Introduction
Cancer is a leading cause of death worldwide, with
millions of individuals succumbing to various types of cancer
annually (1). Therefore, it is of
utmost importance to identify anticancer prevention and treatment
strategies. According to epidemiology, cell apoptosis plays a role
in the incidence of cancers. Apoptosis, also known as programmed
cell death, is a fundamentally important biological process
triggered by a variety of stimuli, including deprivation of
growth/survival factors, exposure to cytotoxic drugs or DNA
damaging agents, activation of death receptors and activity of
cytotoxic cells, that is involved in controlling cell number and
eliminating harmful or virus-infected cells to maintain cell
homeostasis (2–4). The inappropriate process of apoptosis
potentially results in various pathological disorders (5). The caspase family (cysteine and
aspartic proteases) is mainly involved in the regulation of cell
apoptosis (6), and has two major
functions: caspase-1, −4, −5 and −11, as initiator caspases, are
primarily involved in the processing and activation of
pro-inflammatory cytokines, while caspase-2, −3, −6, −7, −8 and −9,
as executor caspases, play a role in the execution phase of
apoptosis (6,7). CASP activation has two dinstinct
albeit converging pathways: the extrinsic or receptor-mediated
pathway, and the intrinsic or mitochondrial pathway. These two
pathways possess an independent group of initiator caspases despite
using the same group of effector caspases (8–10).
Caspase-8 (CASP-8) is essential for the extrinsic cell death
pathways initiated by the TNF family members with the formation of
the death-inducing signaling complex (11).
Single-nucleotide polymorphisms (SNPs) are the most
common form of human genetic variation, leading to susceptibility
to cancer. Findings of previous studies showed that some variants
in CASP-8 gene are associated with susceptibility to various human
cancers (12,13). A case-control study in a Chinese
population found that CASP-8 −652 6N del/del genotypes showed a
multiplicative joint effect with FasL and Fas in attenuating
susceptibility to pancreatic cancer (14). However, relevant studies on −652 6N
del in CASP-8 are inconclusive and inconsistent. Therefore, a human
genome epidemiology (HuGE) review and meta-analysis were conducted,
including the most recent and relevant articles in order to
identify statistical evidence of the association between the CASP-8
−652 6N ins/del polymorphism and cancer risk that have been
investigated.
Materials and methods
Literature search
An extensive electronic search of the PubMed,
Cochrane Library, Embase, Web of Science, SpringerLink, CNKI and
CBM databases was performed to identify relevant studies available
up to May 1, 2012. The search terms used included [‘caspase-8’,
‘CASP-8’ or ‘Caspase 8’ (Mesh)] and [‘SNPs’, ‘SNP’ or
‘polymorphism, genetic’ (Mesh)] and [‘cancer’, ‘tumor’ or
‘Neoplasms’ (Mesh)]. The references in the eligible studies or
textbooks were also reviewed to check through manual searches to
find other potentially eligible studies.
Inclusion and exclusion criteria
The included studies had to meet the following
criteria: i) case-control study focused on the associations between
CASP-8 −652 6N ins/del polymorphism and cancer risk; ii) all
patients diagnosed with a malignant tumor confirmed by pathological
examination of the surgical specimen; iii) the frequencies of
alleles or geno-types in case and control groups could be
extracted; iv) the publication was in English or Chinese. Studies
were excluded when they were: i) not case-control studies about
CASP-8 −652 6N ins/del polymorphism and cancer risk; ii) based on
incomplete data; iii) useless or overlapping data were reported;
iv) meta-analyses, letters, reviews or editorial articles.
Data extraction
Using a standardized form, data from published
studies were extracted independently by two reviewers to populate
the necessary information. The information extracted from each of
the articles included: first author, year of publication, country,
language, ethnicity, study design, source of cases and controls,
number of cases and controls, mean age, sample, cancer type,
genotype method, allele and genotype frequency, and evidence of
Hardy-Weinberg equilibrium (HWE) in controls. In case of
conflicting evaluations, an agreement was reached following a
discussion with a third reviewer.
Quality assessment of included
studies
Two reviewers independently assessed the quality of
papers according to modified STROBE quality score systems (15,16).
Forty assessment items associated with the quality appraisal were
used in this meta-analysis, scores ranging from 0 to 40. Scores of
0–20, 20–30 and 30–40 were defined as low, moderate and high
quality, respectively. Disagreement was resolved by discussion.
Statistical analysis
The odds ratio (OR) and 95% confidence interval (95%
CI) were calculated using Review Manager Version 5.1.6 (provided by
the Cochrane Collaboration, available at: http://ims.cochrane.org/revman/download) and STATA
Version 12.0 (Stata Corp., College Station, TX, USA) software.
Between-study variations and heterogeneities were estimated using
Cochran’s Q-statistic (17,18)
(P≤0.05 was considered to be a manifestation of statistically
significant heterogeneity). The effect of heterogeneity, ranging
from 0 to 100% and representing the proportion of inter-study
variability that can be contributed to heterogeneity rather than to
chance, was quantified using the I2 test. When a significant Q-test
(P≤0.05) or I2>50% indicated that heterogeneity among
studies existed, the random-effects model was employed for the
meta-analysis. Otherwise, the fixed-effects model was used. To
establish the effect of heterogeneity on conclusions of the
meta-analyses, a subgroup analysis was carried out. We also tested
whether genotype frequencies of controls were in HWE using the
χ2 test. Funnel plots are often used to detect
publication bias. However, due to its limitations caused by varied
sample sizes and subjective reviews, Egger’s linear regression
test, which measures the funnel plot’s asymmetry using a natural
logarithmic scale of OR, was used to evaluate the publication bias
(19). When the P-value is
<0.1, publication bias is considered significant. All the
P-values were two-sided. To ensure the reliability and accuracy of
the results, two reviewers populated the data in the statistical
software programs independently and obtained identical results.
