Introduction
Antineutrophil cytoplasmic antibody
(ANCA)-associated vasculitis (AAV) is an autoimmune disease
characterized by necrotizing inflammation of small and medium-sized
blood vessels and the presence of ANCAs in circulation. Based on
their targeted antigen, ANCAs are classified as proteinase 3-ANCA
(PR3-ANCA) or myeloperoxidase-ANCA (MPO-ANCA) (1-3).
Among the organs affected by AAV, the kidney is one of the most
severely impacted. The pathogenesis of AAV remains unclear. It is
currently hypothesized that environmental factors, infection and
immune status lead to the development of this disease. A genetic
predisposition to AAV has been reported in familial cases and large
genome-wide association studies have revealed that susceptibility
to AAV is linked to gene variants (4-6).
Autophagy is the process by which intracellular
macromolecules, organelles and invasive microorganisms are
transported to the lysosome for degradation under regulation of
autophagy-related genes (ATGs) (7).
Autophagy is key for the realization of cellular metabolic needs,
the maintenance of cell and tissue homeostasis and innate and
adaptive immunity (8). Autophagy
has been implicated in a variety of diseases, including malignant
tumours, neurodegenerative disease, metabolic syndrome and
autoimmune diseases (9-12).
Neutrophil extracellular traps (NETs) are reticular DNA structures
combined with immunogenic proteins released by activated
neutrophils that not only capture and kill pathogenic
microorganisms but also induce tissue damage (13), serving a key role in the initiation
and progression of AAV (14). Sha
et al (15) reported that
human neutrophils treated with ANCA-positive IgG exhibit higher
levels of autophagy and release more NETs, which can be enhanced by
autophagy inducers, suggesting that autophagic activity may affect
ANCA-induced NET formation and release and thus participate in the
pathogenesis of AAV.
Autophagy-related 16-like 1 (ATG16L1) is a key
component of ATG12-ATG5/ATG16L1, a large protein complex essential
for all stages of autophagy (16).
ATG16L1 mediates recognition of cellular cargo and activates
autophagy-associated enzyme activity required for recruitment to
lysosomes (17). ATG16L1 gene
polymorphisms [rs2241880(T300A) and rs4663421] may be associated
with rheumatoid arthritis, ankylosing spondylitis and inflammatory
bowel disease (18-20).
The present analysed the associations between ATG16L1
rs2241880(T300A) and rs4663421 polymorphisms and susceptibility in
AAV using propensity score matching (PSM) to control for
confounding factors. The aim of the present study was to provide a
theoretical basis for pathogenesis and therapeutic targets of
AAV.
Materials and methods
Subjects
Patients with AAV were recruited from Department of
Nephrology, Second Affiliated Hospital of Guangxi Medical
University (Guangxi, China) from January 2005 to April 2018. A
total of 177 patients were included, 68 male (38.4%) and 109 female
(61.6%), with an age range of 18-82 years and a median (IQR) age of
58.0 (43.0-64.0) years. Inclusion criteria were as follows: i)
Diagnosis of AAV in strict accordance with the diagnostic criteria
for vasculitis formulated at the International Chapel Hill
Consensus Conference in 2012(21)
and ii) have been born in the Guangxi Zhuang Autonomous Region with
no blood relationship with any other subject in the present study.
The exclusion criteria were diagnosis of secondary vasculitis,
other autoimmune or disease and malignant tumours. The control
group consisted of healthy individuals from the Physical
Examination Centre of the same hospital during the same period. A
total of 216 healthy individuals were included, 84 male (38.9%) and
132 female (61.1%), with an age range of 19-81 years and a median
(IQR) age of 51.0 (44.0-59.0) years. Inclusion criteria were as
follows: i) Does not meet any of the above classification
diagnostic criteria; and ii) have been born in the Guangxi Zhuang
Autonomous Region with no blood relationship with any other subject
who has participated in this study. The exclusion criteria were
diagnosis of autoimmune diseases, hereditary diseases, and other
serious diseases. The study was conducted in accordance with the
Declaration of Helsinki and approved by the Ethics Committee of the
Second Affiliated Hospital of Guangxi Medical University (approval
no. 2018 KY-0100). Verbal informed consent was obtained from all
participants.
Clinical data
The clinical data collected included sex, age, white
blood cell count, 24-h urine protein, albumin and serum creatinine
(SCR) levels, estimated glomerular filtration rate (eGFR),
C-reactive protein (CRP) levels, erythrocyte sedimentation rate
(ESR), MPO-ANCA and PR3-ANCA titre, Birmingham Vasculitis Activity
Score (BVAS) and renal pathology (including renal pathological
type, normal glomeruli, sclerotic glomeruli, global sclerosis,
segmental sclerosis, crescents, cellular crescents, fibrous
crescents and cellular fibrous crescents). The renal samples were
fixed at 35˚C with 10% buffered formalin for 5 min, embedded in
paraffin, sectioned at a thickness of 1.5-2 µm, then stained with
hematoxylin and eosin for 10 min and 31 min respectively at room
temperature. Finally, observations were made using an light
microscope (Olympus Corporation) with x40. Disease activity was
assessed using the Birmingham Vasculitis Activity Score (version 3)
(22). eGFR was calculated using
the Chronic Kidney Disease Epidemiology Collaboration equation
(23). ANCA-associated
glomerulonephritis was classified according to the
histopathological classification proposed by Berden et al
(24). The clinical data collected
were obtained at the time of diagnosis of patients or during
physical examination of healthy individuals.
Single nucleotide polymorphism (SNP)
selection
Locus information for the ATG16L1 gene was
downloaded from 1000 Genomes database (grch37.ensembl.org/), and SNP loci were filtered out
using Haploview v.4.2 software (25). SNP loci meeting the following
criteria were selected: i) Located in a functional region; ii)
previously reported to be associated with autoimmune or
inflammatory disease and iii) having a minor allele frequency
≥0.05. National Center for Bioinformatics (ncbi.nlm.nih.gov/snp/) was used to confirm functional
consequence of SNPs and ATG16L1 marker SNPs [rs2241880(T300A) and
rs4663421] were selected.
Gene polymorphism detection
Blood samples were collected from all the enrolled
patients with AAV and controls between January 2005 to April 2018.
EDTA tubes were used for the collection of 5 ml peripheral venous
blood samples and genomic DNA was extracted using the Blood DNA
Extraction kit [cat. no. DP319-02; Beijing Tiangen Biotech
(Beijing) Co., Ltd.] according to the manufacturer's instructions.
Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific, Inc.) to
measure the concentration of DNA to ensure the adequate amounts of
high-quality genomic DNA. The samples with an absorbance value
(A260/A280) of 1.7-1.9 and a DNA concentration >25 mg/l were
included and the isolated DNA was stored at -80˚C for further
study.
Genotyping was performed by Sangon Biotech
(Shanghai) Co., Ltd. The sequences of primers were as follows:
rs2241880 forward, 5'-TCTCATTTGAGTGAGGGTGCTTTT-3' and reverse,
5'-GTAGCTGGTACCCTCACTTCTTTAC-3' (product size, 274 bp); and
rs4663421 forward, 5'-CCCTTCTTCCATGTATCCTGCTT-3' and reverse
5'-CTTCCAGCCAAATCTGCTTTTCC-3'(product size, 274 bp). Library
preparation was performed by two-step PCR. First round PCR reaction
was set up as follows: DNA (10 ng/µl) 2 µl; amplicon PCR forward
primer mix (10 µM) 1 µl; amplicon PCR reverse primer mix (10 µM) 1
µl; 2xPCR Ready Mix 15 µl (total 25 µl). This step was performed
with Kapa HiFi Ready Mix [Roche Dianostics (Shanghai) Co., Ltd.).
PCR was performed in a thermal instrument (Bio-Rad Laboratories,
Inc.; T100TM) using the following program: 1 cycle of denaturing at
98˚C for 5 min, first 8 cycles of denaturing at 98˚C for 30 sec,
annealing at 50˚C for 30 sec, elongation at 72˚C for 30 sec, then
25 cycles of denaturing at 98˚C for 30 sec, annealing at 66˚C for
30 sec, elongation at 72˚C for 30 sec and a final extension at 72˚C
for 5 min. Finally hold at 4˚C.
The PCR products were checked using electrophoresis
in 1 % (w/v) agarose gels in TBE buffer, stained with ethidium
bromide and visualized under UV light. Then we used AMPure XP beads
to purify the amplicon product. After that, the second round of PCR
was performed. The PCR reaction was set up as follows: DNA (10
ng/µl) 2 µl; universal P7 primer with barcode (10 µM) 1 µl;
universal P5 primer (10 µM) 1 µl; 2X PCR Ready Mix 15 µl (total 30
µl)(Kapa HiFi Ready Mix). The plate was sealed and PCR was
performed as follows: Initial denaturing at 98˚C for 3 min, then 5
cycles of denaturing at 94˚C for 30 sec, annealing at 55˚C for 20
sec, elongation at 72˚C for 30 sec, and a final extension at 72˚C
for 5 min. Then we used AMPure XP beads to purify the amplicon
product. The libraries were then quantified and pooled. Paired-end
sequencing of the library was performed on the HiSeq XTen
sequencers (Illumina, Inc.). The samples were sequenced using
NovaSeq 6000 S4 Reagent kit v1.5 (300 cycles) (cat. no. 20028312;
Illumina Inc.). The loading concentration of the final library was
100 pM. Samtools v 1.18software (htslib.org/) was
used to calculate each genotype of target site.
PSM
IBM SPSS Statistics for Windows, Version 26.0 (IBM
Corp.) and R 3.5.1 software (26)
was used to match sex, age and ethnicity with a matching tolerance
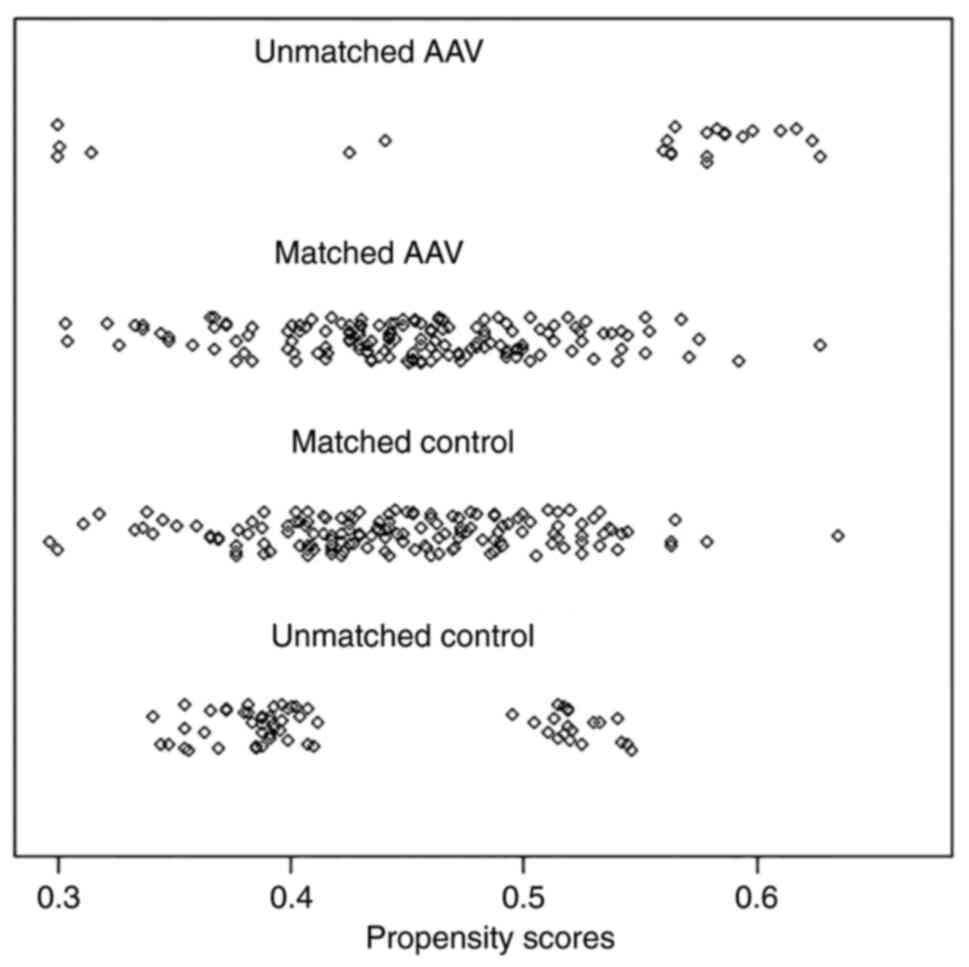
of 0.2(27). Jetter plot of
propensity score matching was obtained.
Statistical analysis
Hardy-Weinberg equilibrium was checked using SHEsis
online software (analysis.biox.cn/myanalysis.php), and linkage
disequilibrium tests were performed. The strength of association
between genetic models and the risk of AAV was evaluated by odds
ratios (ORs) and 95% confidence intervals (CIs) through online
SNPstats (https://www.snpstats.net/start.htm). SNP-SNP
interactions were assessed using generalized multifactor
dimensionality reduction (GMDR). Expression quantitative trait loci
(eQTL) database (gtexportal.org/home/eqtlDashboardPage) was used to
assess the effect of different genotypes of candidate loci on
tissue-specific gene expression. All statistical analyses were
performed with IBM SPSS Statistics for Windows, Version 26.0 (IBM
Corp.). Normally distributed variables are presented as the mean ±
SD of 3 independent experimental repeats and non-normally
distributed variables are presented as median and interquartile
range (IQR). The categorical variables are expressed as frequency
and percentage and comparisons between groups were made using
Pearson's χ2 test or Fisher's exact test. P<0.05 was
considered to indicate a statistically significant difference.
Results
Demographic characteristics
A total of 177 patients and 216 healthy individuals
were included. There was a significant age gap between the AAV and
control groups before PSM (Table I)
but no apparent difference in sex or ethnicity between the groups.
Following PSM, 154 patients were successfully matched. There was no
significant difference between age of groups and the balance of
each covariate was significantly increased (Figs. 1 and 2).
 | Table IBaseline influencing factors before
and after PSM. |
Table I
Baseline influencing factors before
and after PSM.
| | Before PSM | After PSM |
|---|
| Characteristic | AAV (n=177) | Control
(n=216) | P-value | AAV (n=154) | Control
(n=154) | P-value |
|---|
| Sex (%) | | | 0.924 | | | 0.483 |
|
Male | 68 (38.4) | 84 (38.9) | | 63 (40.9) | 57 (37.0) | |
|
Female | 109 (61.6) | 132 (61.1) | | 91 (59.1) | 97 (63.0) | |
| Ethnicity (%) | | | 0.099 | | | 0.611 |
|
Zhuang | 62 (35.0) | 58 (26.9) | | 45 (29.2) | 41 (26.6) | |
|
Han | 115 (65.0) | 158 (73.1) | | 109 (70.8) | 113 (73.4) | |
| Median age (IQR),
years | 58.0
(43.0-64.0) | 51.0
(44.0-59.0) | 0.005 | 56.0
(42.0-63.0) | 53.0
(45.8-61.0) | 0.445 |
Genotype distribution, allele
frequency, linkage disequilibrium and Hardy-Weinberg equilibrium
analysis
The genotype distribution of rs2241880(T300A) and
rs4663421 of ATG16L1 were in accordance with the genetic law of
Hardy-Weinberg equilibrium. The linkage disequilibrium was weak
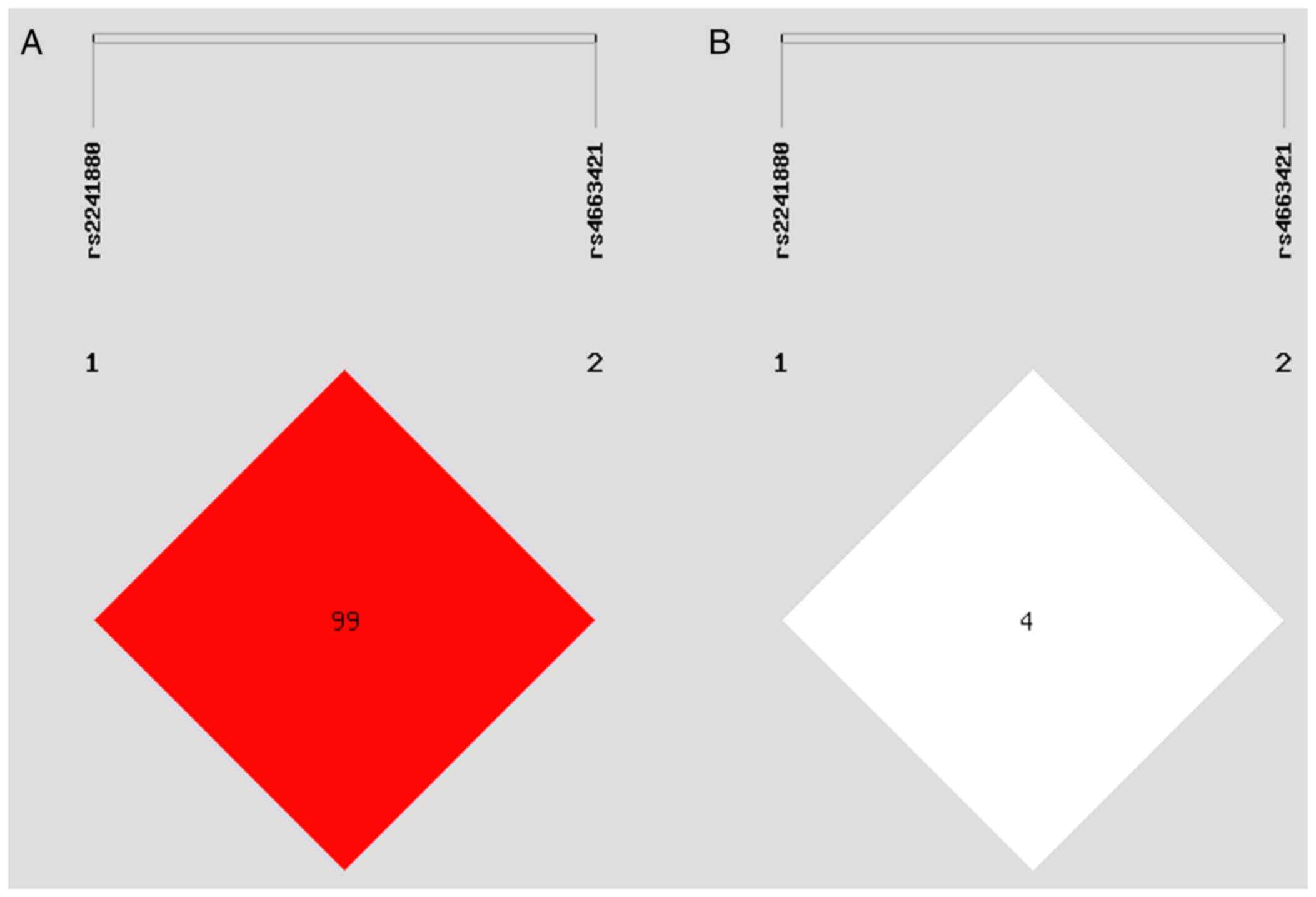
between rs2241880(T300A) and rs4663421 loci (Fig. 3; D'=0.998, R2=0.045).
Associations between ATG16L1 SNPs and AAV risk were evaluated under
different genetic models (Tables
II and III). Genotypes of
rs2241880 and rs4663421 under different genetic models did not
significantly difference between AAV and control groups. And there
was no significant difference in the alleles of the two loci
between AAV and control groups.
 | Table IIAssociation between the risk of AAV
and single nucleotide polymorphism rs2241880. |
Table II
Association between the risk of AAV
and single nucleotide polymorphism rs2241880.
| Model |
Genetype/Allele | Control (%) | AAV (%) | OR (95% CI) | P-value |
|---|
| Allele | G | 98 (31.8) | 88 (28.6) | Reference | 0.38 |
| | A | 210 (68.2) | 220 (71.4) | 1.17
(0.83-1.65) | |
| Codominant | AA | 71 (46.1) | 78 (50.6) | Reference | 0.68 |
| | AG | 68 (44.2) | 64 (41.6) | 0.86
(0.54-1.37) | |
| | GG | 15 (9.7) | 12 (7.8) | 0.73
(0.32-1.66) | |
| Dominant | AA | 71 (46.1) | 78 (50.6) | Reference | 0.42 |
| | AG-GG | 83 (53.9) | 76 (49.4) | 0.83
(0.53-1.30) | |
| Recessive | AA-AG | 139 (90.3) | 142 (92.2) | Reference | 0.55 |
| | GG | 15 (9.7) | 12 (7.8) | 0.78
(0.35-1.73) | |
| Overdominant | AA-GG | 86 (55.8) | 90 (58.4) | Reference | 0.65 |
| | AG | 68 (44.2%) | 64 (41.6%) | 0.90
(0.57-1.41) | |
 | Table IIIAssociation between risk of AAV and
single nucleotide polymorphism rs4663421 in different genotypic
models. |
Table III
Association between risk of AAV and
single nucleotide polymorphism rs4663421 in different genotypic
models.
| Model |
Genetype/Allele | Control (%) | AAV (%) | OR (95% CI) | P-value |
|---|
| Allele | G | 279 (90.6) | 282 (91.6) | Reference | 0.67 |
| | C | 29 (9.4) | 26 (8.4) | 0.89
(0.51-1.55) | |
| Codominant | GG | 125 (81.2) | 129 (83.8) | Reference | 0.38 |
| | GC | 29 (18.8) | 24 (15.6) | 0.80
(0.44-1.45) | |
| | CC | 0 (0.0) | 1 (0.6) | NA (0.00-NA) | |
| Dominant | GG | 125 (81.2) | 129 (83.8) | Reference | 0.55 |
| | GC-CC | 29 (18.8) | 25 (16.2) | 0.84
(0.46-1.51) | |
| Recessive | GG-GC | 154(100) | 153 (99.3) | Reference | 0.24 |
| | CC | 0 (0) | 1 (0.6) | NA (0.00-NA) | |
| Overdominant | GG-CC | 125 (81.2) | 130 (84.4) | Reference | 0.45 |
| | GC | 29 (18.8) | 24 (15.6) | 0.80
(0.44-1.44) | |
SNP interaction analysis
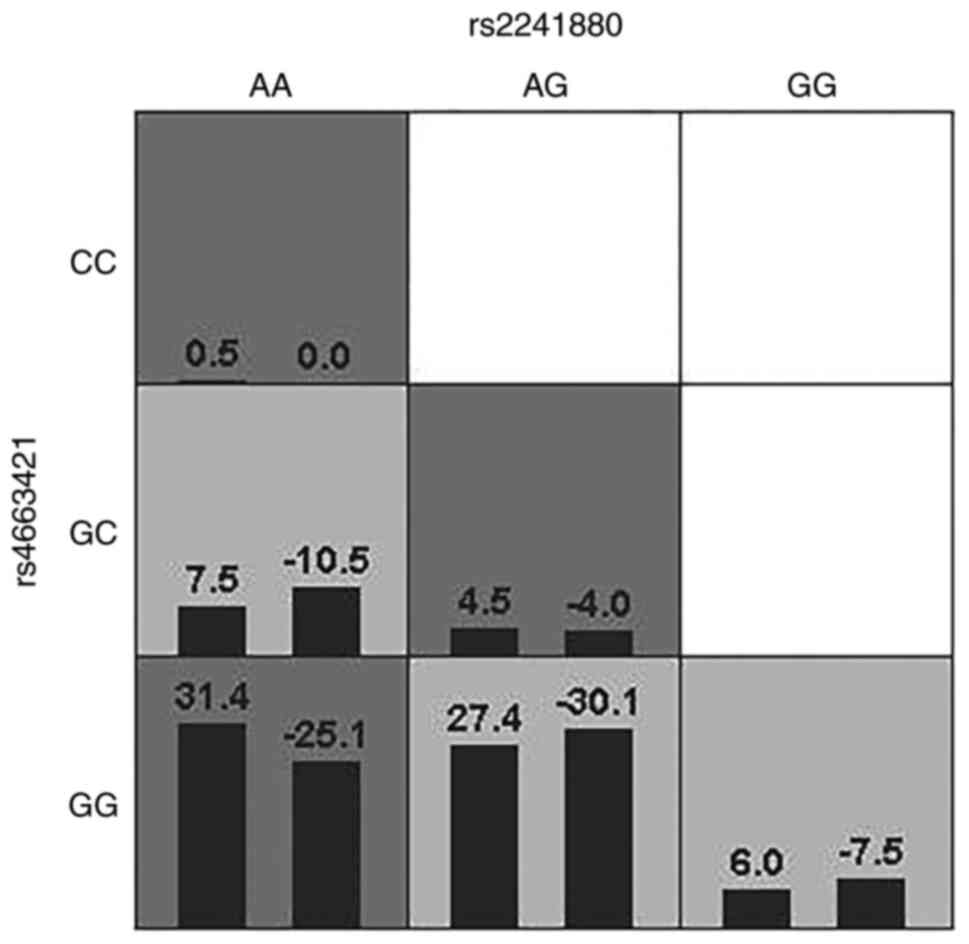
GMDR obtains the best model combination from
multiple genes or SNPs and behavioural indicators to analyse
gene-gene or SNP-SNP interactions. The interaction between
rs2241880(T300A) and rs4663421 was significant. The cross-verifying
consistency was 8/10 and the training balance accuracy was 0.5844
after 1,000 permutation tests. The highest risk was found when the
AA genotype of rs2241880(T300A) combined with the GG genotype of
rs4663421 (Fig. 4).
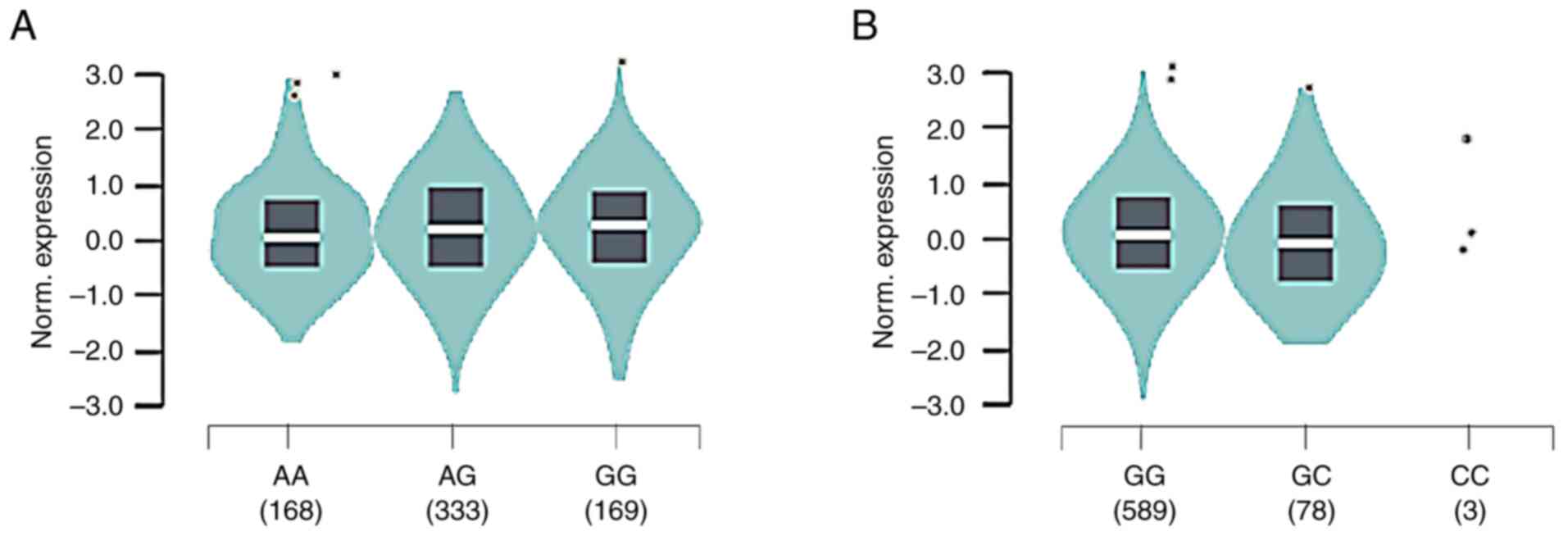
eQTL analysis
rs2241880/T300A G allele was associated with
increased ATG16L1 mRNA in whole-blood samples from 670 healthy
donors (median relative expression AA: -0.1331, AG: -0.02428, GG:
0.08790, P=0.0000037; Fig. 5).
rs4663421 G allele was associated with increased ATG16L1 mRNA
(median relative expression: GC: -0.1746, GG: 0.03176). Therefore,
the recessive model of rs2241880(T300A) (AA + AG, GG) and the
dominant model of rs4663421 (CC + CG, GG) were used to examine the
association between SNPs and clinical characteristics.
Association between ATG16L1 gene
polymorphisms and AAV clinical characteristics
CRP level and BVAS were higher, whereas the 24-h
urinary protein level was significantly lower for patients with
rs2241880(T300A) AA+AG genotype than for patients with the GG
genotype (Table IV). There was no
significant association between the rs4663421 gene polymorphism and
CRP and 24-h urine protein level, BVAS, ANCA type, white blood cell
count, serum creatinine level, ESR, eGFR or serum albumin level in
patients with AAV.
 | Table IVGenotypes and clinical
characteristics at two single nucleotide polymorphisms of
autophagy-related protein 16 like 1 gene. |
Table IV
Genotypes and clinical
characteristics at two single nucleotide polymorphisms of
autophagy-related protein 16 like 1 gene.
| | rs2241880 | rs4663421 |
|---|
| Variable | AA + AG
(n=163) | GG (n=14) | P-value | CC + CG (n=30) | GG (n=147) | P-value |
|---|
| ANCA specificity
(%) | | | 0.824 | | | 0.206 |
|
MPO | 102 (77.86) | 10 (76.92) | | 16 (69.57) | 96 (79.34) | |
|
PR3 | 15 (11.45) | 1 (7.69) | | 2 (8.70) | 14 (11.57) | |
|
None | 14 (10.69) | 2 (15.38) | | 5 (21.74) | 11 (9.09) | |
| Median WBC count,
x109/l (IQR) | 8.16
(5.84-10.90) | 8.01
(6.45-10.70) | 0.900 | 7.49
(5.94-11.61) | 8.19
(6.10-10.70) | 0.815 |
| Median 24-h UPR, mg
(IQR) | 1,011.90
(352.45-1759.40) | 2116.20
(694.80-4393.00) | 0.024 | 1,369.37
(759.19-2875.45) | 912.10
(371.48-1,786.45) | 0.157 |
| Mean albumin,
g/l | 29.99±6.51 | 31.28±8.10 | 0.504 | 28.71±8.11 | 30.37±6.33 | 0.274 |
| Median SCR, µmol/l
(IQR) | 259.00
(99.00-486.00) | 317
(141.00-607.00) | 0.378 | 249.00
(68.00-450.00) | 267.00
(110.00-511.00) | 0.597 |
| Median eGFR, ml/min
x 1.73 m2 (IQR) | 19.08
(8.46-63.37) | 12.66
(7.79-49.11) | 0.439 | 21.41
(8.96-92.15) | 18.17
(8.03-60.29) | 0.626 |
| Median CRP, mg/l
(IQR) | 20.80
(9.10-70.60) | 5.80
(3.14-27.45) | 0.010 | 13.58
(9.69-72.20) | 20.60
(6.67-63.34) | 0.941 |
| Median ESR, mm/h
(IQR) | 81.00
(52.00-109.00) | 70.00
(27.50-92.00) | 0.189 | 78.50
(38.50-96.75) | 81.00
(44.50-109.75) | 0.542 |
| Mean BVAS | 16.81±0.39 | 13.50±1.41 | 0.017 | 18.23±4.58 | 16.22±4.56 | 0.060 |
Analysis of the association of ATG16L1
gene polymorphisms with AAV renal pathology
A total of 68 patients with renal pathology data
were included. Based on pathological classification of glomerular
burden, 35 patients (51.47%) were classified as having focal-type
AAV, 20 (29.41%) as having sclerosing-type AAV, 5 (7.35%) as having
crescent-type AAV and 8 (11.76%) as having mixed-type AAV. Compared
with the AA + AG genotype, GG genotype of rs2241880(T300A) was
significantly associated with more severe glomerulosclerosis and
global sclerosis (Table V). For
rs4663421, segmental sclerosis was more severe in patients with the
CC + CG and GG genotypes. SNPs were not associated with renal
pathological type, percentage of normal glomeruli, crescents,
cellular crescents, fibrous crescents and cellular fibrous
crescents in AAV.
 | Table VGenotypes and renal pathology at two
single nucleotide polymorphisms of autophagy-related protein
16-like 1 gene. |
Table V
Genotypes and renal pathology at two
single nucleotide polymorphisms of autophagy-related protein
16-like 1 gene.
| | rs2241880 | rs4663421 |
|---|
| Variable | AA + AG (n=61) | GG (n=7) | P-value | CC + CG (n=10) | GG (n=58) | P-value |
|---|
| Berden
classificatio, n (%) | | | 0.302 | | | 0.268 |
|
Focal | 32 (52.46) | 3 (42.86) | | 5 (50.00) | 30 (51.72) | |
|
Sclerotic | 16 (26.23) | 4 (57.14) | | 5 (50.00) | 15 (25.86) | |
|
Crescent | 5 (8.20) | 0 (0.00) | | 0 (0.00) | 5 (8.62) | |
|
Mixed | 8 (13.11) | 0 (0.00) | | 0 (0.00) | 8 (13.79) | |
| Median normal
glomeruli (IQR) | 52.63
(18.34,81.67) | 20.00
(0.00,81.25) | 0.210 | 43.18
(14.58,66.67) | 51.32
(15.63,81.77) | 0.801 |
| Sclerotic
glomeruli, n (%) | 47 (77.05) | 7 (100.00) | 0.330 | 9 (90.00) | 45 (77.59) | 0.370 |
| Degree of disease,
% (median, IQR) | 21.43
(4.12,52.28) | 62.50
(18.75,80.00) | 0.042 | 55.00
(19.41,65.42) | 18.47
(4.56,51.14) | 0.082 |
| Global sclerosis, n
(%) | 46 (75.41) | 7 (100.00) | 0.334 | 9 (90.00) | 44 (75.86) | 0.319 |
|
Degree of
disease, % (median, IQR) | 17.43
(3.67,36.20) | 37.50
(18.75,75.00) | 0.040 | 31.19
(12.39,46.59) | 18.18
(4.12,36.93) | 0.185 |
| Segmental
sclerosis, n (%) | 22 (36.07) | 3 (42.86) | 0.724 | 8 (80.00) | 17 (29.31) | 0.002 |
|
Degree of
disease, % (median, IQR) | 0.00
(0.00,7.14) | 0.00
(0.00,25.00) | 0.496 | 10.72
(5.00,22.50) | 0.00
(0.00,5.61) | 0.002 |
| Crescents, n
(%) | 37 (60.66) | 2 (28.57) | 0.104 | 5 (50.00) | 34 (58.62) | 0.611 |
|
Degree of
disease, % (median, IQR) | 15.38
(0.00,27.62) | 0.00
(0.00,25.00) | 0.204 | 3.34
(0.00,21.63) | 13.25
(0.00,29.41) | 0.260 |
| Cellular crescents,
n (%) | 12 (19.67) | 0 (0.00) | 0.196 | 2 (20.00) | 10 (17.24) | 0.833 |
|
Mean degree
of disease | 2.20±5.51 | 0.00±0.00 | 0.202 | 1.38±2.91 | 2.08±5.58 | 0.865 |
| Cellular fibrous
crescents, n (%) | 24 (39.34) | 2 (28.57) | 0.579 | 2 (20.00) | 24 (41.38) | 0.199 |
|
Degree of
disease, % (median, IQR) | 0.00
(0.00,17.92) | 0.00
(0.00,15.00) | 0.541 | 0.00
(0.00,3.57) | 0.00
(0.00,18.64) | 0.166 |
| Fibrous crescents,
n (%) | 18 (29.51) | 2 (28.57) | 0.959 | 3 (30.00) | 17 (29.31) | 0.965 |
|
Degree of
disease, % (median, IQR) | 0.00
(0.00,15.00) | 0.00
(0.00,10.00) | 0.880 | 0.00
(0.00,10.00) | 0.00
(0.00,5.73) | 0.914 |
Discussion
The exact cause of AAV remains uncertain; however,
genetic factors serve a significant role in AAV (28). The present study explored the
relationship between polymorphisms in ATG16L1 and AAV
susceptibility, laboratory examination results and renal pathology
in AAV. PSM was used to match AAV and control groups. Before
matching, some baseline clinical data differed between the groups,
which may have affected the distribution of genes. Following 1:1
matching for sex, age and ethnicity, both groups presented similar
demographic characteristics, with no significant differences,
indicating PSM significantly increased the balance of
covariates.
