1. Introduction
The role of nutrition as an integral component in
human health and disease management is well established. However,
dietary responses are markedly influenced by inter-individual
metabolic variability which challenges the one-size-fits-all
approach to dietary advice. This variability is multifactorial,
influenced by genetic, epigenetic and environmental factors
(1,2). The refinement of nutritional advice to
meet the needs of distinct population subgroups, based on optimal
patient stratification is a defining characteristic of Precision
Nutrition (PN). This approach is made feasible by integrating deep
molecular profiling of biological samples (for instance, whole
blood, urine and saliva) from target groups with individual
characteristics that contribute to heterogeneity both within and
between populations. These characteristics include demographic
factors (for instance, age, sex and ethnicity), psychosocial and
cultural influences, health status, dietary patterns and behaviors
such as adherence to or deviation from a healthy lifestyle
(3,4).
Dietary interventions formulated on the principles
of PN are well-suited to address metabolic perturbations associated
with the onset and progression of non-communicable diseases, while
illuminating diet-disease interactions (5-10).
For instance, in prediabetic and diabetic subjects supplementation
with inulin, a naturally occurring fiber, has been associated with
improvements in clinical outcomes including reductions in blood
glucose, total cholesterol and triglycerides (11). Additionally, dietary iron intake has
been linked to lung cancer oncogenesis through epigenetic
mechanisms (12). Moreover,
proteomic and metabolomic profiling in the context of nutritional
studies may assist in deciphering the molecular signatures
associated with dietary patterns, providing greater insight on the
onset of diet related diseases (8).
In this context, the effects of precision based dietary
interventions for weight loss in overweight and obese adults, were
evaluated in a double blind intervention trial aiming to facilitate
the adoption of health promoting behaviors (9). Therefore, PN plays a crucial role
within precision health (PH), which incorporates a broader
framework of disease prevention and treatment strategies. PH, which
encompasses both precision medicine and PN, integrates molecular
and clinical data to stratify populations, ultimately aiming to
deliver targeted lifestyle and therapeutic interventions (13,14).
Aimed at deciphering the multifaceted relationship
between nutrition and health at the molecular level, omics
platforms represent a driving force in the development of PN. These
platforms adopt a holistic approach to the precise qualitative and
quantitative characterization of genes, proteins and metabolites
present in biological materials (6). The integration of distinct omics
layers, genomics, proteomics, transcriptomics and metabolomics,
known as multi-omics, has gained prominence in PN, especially in
tandem with advances in bioinformatics (15). Nevertheless, due to the elaborate
nature of the raw data obtained from these technologies,
significant computational power is required for their integration
and interpretation, often utilizing artificial intelligence (for
instance, deep learning algorithms). Despite these advancements,
the availability of the necessary infrastructure is still in its
early stages (16).
The aim of the present study is twofold; to provide
a detailed overview of omics technologies and their application
towards the realization of PN, with an emphasis on challenges and
opportunities and to introduce an integrative bioinformatics
pipeline for omics data analysis and interpretation within the same
context.
2. Materials and methods
A literature review and computational workflow
emphasizing the integration of multiple omics layers towards the
realization of PN are presented.
Literature review
The present study conducted a comprehensive search
of publications addressing advances in PN, including challenges and
opportunities. An exhaustive literature search was conducted across
three academic databases [PubMed (https://subread.sourceforge.net/featureCounts.html),
Scopus (www.scopus.com) and Google Scholar
(https://scholar.google.com/)] using the
following keywords: ‘Precision Nutrition’, ‘Precision Health’,
‘multi-omics’, ‘omics’, ‘Systems Biology’, ‘data integration’ and
‘personalized nutritional advice’. Among identified publications,
duplicate records were manually removed and article screening was
guided by consensus among all authors.
To be considered as eligible for inclusion retrieved
records were required to meet specific criteria including
publication in the English language and integrative use of omics
technologies within the framework of PN. Articles reporting data
from animal models, isolated proteomic, genomic or metabolomic and
personalized nutrition studies were excluded from the review
process.
Construction of an integrative
multi-omic data analysis pipeline
Data handling protocols for different omics layers
were compiled to develop a multi-omics pipeline applicable to PN.