Results
Characteristics of included studies
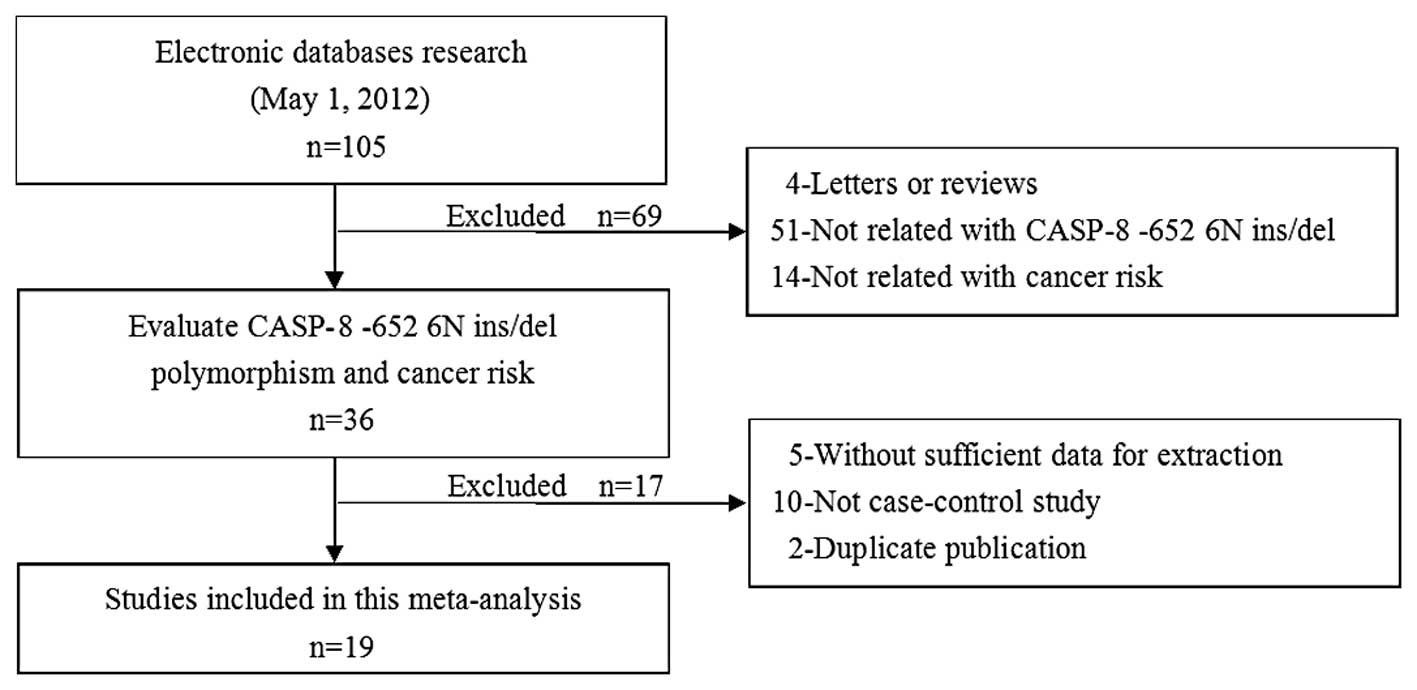
Subsequent to the initial screening a total of 105
relevant publications were identified. Nineteen studies (20–37)
appeared to have met the inclusion criteria and were subjected to
further examination. The flow chart of study selection is shown in
Fig. 1. In the pooled analysis, a
total of 23,172 cancer cases and 26,532 healthy controls from 19
studies were included and addressed. The publication year of
involved studies ranged from 2006 to 2011. Twelve of these studies
were conducted in Asian populations, 6 in Caucasian populations and
1 in African populations. The HWE test was performed on the
genotype distribution of the controls in all the included studies,
2 of these studies were out of HWE (34,37)
and the remaining studies showed to be in HWE (P>0.05). Quality
scores of included studies were >20 (moderate-high quality). The
characteristics and methodological quality of the included studies
are shown in Table I. The genotype
distribution of the CASP-8 −652 6N ins/del polymorphism in the case
and control groups is shown in Table
II.
 | Table ICharacteristics of individual studies
in this meta-analysis. |
Table I
Characteristics of individual studies
in this meta-analysis.
| Authors (Refs.) | Year | Country | Case no.
| Sample | Genotype method | Cancer type | Quality scores |
|---|
| AS | Control |
|---|
| Son et al
(20) | 2006 | Korea | 432 | 432 | Blood | PCR-RFLP | Lung cancer | 27 |
| Sun et al
(21) | 2007 | China | 4995 | 4972 | Blood | PCR-RFLP | Mixed cancer | 23 |
| Cybulski et al
(22) | 2008 | Poland | 618 | 531 | Blood | AS-PCR | Mixed cancer | 20 |
| Frank et al
(23) | 2008 | Germany | 7753 | 7921 | Blood | FFA | Breast cancer | 23 |
| