101:13–36. 2018.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Xue A, Zhao D, Zhao C, Li X, Yang M, Zhao
H, Zhao C, Lei X, Wu J and Zhang N: Study on the neuroprotective
effect of Zhimu-Huangbo extract on mitochondrial dysfunction in
HT22 cells induced by D-galactose by promoting mitochondrial
autophagy. J Ethnopharmacol. 318(117012)2024.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Kwon HJ, Hahn KR, Nam SM, Yoon YS, Moon
SM, Hwang IK and Kim DW: Purpurin ameliorates D-galactose-induced
aging phenotypes in mouse hippocampus by reducing inflammatory
responses. Neurochem Int. 167(105552)2023.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Jabir NR, Firoz CK, Zughaibi TA, Alsaadi
MA, Abuzenadah AM, Al-Asmari AI, Alsaieedi A, Ahmed BA, Ramu AK and
Tabrez S: A literature perspective on the pharmacological
applications of yohimbine. Ann Med. 54:2861–2875. 2022.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Bremer AM, Yamada K and West CR: Ischemic
cerebral edema in primates: effects of acetazolamide, phenytoin,
sorbitol, dexamethasone, and methylprednisolone on brain water and
electrolytes. Neurosurgery. 6:149–154. 1980.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Mailloux A, Deslandes B, Vaubourdolle M
and Baudin B: Captopril and enalaprilat decrease antioxidant
defences in human endothelial cells and are unable to protect
against apoptosis. Cell Biol Int. 27:825–830. 2003.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Gomez HJ, Cirillo VJ and Irvin JD:
Enalapril: A review of human pharmacology. Drugs. 30 (Suppl
1):S13–S24. 1985.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Niu JJ, Bai Lf and Hou CN: Comparison of
therapeutic effects of captopril, enalapril and sodium
nitroprusside on hypertension, 2018.
|
|
47
|
Turovsky EA, Varlamova EG, Gudkov SV and
Plotnikov EY: The protective mechanism of deuterated linoleic acid
involves the activation of the Ca2+ signaling system of
astrocytes in ischemia in vitro. Int J Mol Sci.
22(13216)2021.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Kenzelmann Broz D and Attardi LD: TRP53
activates a global autophagy program to promote tumor suppression.
Autophagy. 9:1440–1442. 2013.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Jacobs WB, Govoni G, Ho D, Atwal JK,
Barnabe-Heider F, Keyes WM, Mills AA, Miller FD and Kaplan DR: p63
is an essential proapoptotic protein during neural development.
Neuron. 48:743–756. 2005.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Wei R, Zhang L, Hu W, Wu J and Zhang W:
Long non-coding RNA AK038897 aggravates cerebral
ischemia/reperfusion injury via acting as a ceRNA for miR-26a-5p to
target DAPK1. Exp Neurol. 314:100–110. 2019.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Markiewicz A, Sigorski D, Markiewicz M,
Owczarczyk-Saczonek A and Placek W: Caspase-14-from biomolecular
basics to clinical approach. A review of available data. Int J Mol
Sci. 22(5575)2021.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Hoefsmit EP, van Royen PT, Rao D,
Stunnenberg JA, Dimitriadis P, Lieftink C, Morris B, Rozeman EA,
Reijers ILM, Lacroix R, et al: Inhibitor of apoptosis proteins
antagonist induces T-cell Proliferation after cross-presentation by
dendritic cells. Cancer Immunol Res. 11:450–465. 2023.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Sasnauskiene A, Kadziauskas J, Vezelyte N,
Jonusiene V and Kirveliene V: Apoptosis, autophagy and cell cycle
arrest following photodamage to mitochondrial interior. Apoptosis.
14:276–286. 2009.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Wensveen FM, Unger PPA, Kragten NAM, Derks
IA, ten Brinke A, Arens R, van Lier RA, Eldering E and van
Gisbergen KP: CD70-driven costimulation induces survival or
Fas-mediated apoptosis of T cells depending on antigenic load. J
Immunol. 188:4256–4267. 2012.PubMed/NCBI View Article : Google Scholar
|



