The proposed computational workflow represents a flexible approach
for data handling and interpretation of genomics, transcriptomics,
proteomics and metabolomics datasets in preparation for multi-omic
integration. In detail, three modules were developed incorporating
the preprocessing and analysis of raw DNA-seq, RNA-seq and liquid
chromatography-mass spectrometry (LC-MS/MS) data.
Quality control and correction of genomic and
transcriptomic sequencing datasets is proposed utilizing the
software packages FastQC, (a specialized tool used for the
detection of low quality control reads and bias in NGS data)
(17) and Trimmomatic, (a flexible,
pair-aware tool used for adapter sequence identification and
quality filtering of NGS datasets) (18). This process is followed by DNA/RNA
sequence alignment to a refence genome with advanced algorithms;
for instance, Magic-BLAST (19) (a
specific tool used for the alignment of RNA-seq data) and STAR (a
specific tool used for the alignment of DNA-seq and RNA-seq data)
(20). Thereafter, downstream
analysis of genomic data incorporates advanced sequence processing)
with the SAMtools toolkit (a tool used to perform variant calling,
error correcting, sorting, merging and indexing) (21). Regarding transcriptomics data, the
implementation of Differential Expression analysis (DEA) with R
language (22) (version 4.4.1)
software packages such as DESeq2 (version 1.44.0; https://bioconductor.org/packages/release/bioc/html/DESeq2.html)
(23), edgeR (version 4.2.2;
https://bioconductor.org/packages/release/bioc/html/edgeR.html)
(24) and limma (version 3.60.6;
https://bioconductor.org/packages/release/bioc/html/limma.html)
(25) (all tools are used to
perform Differential Expression analysis), to evaluate gene
expression, is recommended.
Furthermore, for the analysis of raw proteomics and
metabolomics data, several data pre-processing steps were proposed.
These included dataset filtration, normalization and missing value
imputation which may be conducted using standard data manipulation
algorithms such as tidyverse (26),
dplyr (27) and advanced modelling
tools including MissForest (Random Forest algorithm) (28) and Multivariate Imputation by Chained
Equations (mice) (29),
respectively. Appropriate selection of imputation techniques
relative to the nature of missing values present in the analyzed
LC-MS/MS datasets, including values missing not at random, missing
completely at random and missing at random, is critical, as this
has been associated with error rate reduction and improved
efficiency regarding downstream data analysis (30-32).
To evaluate disparities in protein/metabolite expression, DEA
outputs can be generated using advanced models including limma
(25) and non-parametric
statistical tests (for instance, Mann-Whitney test).
Finally, to aid the process of data interpretation,
functional annotation by means of enrichment analysis, is
suggested. To this end several core analyses may be employed
towards the identification of key biological processes, molecular
functions, topology and molecular pathways (33), including Gene Ontology (34,35)
analysis, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway
enrichment (36) and Gene Set
Enrichment analysis (37). A core
component of all aforementioned steps is data visualization, which
can be achieved with data visualization software packages available
through the R language (for instance, ggplot2 and lattice)
(38,39).