ANCA is a serum marker for AAV. However, ANCA is
also found in patients with inflammatory bowel disease (29) and PR3-ANCA-positive patients with
ulcerative colitis have more extensive disease than those who are
PR3-ANCA negative (30). The
pathogenicity of high-titre ANCA has also been highlighted in case
reports of AAV in patients with Crohn's disease (CD) (31,32).
Therefore, there may be a common pathogenic mechanism between AAV
and inflammatory bowel disease. ATG16L1 is associated with elevated
levels of inflammatory factors IL-6, IL-8 and IL-1β (33-34).
The release of cytokines IL-8, IL-6 and TNF-α by immune cells leads
to excessive activation of neutrophils to induce the formation of
NETs, resulting in occurrence of AAV (35,36)
and an association with AAV disease activity (37). Here, it was hypothesized that
ATG16L1 rs2241880(T300A) and rs4663421 may be involved in the
pathogenesis of AAV through effects on NETs and cytokines.
rs2241880(T300A) and rs4663421 are associated with inflammatory
bowel disease and several studies of polymorphisms in the gene
encoding ATG16L1 have revealed that the T300A G allele is
associated with a greater risk of IBD development (38-41).
Almost all CD risk imposed by the ATG16L1 locus is associated with
carrying the T300A variant (42).
The ATG16L1 variant T300A affects secretion of TNF-α and IL-1β,
promotes inflammatory responses and increases susceptibility to CD
(43). Here, the AA genotype (50.6
vs. 46.1) and AG genotype (41.6 vs. 44.2%) of the ATG16L1
rs2241880(T300A) polymorphism were more frequent in patients with
AAV than in healthy controls, whereas the GG genotype was less
frequent (7.8 vs. 9.7) and the G allele was present in a minority
of patients and controls (7.8 vs. 9.7%). A meta-analysis by Zhang
et al (44) confirmed that
the T300A G allele is positively associated with CD occurrence in
Caucasians, but no significant association was found in Asians and
the frequency of the G allele differed between Asians and
Caucasians, potentially because racial differences play a role in
the genetic background. The present study did not find a
significant difference in the ATG16L1 T300A genotype or allele
frequency between patients and healthy controls (P>0.05), which
may be due to the low frequency of G allele and GG genotype. The GG
genotype of rs4663421 was predominant in healthy individuals and
patients with AAV. By contrast, the CC genotype has rarely been
reported, which is consistent with findings in other populations
(20,38), but an association between the
rs4663421 polymorphism and AAV has not been confirmed. No
significant difference was found between cases and controls.
Associations between codominant, dominant, recessive and
overdominant models with AAV were assessed. However, there was no
significant association between these models of ATG16L1
rs2241880(T300A) and rs4663421 and risk of developing AAV. AAV is
relatively rare, is not the result of a single gene mutation but is
associated with multiple genetic variants and may be associated
with gene-gene or SNP-SNP interactions (45). SNP-SNP interaction analysis between
ATG16L1 rs2241880(T300A) and rs4663421 found that the combination
of the AA genotype of rs2241880 and the GG genotype of rs4663421
increased risk of AAV. Thus, although rs2241880 and rs4663421 did
not show any association with AAV individually, the interaction of
these two SNPs may promote the development of AAV.
Due to diverse clinical manifestations, courses and
outcomes of AAV, the present study investigated the impact of gene
polymorphisms on the clinical manifestations of AAV. Higher BVAS
indicates more active disease (46). The CRP level is often used as a
non-specific marker of inflammation and is associated with disease
activity in AAV (47). ATG16L1
T300A variant can affect autophagy (48). rs2241880(T300A), when carried by a
GG homozygote, impairs autophagic flux and associated vesicle
trafficking (49). Here, GG
genotype of rs2241880(T300A) was associated with lower BVAS and CRP
level than the AA + AG genotype was. Autophagy regulates formation
and release of NETs. The PI3K/AKT/mTOR pathway connects autophagy
and NETs, and mTOR inhibitor rapamycin increases the formation of
NETs by promoting autophagy (50,51).
Therefore, rs2241880(T300A) gene polymorphism may influence the
disease activity of AAV by affecting the production and release of
NETs and ANCAs. ATG16L1 maintains cell homeostasis by inhibiting
inflammatory responses (52). eQTL
analysis demonstrated that the rs2241880(T300A) G allele was
associated with higher mRNA levels of ATG16L1 than the
rs2241880(T300A) A allele. rs2241880(T300A) GG genotype may reduce
disease activity and CRP levels due to the upregulation of ATG16L1
expression. Further studies are needed to distinguish the high
disease activity and CRP levels in patients with AAV from those
with pulmonary infection as there were many confounding factors in
the present study.
Renal involvement in AAV can manifest as active
lesions, including glomerular crescent formation and necrosis, or
as chronic lesions, including glomerulosclerosis. Patients with the
GG genotype of rs2241880(T300A) presented more pronounced
glomerulosclerosis and global sclerosis than those with the AA + AG
genotype. Additionally, 24-h urine protein level was greater in
patients with the GG genotype, indicating more severe damage to the
glomeruli. When autophagy was impaired, patients exhibited more
severe glomerular damage. Patients with AAV carrying CC + CG
genotype of rs4663421 had more severe segmental sclerosis. Further
investigation of the association between the rs4663421 gene
polymorphism and the level of autophagy is needed. Proteinuria
specifically reflects renal involvement of AAV and podocyte injury
serves a pivotal role in the development of proteinuria. When ATGs
are specifically knocked out in podocytes, mice develop
proteinuria, progressive glomerulosclerosis, severe renal
interstitial lesions and renal failure, thus showing focal
segmental glomerulosclerosis (53).
These findings indicate the role of autophagy as a key mechanism
for maintaining the homeostasis of podocytes and renal epithelial
cells, thus preserving their integrity. The present study revealed
a connection between autophagy gene variants and glomerulosclerosis
and proteinuria in AAV. This association may be connected to the
aforementioned mechanism, but further investigation is necessary to
develop appropriate animal models.
The present study has numerous limitations. First,
some patients had missing data, and the single-centre, small sample
size study should be further validated by a larger, more diverse
cohort. Second, due to difficulties in sample collection and
storage, assays for mRNA expression to complement the genetic
association studies were not performed. In vitro experiments
or animal models should be used in future to validate the effects
of these genetic variants on ATG16L1 expression and other
biological processes such as renal pathological changes and disease
activity. In addition, the present study only studied the
interaction with SNPs in the same gene. In further studies,
interactions with SNPs in different genes should be included to
provide more insight into the effects of gene interactions in AAV
progression.