3. Results and discussion
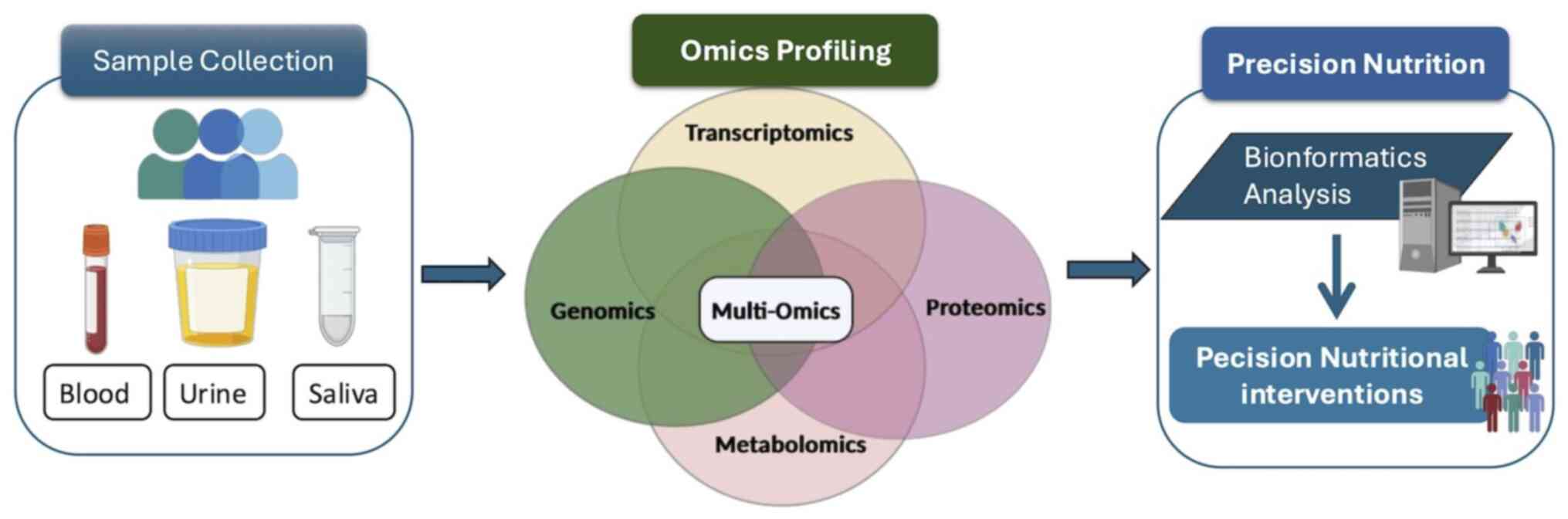
The currently available PN pipelines consist of
multiple tandem bioanalytical steps. Specifically, following sample
collection, biological materials are analyzed using omics
approaches, resulting in the production of large data arrays, which
are subsequently processed using relevant computational tools
(Fig. 1). Data analysis and
interpretation using bioinformatics delivers key information
pertaining to the exact molecular profile of examined samples
(whole blood, urine and saliva) (40). This procedure aims to stratify the
populations under study into appropriate clusters based their
distinct molecular profile and ultimately deliver tailor-made
dietary interventions adjusted to their characteristics (41).
For PN datasets to be populated, efficient
analytical handling, adjusted to the nature of biological samples
under investigation, is required. Genomic and transcriptomic
analytical workflows encompass the preparation of DNA and RNA
libraries for sequencing, using advanced NGS platforms. In
addition, proteomic and metabolomic pipelines involve the analysis
of tissues and other biological samples using LC-MS/MS. For
proteomics, prior to LC-MS/MS analysis, protein extraction and
peptide generation steps are required, while appropriate metabolite
extraction is an essential component in metabolic profiling. The
generated omics data are analyzed using bioinformatics algorithms
in order to achieve multi-omic data integration and ultimately
create predictive models capable of delivering dietary advice based
on the principles of PN. For this purpose, raw data is processed
using algorithms, including machine-learning models and statistical
tools, such as STAR aligner for sequence alignment of RNA-seq data,
DESeq2 for differential expression analysis in transcriptomics and
proteomics and mice for the imputation of missing values in
proteomics and metabolomics (42).
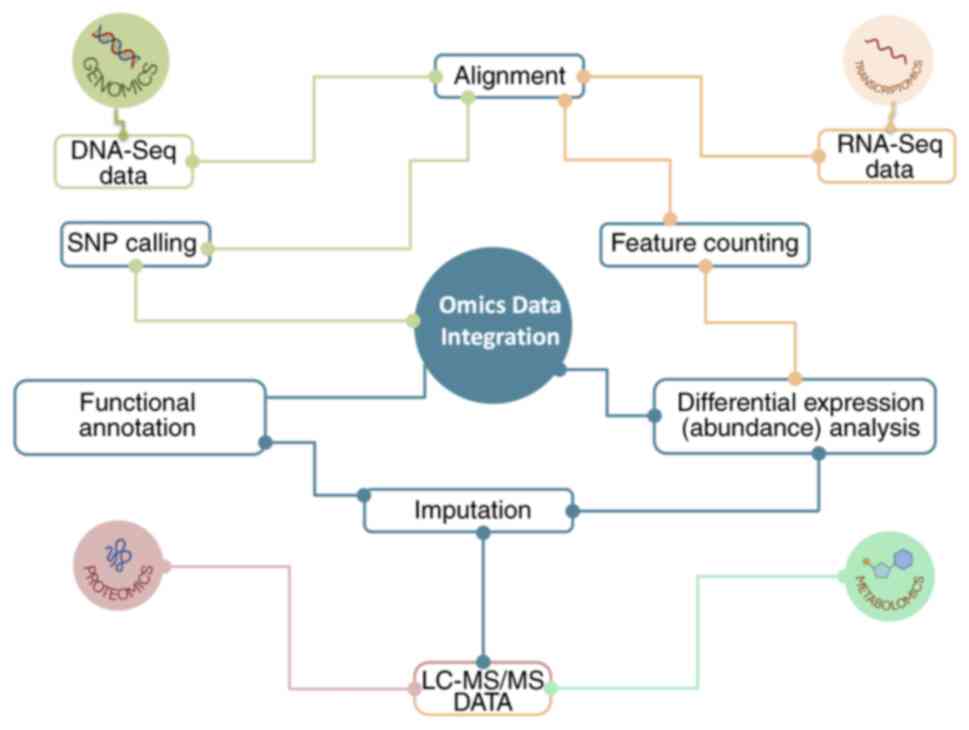
The present study constructed a pipeline consisting
of multiple bioinformatic analysis steps/nodes for the processing
of multi-modal omics data with a view to their integration
(Fig. 2). The following
bioinformatic steps were introduced: i) Alignment of DNA- and RNA-
seq data (for instance, utilizing BWA or Bowtie2 and Magic, BLAST,
STAR and HISAT2 respectively). ii) Determination of polymorphism
topology in genomics data using single nucleotide polymorphism
calling [SAMtools (version 1.21; https://github.com/samtools/samtools/releases/tag/1.21),
BCFtools (version 1.21; https://github.com/samtools/bcftools/releases/tag/1.21)].
iii) Gene quantification/gene expression count generation (feature
counting) from RNA-Seq data using tools such as HT-Seq count
(version 2.0; https://htseq.readthedocs.io/en/latest/) and
featureCounts (version 2.20.0; https://subread.sourceforge.net/featureCounts.html).
iv) Imputation of missing values in LC-MS/MS data derived from
proteomics and metabolomics. v) Differential expression (abundance)
analysis to discern expression patterns in transcriptomics,
proteomics and metabolomics data (DESeq2, limma and SDAMS)
(43-45).
vi) Functional annotation of proteomics and transcriptomics data
(for instance, KEGG pathway, Gene Ontology analysis, Gene Set
Enrichment Analysis). The integration of data generated by the
aforementioned procedures was achieved using artificial
intelligence based models, for instance, machine learning tools
based on multivariate techniques, such as the mixOmics framework
(46) and deep-learning approaches
including large language models, convolutional neural networks.
Nevertheless, despite the rapid technological
advancements supporting progress in the biomedical field, a number
of critical gaps need to be bridged before PN is realized (47-49).
Crucially, the creation of relevant repositories enriched with data
from additional nutritional epidemiological studies will play a
pivotal role towards the realization of PN. Efficient adoption of
such strategies must be based upon sound study design, encompassing
sample size among other parameters (50). Furthermore, harmonization of data
storage and retrieval protocols should be prioritized in parallel
with the much-needed enrichment of publicly available omics data
repositories (51,52).
Breakthroughs achieved by artificial intelligence
can be leveraged to address limitations across the entire PN
pipeline (53). When implemented
under human supervision, artificial intelligence can be leveraged
towards efficient study design and subsequent data gathering and
processing. As such, use of artificial intelligence models as
catalysts for the development of predictive models adapted to PN
(16). Finally, active learning
based models use feedback to refine their predictive accuracy, thus
facilitating patient guided study design and data interpretation
(54).
Overall, the process leading up to the adoption of
PN gives rise to several practical and ethical concerns. The
large-scale analytical accumulation of molecular, personal data
requires that an adequate degree of data protection is ensured to
maintain patient privacy and reduce healthcare disparities
(55). Moreover, firm understanding
of PN and omics concepts by targeted groups should be ensured prior
to recruitment in nutritional epidemiological studies so that
informed consent is obtained (56).
Thus, the necessary regulatory framework for the protection of
patient rights must be established (57-59),
taking present legislature such as the General Data Protection
Regulation into account.
While valuable insights have been gained by the
progress achieved in the field of PN through the application of
omics technologies, its widespread implementation may currently
seem far-fetched due to existing limitations with regard to the
organization of such efforts at a global scale. To this end,
further nutritional epidemiological studies in tandem with
standardization of omics pipelines, including analytical and
bioinformatics workflows, are required (15,58,60,61).
Furthermore, the capacity to integrate epigenetic omics datasets in
future PN multi-omics workflows is something to be taken under
consideration. The lack of its integration is a limitation of the
present work. The establishment of international networks fostering
cross-disciplinary collaboration will facilitate the gradual
creation of new and enrichment of existing biological databases,
whilst addressing ethical concerns.
4. Conclusion
Individual responses to dietary exposures are
characterized by a high degree of heterogeneity driven by
demographic, nutritional and psychosocial factors (1,62). PN
is an emerging discipline aiming to provide optimized dietary
advice to relevant population subgroups in the context of disease
prevention and management (6). The
successful realization of PN requires a systems-level understanding
of human physiology and individual characteristics associated with
metabolic variability.
To this end, the present review offered for the
first time, to the best of the authors' knowledge, a detailed
overview of omics technologies and their application towards the
realization of PN. Furthermore, an integrative bioinformatics
pipeline that can be used as a model for omics data analysis and
interpretation was introduced in the present study. The holistic
adoption of omics technologies in parallel with large scale
epidemiological studies is anticipated to give rise to molecular
biomarkers linking nutrition to the onset and/or progression of
chronic diseases. Embracing such efforts will bring us one step
closer to bridging the gap between PN and its implementation in
clinical practice within the broader framework of precision health
(63-65).
Acknowledgements
The present study was presented, in part, at the 4th
International Congress on Pharmacogenomics and Personalized
Diagnosis and Therapy, 1-3rd December 2023, Athens, Greece.
Funding
Funding: No funding was received.
Availability of data and materials
Not applicable.
Authors' contributions
Conceptualization was by AA and investigation by MP,
HT, AP, OM, MK, IV and IG. MP, HT, AP, OM, MK, IV, IG and AA were
responsible for writing the original draft of the manuscript and AA
and KS for writing, review and editing the manuscript. All authors
read and approved the final manuscript. Data authentication is not
applicable.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Zeisel SH: A conceptual framework for
studying and investing in precision nutrition. Front Genet.
10(200)2019.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Matusheski NV, Caffrey A, Christensen L,
Mezgec S, Surendran S, Hjorth MF, McNulty H, Pentieva K, Roager HM,
Seljak BK, et al: Diets, nutrients, genes and the microbiome:
recent advances in personalised nutrition. Br J Nutr.
126:1489–1497. 2021.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Livingstone KM, Ramos-Lopez O, Pérusse L,
Kato H, Ordovas JM and Martínez JA: Precision nutrition: A review
of current approaches and future endeavors. Trends Food Sci
Technol. 128:253–264. 2022.
|
|
4
|
De Toro-Martín J, Arsenault BJ, Després JP
and Vohl MC: Precision nutrition: A review of personalized
nutritional approaches for the prevention and management of
metabolic syndrome. Nutrients. 9(913)2017.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Cuyàs E, Verdura S, Martin-Castillo B,
Alarcón T, Lupu R, Bosch-Barrera J and Menendez JA: Tumor
cell-intrinsic immunometabolism and precision nutrition in cancer
immunotherapy. Cancers (Basel). 12(1757)2020.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Ramos-Lopez O, Martinez JA and Milagro FI:
Holistic integration of omics tools for precision nutrition in
health and disease. Nutrients. 14(4074)2022.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Morin-Bernier J, De Toro-Martín J, Barbe
V, San-Cristobal R, Lemieux S, Rudkowska I, Couture P, Barbier O
and Vohl MC: Revisiting multi-omics-based predictors of the plasma
triglyceride response to an omega-3 fatty acid supplementation.
Front Nutr. 11(1327863)2024.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Walker ME, Song RJ, Xu X, Gerszten RE, Ngo
D, Clish CB, Corlin L, Ma J, Xanthakis V, Jacques PF and Vasan RS:
Proteomic and metabolomic correlates of healthy dietary patterns:
The framingham heart study. Nutrients. 12(1476)2020.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Aldubayan MA, Pigsborg K, Gormsen SMO,
Serra F, Palou M, Mena P, Wetzels M, Calleja A, Caimari A, Del Bas
J, et al: Empowering consumers to PREVENT diet-related diseases
through OMICS sciences (PREVENTOMICS): Protocol for a parallel
double-blinded randomised intervention trial to investigate
biomarker-based nutrition plans for weight loss. BMJ Open.
12(e051285)2022.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Ďásková N, Modos I, Krbcová M, Kuzma M,
Pelantová H, Hradecký J, Heczková M, Bratová M, Videňská P,
Šplíchalová P, et al: Multi-omics signatures in new-onset diabetes
predict metabolic response to dietary inulin: Findings from an
observational study followed by an interventional trial. Nutr
Diabetes. 13(7)2023.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Li L, Li P and Xu L: Assessing the effects
of inulin-type fructan intake on body weight, blood glucose, and
lipid profile: A systematic review and meta-analysis of randomized
controlled trials. Food Sci Nutr. 9:4598–4616. 2021.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Zhang L, Ye Y, Tu H, Hildebrandt MA, Zhao
L, Heymach JV, Roth JA and Wu X: MicroRNA-related genetic variants
in iron regulatory genes, dietary iron intake, microRNAs and lung
cancer risk. Ann Oncol. 28:1124–1129. 2017.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Roberts MC, Holt KE, Del Fiol G,
Baccarelli AA and Allen CG: Precision public health in the era of
genomics and big data. Nat Med. 30:1865–1873. 2024.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Mathur S and Sutton J: Personalized
medicine could transform healthcare. Biomed Rep. 7:3–5.
2017.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Karczewski KJ and Snyder MP: Integrative
omics for health and disease. Nat Rev Genet. 19:299–310.
2018.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Theodore Armand TP, Nfor KA, Kim JI and
Kim HC: Applications of artificial intelligence, machine learning,
and deep learning in nutrition: A systematic review. Nutrients.
16(1073)2024.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Andrews S: FastQC: A Quality Control Tool
for High Throughput Sequence Data, 2010. http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
|
|
18
|
Bolger AM, Lohse M and Usadel B:
Trimmomatic: A flexible trimmer for Illumina sequence data.
Bioinformatics. 30:2114–2120. 2014.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Boratyn GM, Thierry-Mieg J, Thierry-Mieg
D, Busby B and Madden TL: Magic-BLAST, an accurate RNA-seq aligner
for long and short reads. BMC Bioinformatics.
20(405)2019.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Dobin A, Davis CA, Schlesinger F, Drenkow
J, Zaleski C, Jha S, Batut P, Chaisson M and Gingeras TR: STAR:
Ultrafast universal RNA-seq aligner. Bioinformatics. 29:15–21.
2013.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Danecek P, Bonfield JK, Liddle J, Marshall
J, Ohan V, Pollard MO, Whitwham A, Keane T, McCarthy SA, Davies RM
and Li H: Twelve years of SAMtools and BCFtools. GigaScience.
10(giab008)2021.PubMed/NCBI View Article : Google Scholar
|
|
22
|
R Core Team: R: A language and environment
for statistical computing. R Foundation for Statistical Computing,
Vienna, 2020. https://www.R-project.org/.
|
|
23
|
Love MI, Huber W and Anders S: Moderated
estimation of fold change and dispersion for RNA-seq data with
DESeq2. Genome Biol. 15(550)2014.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Robinson MD, McCarthy DJ and Smyth GK:
edgeR : A Bioconductor package for differential expression analysis
of digital gene expression data. Bioinformatics. 26:139–140.
2010.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43(e47)2015.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Wickham H, Averick M, Bryan J, Chang W,
McGowan LD, François R, Grolemund G, Hayes A, Henry L, Hester J, et
al: Welcome to the Tidyverse. J Open Source Softw. 4(1686)2019.
|
|
27
|
Whickham H, François R, Lionel H, Müller K
and Vaughan D: dplyr: A Grammar of Data Manipulation. Version
1.1.0, 2023. https://github.com/tidyverse/dplyr.
|
|
28
|
Stekhoven DJ and Bühlmann P:
MissForest-non-parametric missing value imputation for mixed-type
data. Bioinformatics. 28:112–118. 2012.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Buuren SV and Groothuis-Oudshoorn K: Mice
: Multivariate Imputation by Chained Equations in R. J Stat
Softw. 45:1–67. 2011.
|
|
30
|
Jin L, Bi Y, Hu C, Qu J, Shen S, Wang X
and Tian Y: A comparative study of evaluating missing value
imputation methods in label-free proteomics. Sci Rep.
11(1760)2021.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Kong W, Hui HWH, Peng H and Goh WWB:
Dealing with missing values in proteomics data. Proteomics.
22(2200092)2022.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Wei R, Wang J, Su M, Jia E, Chen S, Chen T
and Ni Y: Missing value imputation approach for mass
spectrometry-based metabolomics Data. Sci Rep.
8(663)2018.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Stamoula E, Sarantidi E, Dimakopoulos V,
Ainatzoglou A, Dardalas I, Papazisis G, Kontopoulou K and
Anagnostopoulos AK: Serum Proteome Signatures of Anti-SARS-CoV-2
Vaccinated Healthcare Workers in Greece Associated with Their Prior
Infection Status. Int J Mol Sci. 23(10153)2022.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene Ontology: Tool for the unification of biology. The Gene
Ontology Consortium. Nat Genet. 25:25–29. 2000.PubMed/NCBI View
Article : Google Scholar
|
|
35
|
The Gene Ontology Consortium. Aleksander
SA, Balhoff J, Carbon S, Cherry JM, Drabkin HJ, Ebert D, Feuermann
M, Gaudet P, Harris NL, et al: The Gene Ontology knowledgebase in
2023. Genetics. 224(iyad031)2023.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES and Mesirov JP: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Wickham H: ggplot2: Elegant Graphics for
Data Analysis [Internet]. Springer-Verlag, New York, NY, 2016.
https://ggplot2.tidyverse.org.
|
|
39
|
Sarkar D: Lattice: Multivariate Data
Visualization with R [Internet]. Springer, New York, NY, 2008.
http://link.springer.com/10.1007/978-0-387-75969-2.
|
|
40
|
Vavilis T, Petre ML, Vatsellas G,
Ainatzoglou A, Stamoula E, Sachinidis A, Lamprinou M, Dardalas I,
Vamvakaris IN, Gkiozos I, et al: Lung cancer proteogenomics:
Shaping the future of clinical investigation. Cancers (Basel).
16(1236)2024.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Ulusoy-Gezer HG and Rakıcıoğlu N: The
future of obesity management through precision nutrition: Putting
the individual at the center. Curr Nutr Rep. 13:455–477.
2024.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Morgenstern JD, Rosella LC, Costa AP, De
Souza RJ and Anderson LN: Perspective: Big data and machine
learning could help advance nutritional epidemiology. Adv Nutr.
12:621–631. 2021.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Rapaport F, Khanin R, Liang Y, Pirun M,
Krek A, Zumbo P, Mason CE, Socci ND and Betel D: Comprehensive
evaluation of differential gene expression analysis methods for
RNA-seq data. Genome Biol. 14(3158)2013.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Peng H, Wang H, Kong W, Li J and Goh WWB:
Optimizing differential expression analysis for proteomics data via
high-performing rules and ensemble inference. Nat Commun.
15(3922)2024.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Huang Z and Wang C: A review on
differential abundance analysis methods for mass spectrometry-based
metabolomic data. Metabolites. 12(305)2022.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Rohart F, Gautier B, Singh A and Lê Cao
KA: mixOmics: An R package for ‘omics feature selection and
multiple data integration. PLoS Comput Biol.
13(e1005752)2017.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Vadiveloo MK, Juul F, Sotos-Prieto M and
Parekh N: Perspective: Novel approaches to evaluate dietary
quality: Combining methods to enhance measurement for dietary
surveillance and interventions. Adv Nutr. 13:1009–1015.