In summary, rs2241880(T300A) and rs4663421
polymorphisms of ATG16L1 demonstrated regional specificity. The
present study did not reveal any connection between these variants
and genetic predisposition to AAV. However, the SNP-SNP interaction
in the model constructed with the two SNPs was significant. The
ATG16L1 rs2241880(T300A) gene polymorphism may be associated with
disease activity and glomerulosclerosis in AAV and the rs4663421
gene polymorphism may be associated with segmental sclerosis.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by Joint Project on
Regional High –Incidence Diseases Research of Guangxi Natural
Science Foundation (grant no. 2024GXNSFAA999092), the Guangxi
Health Commission self-financial project (No. Z-A20230648), Guangxi
Natural Science Foundation Program (grant no. 2018GXNSFAA281122)
and the National Natural Science Foundation of China (grant no.
81360117).
Availability of data and materials
The data generated in the present study are not
publicly available due to legal constraints but may be requested
from the corresponding author.
Authors' contributions
WLT, YRZ and CX conceived the study and interpreted
data. WLT and YRZ designed the experiments and wrote the
manuscript. WLT and SRL performed experiments. SRL analysed and
interpreted data. WLT and YRZ confirm the authenticity of all the
raw data. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was conducted according to the
Declaration of Helsinki and approved by the Ethics Committee of the
Second Affiliated Hospital of Guangxi Medical University (approval
no. 2018 KY-0100). Verbal informed consent for participation was
obtained from all participants.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Cornec D, Cornec-Le Gall E, Fervenza FC
and Specks U: ANCA-associated vasculitis-clinical utility of using
ANCA specificity to classify patients. Nat Rev Rheumatol.
12:570–579. 2016.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Geetha D and Jefferson JA: ANCA-Associated
Vasculitis: Core Curriculum 2020. Am J Kidney Dis. 75:124–137.
2020.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Kitching AR, Anders HJ, Basu N, Brouwer E,
Gordon J, Jayne DR, Kullman J, Lyons PA, Merkel PA, Savage COS, et
al: ANCA-associated vasculitis. Nat Rev Dis Primers.
6(71)2020.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Lyons PA, Peters JE, Alberici F, Liley J,
Coulson RMR, Astle W, Baldini C, Bonatti F, Cid MC, Elding H, et
al: Genome-wide association study of eosinophilic granulomatosis
with polyangiitis reveals genomic loci stratified by ANCA status.
Nat Commun. 10(5120)2019.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Prendecki M, Cairns T and Pusey CD:
Familial vasculitides: Granulomatosis with polyangiitis and
microscopic polyangiitis in two brothers with differing
anti-neutrophil cytoplasm antibody specificity. Clin Kidney J.
9:429–431. 2016.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Tanna A, Salama AD, Brookes P and Pusey
CD: Familial granulomatosis with polyangiitis: Three cases of this
rare disorder in one Indoasian family carrying an identical HLA
DPB1 allele. BMJ Case Rep. 2012(bcr0120125502)2012.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Matoba K and Noda NN: A structural catalog
of core Atg proteins opens a new era of autophagy research. J
Biochem. 169:517–525. 2021.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Yin Z, Pascual C and Klionsky DJ:
Autophagy: Machinery and regulation. Microb Cell. 3:588–596.
2016.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Arias E and Cuervo AM: Pros and cons of
chaperone-mediated autophagy in cancer biology. Trends Endocrinol
Metab. 31:53–66. 2020.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Celia AI, Colafrancesco S, Barbati C,
Alessandri C and Conti F: Autophagy in Rheumatic Diseases: Role in
the pathogenesis and therapeutic approaches. Cells.
11(1359)2022.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Pei Y, Chen S, Zhou F, Xie T and Cao H:
Construction and evaluation of Alzheimer's disease diagnostic
prediction model based on genes involved in mitophagy. Front Aging
Neurosci. 15(1146660)2023.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Yang M, Luo S, Chen W, Zhao L and Wang X:
Chaperone-Mediated Autophagy: A potential target for metabolic
diseases. Curr Med Chem. 30:1887–1899. 2023.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Masucci MT, Minopoli M, Del Vecchio S and
Carriero MV: The emerging role of neutrophil extracellular traps
(NETs) in tumor progression and metastasis. Front Immunol.
11(1749)2020.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Arneth B and Arneth R: Neutrophil
Extracellular Traps (NETs) and Vasculitis. Int J Med Sci.
18:1532–1540. 2021.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Sha LL, Wang H, Wang C, Peng HY, Chen M
and Zhao MH: Autophagy is induced by anti-neutrophil cytoplasmic
Abs and promotes neutrophil extracellular trap formation. Innate
Immun. 22:658–665. 2016.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Romanov J, Walczak M, Ibiricu I, Schuchner
S, Ogris E, Kraft C and Martens S: Mechanism and functions of
membrane binding by the Atg5-Atg12/Atg16 complex during
autophagosome formation. EMBO J. 31:4304–4317. 2012.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Gammoh N: The multifaceted functions of
ATG16L1 in autophagy and related processes. J Cell Sci.
133(jcs249227)2020.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Salem M, Ammitzboell M, Nys K, Seidelin JB
and Nielsen OH: ATG16L1: A multifunctional susceptibility factor in
Crohn's disease. Autophagy. 11:585–594. 2015.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Mo YJ, Zhang W, Wen QW, Wang TH, Qin W,
Zhang Z, Huang H, Cen H and Wu XD: Corrigendum to ‘Genetic
association analysis of ATG16L1 rs2241880, rs6758317 and ATG16L2
rs11235604 polymorphisms with rheumatoid arthritis in a Chinese
population’ [Int. Immunopharmacol 93 (2021) 107378]. Int.
Immunopharmacol 104: 108511, 2022.
|
|
20
|
Li X, Chen M, Zhang X, Wang M, Yang X, Xia
Q, Han R, Liu R, Xu S, Xu J, et al: Single nucleotide polymorphisms
of autophagy-related 16-like 1 gene are associated with ankylosing
spondylitis in females: A case-control study. Int J Rheum Dis.
21:322–329. 2018.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Jennette JC, Falk RJ, Bacon PA, Basu N,
Cid MC, Ferrario F, Flores-Suarez LF, Gross WL, Guillevin L, Hagen
EC, et al: 2012 revised International Chapel Hill Consensus
Conference Nomenclature of Vasculitides. Arthritis Rheum. 65:1–11.
2013.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Mukhtyar C, Lee R, Brown D, Carruthers D,
Dasgupta B, Dubey S, Flossmann O, Hall C, Hollywood J, Jayne D, et
al: Modification and validation of the Birmingham Vasculitis
Activity Score (version 3). Ann Rheum Dis. 68:1827–1832.
2009.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Stevens LA, Schmid CH, Zhang YL, Coresh J,
Manzi J, Landis R, Bakoush O, Contreras G, Genuth S, Klintmalm GB,
et al: Development and validation of GFR-estimating equations using
diabetes, transplant and weight. Nephrol Dial Transplant.
25:449–457. 2010.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Berden AE, Ferrario F, Hagen EC, Jayne DR,
Jennette JC, Joh K, Neumann I, Noël LH, Pusey CD, Waldherr R, et
al: Histopathologic classification of ANCA-associated
glomerulonephritis. J Am Soc Nephrol. 21:1628–1636. 2010.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Barrett JC, Fry B, Maller J and Daly MJ:
Haploview: Analysis and visualization of LD and haplotype maps.