2022.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Singh VK, Hu XH, Singh AK, Solanki MK,
Vijayaraghavan P, Srivastav R, Joshi NK, Kumari M, Singh SK, Wang Z
and Kumar A: Precision nutrition-based strategy for management of
human diseases and healthy aging: Current progress and challenges
forward. Front Nutr. 11(1427608)2024.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Voruganti VS: Precision nutrition: Recent
advances in obesity. Physiology (Bethesda). 38(0)2023.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Fiocchi C: Omics and Multi-Omics in IBD:
No Integration, No Breakthroughs. Int J Mol Sci.
24(14912)2023.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Anagnostopoulos AK, Gaitanis A, Gkiozos I,
Athanasiadis EI, Chatziioannou SN, Syrigos KN, Thanos D,
Chatziioannou AN and Papanikolaou N: Radiomics/Radiogenomics in
Lung Cancer: Basic principles and initial clinical results. Cancers
(Basel). 14(1657)2022.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Aleksandrova K, Egea Rodrigues C, Floegel
A and Ahrens W: Omics Biomarkers in Obesity: Novel etiological
insights and targets for precision prevention. Curr Obes Rep.
9:219–230. 2020.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Wang H, Fu T, Du Y, Gao W, Huang K, Liu Z,
Chandak P, Liu S, Van Katwyk P, Deac A, et al: Scientific discovery
in the age of artificial intelligence. Nature. 620:47–60.
2023.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Topol EJ: High-performance medicine: The
convergence of human and artificial intelligence. Nat Med.
25:44–56. 2019.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Verma M, Hontecillas R, Tubau-Juni N,
Abedi V and Bassaganya-Riera J: Challenges in Personalized
Nutrition and Health. Front Nutr. 5(117)2018.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Kohlmeier M, De Caterina R, Ferguson LR,
Görman U, Allayee H, Prasad C, Kang JX, Nicoletti CF and Martinez
JA: Guide and position of the international society of
nutrigenetics/nutrigenomics on personalized nutrition: Part 2 -
ethics, challenges and endeavors of precision nutrition. J
Nutrigenet Nutrigenomics. 9:28–46. 2016.PubMed/NCBI View Article : Google Scholar
|
|
57
|
World Health Organization (WHO): WHO
Guideline: Recommendations on digital interventions for health
system strengthening. WHO, Geneva, 2019.
|
|
58
|
Lee BY, Ordovás JM, Parks EJ, Anderson CA,
Barabási AL, Clinton SK, de la Haye K, Duffy VB, Franks PW, Ginexi
EM, et al: Research gaps and opportunities in precision nutrition:
An NIH workshop report. Am J Clin Nutr. 116:1877–1900.
2022.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Röttger-Wirtz S and De Boer A:
Personalised nutrition: The EU's fragmented legal landscape and the
overlooked implications of EU food law. Eur J Risk Regul.
12:212–235. 2021.
|
|
60
|
Aldubayan MA, Pigsborg K, Gormsen SMO,
Serra F, Palou M, Galmés S, Palou-March A, Favari C, Wetzels M,
Calleja A, et al: A double-blinded, randomized, parallel
intervention to evaluate biomarker-based nutrition plans for weight
loss: The PREVENTOMICS study. Clin Nutr. 41:1834–1844.
2022.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Sawicki C, Haslam D and Bhupathiraju S:
Utilising the precision nutrition toolkit in the path towards
precision medicine. Proc Nutr Soc. 82:359–369. 2023.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Berciano S, Figueiredo J, Brisbois TD,
Alford S, Koecher K, Eckhouse S, Ciati R, Kussmann M, Ordovas JM,
Stebbins K and Blumberg JB: Precision nutrition: Maintaining
scientific integrity while realizing market potential. Front Nutr.
9(979665)2022.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Mohr AE, Ortega-Santos CP, Whisner CM,
Klein-Seetharaman J and Jasbi P: Navigating challenges and
opportunities in multi-omics integration for personalized
healthcare. Biomedicines. 12(1496)2024.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Wu Y, Perng W and Peterson KE: Precision
nutrition and childhood obesity: A scoping review. Metabolites.
10(235)2020.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Özdemir V and Kolker E: Precision
nutrition 4.0: A big data and ethics foresight analysis-convergence
of agrigenomics, nutrigenomics, nutriproteomics, and
nutrimetabolomics. OMICS J Integr Biol. 20:69–75. 2016.PubMed/NCBI View Article : Google Scholar
|