Bioinformatics. 21:263–265. 2005.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Huang F, DU C, Sun M, Ning B, Luo Y and An
S: Propensity score matching in SPSS. Nan Fang Yi Ke Da Xue Xue
Bao. 35:1597–1601. 2015.PubMed/NCBI(In Chinese).
|
|
27
|
Austin PC, Yu AYX, Vyas MV and Kapral MK:
Applying propensity score methods in clinical research in
neurology. Neurology. 97:856–863. 2021.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Lyons PA, Rayner TF, Trivedi S, Holle JU,
Watts RA, Jayne DR, Baslund B, Brenchley P, Bruchfeld A, Chaudhry
AN, et al: Genetically distinct subsets within ANCA-associated
vasculitis. N Engl J Med. 367:214–223. 2012.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Tang H, Tan B, Shen BB, Zhang SL and Qian
JM: Diagnostic value of different serological markers and
correlation analysis with disease phenotype in inflammatory bowel
disease. Zhonghua Yi Xue Za Zhi. 102:3743–3748. 2022.PubMed/NCBI View Article : Google Scholar : (In Chinese).
|
|
30
|
Laass MW, Ziesmann J, de Laffolie J, Röber
N and Conrad K: Anti-Proteinase 3 Antibodies as a Biomarker for
Ulcerative Colitis and Primary Sclerosing Cholangitis in Children.
J Pediatr Gastroenterol Nutr. 74:463–470. 2022.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Plafkin C, Zhong W and Singh T: ANCA
vasculitis presenting with acute interstitial nephritis without
glomerular involvement. Clin Nephrol Case Stud. 7:46–50.
2019.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Freeman HJ: Atypical perinuclear
antineutrophil cytoplasmic antibodies in patients with Crohn's
disease. Can J Gastroenterol. 11:689–693. 1997.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Mommersteeg MC, Simovic I, Yu B, van
Nieuwenburg SAV, Bruno IMJ, Doukas M, Kuipers EJ, Spaander MCW,
Peppelenbosch MP, Castaño-Rodríguez N and Fuhler GM: Autophagy
mediates ER stress and inflammation in Helicobacter pylori-related
gastric cancer. Gut Microbes. 14(2015238)2022.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Plantinga TS, Crisan TO, Oosting M, van de
Veerdonk FL, de Jong DJ, Philpott DJ, van der Meer JW, Girardin SE,
Joosten LA and Netea MG: Crohn's disease-associated ATG16L1
polymorphism modulates pro-inflammatory cytokine responses
selectively upon activation of NOD2. Gut. 60:1229–1235.
2011.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Hao J, Lv TG, Wang C, Xu LP and Zhao JR:
Macrophage migration inhibitory factor contributes to
anti-neutrophil cytoplasmic antibody-induced neutrophil activation.
Hum Immunol. 77:1209–1214. 2016.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Takeda S, Watanabe-Kusunoki K, Nakazawa D,
Kusunoki Y, Nishio S and Atsumi T: The Pathogenicity of BPI-ANCA in
a Patient with Systemic Vasculitis. Front Immunol.
10(1334)2019.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Matsumoto K, Suzuki K, Yoshimoto K, Seki
N, Tsujimoto H, Chiba K and Takeuchi T: Longitudinal immune cell
monitoring identified CD14(++) CD16(+) intermediate monocyte as a
marker of relapse in patients with ANCA-associated vasculitis.
Arthritis Res Ther. 22(145)2020.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Pugazhendhi S, Baskaran K, Santhanam S and
Ramakrishna BS: Association of ATG16L1 gene haplotype with
inflammatory bowel disease in Indians. PLoS One.
12(e0178291)2017.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Kee BP, Ng JG, Ng CC, Hilmi I, Goh KL and
Chua KH: Genetic polymorphisms of ATG16L1 and IRGM genes in
Malaysian patients with Crohn's disease. J Dig Dis. 21:29–37.
2020.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Teimoori-Toolabi L, Samadpoor S, Mehrtash
A, Ghadir M and Vahedi H: Among autophagy genes, ATG16L1 but not
IRGM is associated with Crohn's disease in Iranians. Gene.
675:176–184. 2018.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Zhang J, Chen J, Gu J, Guo H and Chen W:
Association of IL23R and ATG16L1 with susceptibility of Crohn's
disease in Chinese population. Scand J Gastroenterol. 49:1201–1206.
2014.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Hampe J, Franke A, Rosenstiel P, Till A,
Teuber M, Huse K, Albrecht M, Mayr G, De La Vega FM, Briggs J, et
al: A genome-wide association scan of nonsynonymous SNPs identifies
a susceptibility variant for Crohn's disease in ATG16L1. Nat Genet.
39:207–211. 2007.PubMed/NCBI View
Article : Google Scholar
|
|
43
|
Salem M, Nielsen OH, Nys K, Yazdanyar S
and Seidelin JB: Impact of T300A Variant of ATG16L1 on
antibacterial response, risk of culture positive infections, and
clinical course of crohn's disease. Clin Transl Gastroenterol.
6(e122)2015.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Zhang HF, Qiu LX, Chen Y, Zhu WL, Mao C,
Zhu LG, Zheng MH, Wang Y, Lei L and Shi J: ATG16L1 T300A
polymorphism and Crohn's disease susceptibility: Evidence from
13,022 cases and 17,532 controls. Hum Genet. 125:627–631.
2009.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Merkel PA, Xie G, Monach PA, Ji X,
Ciavatta DJ, Byun J, Pinder BD, Zhao A, Zhang J, Tadesse Y, et al:
Identification of functional and expression polymorphisms
associated with risk for antineutrophil cytoplasmic
autoantibody-associated vasculitis. Arthritis Rheumatol.
69:1054–1066. 2017.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Luqmani RA, Bacon PA, Moots RJ, Janssen
BA, Pall A, Emery P, Savage C and Adu D: Birmingham Vasculitis
Activity Score (BVAS) in systemic necrotizing vasculitis. QJM.
87:671–678. 1994.PubMed/NCBI
|
|
47
|
Hind CR, Winearls CG, Lockwood CM, Rees AJ
and Pepys MB: Objective monitoring of activity in Wegener's
granulomatosis by measurement of serum C-reactive protein
concentration. Clin Nephrol. 21:341–345. 1984.PubMed/NCBI
|
|
48
|
Lassen KG, Kuballa P, Conway KL, Patel KK,
Becker CE, Peloquin JM, Villablanca EJ, Norman JM, Liu TC, Heath
RJ, et al: Atg16L1 T300A variant decreases selective autophagy
resulting in altered cytokine signaling and decreased antibacterial
defense. Proc Natl Acad Sci USA. 111:7741–7746. 2014.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Le Naour J, Sztupinszki Z, Carbonnier V,
Casiraghi O, Marty V, Galluzzi L, Szallasi Z, Kroemer G and
Vacchelli E: A loss-of-function polymorphism in ATG16L1 compromises
therapeutic outcomes in head and neck carcinoma patients.
Oncoimmunology. 11(2059878)2022.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Guo Y, Gao F, Wang X, Pan Z, Wang Q, Xu S,
Pan S, Li L, Zhao D and Qian J: Spontaneous formation of neutrophil
extracellular traps is associated with autophagy. Sci Rep.
11(24005)2021.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Yu Y and Sun B: Autophagy-mediated
regulation of neutrophils and clinical applications. Burns Trauma.
8(tkz001)2020.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Hamaoui D and Subtil A: ATG16L1 functions
in cell homeostasis beyond autophagy. FEBS J. 289:1779–1800.
2022.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Kawakami T, Gomez IG, Ren S, Hudkins K,
Roach A, Alpers CE, Shankland SJ, D'Agati VD and Duffield JS:
Deficient autophagy results in mitochondrial dysfunction and FSGS.
J Am Soc Nephrol. 26:1040–1052. 2015.PubMed/NCBI View Article : Google Scholar
|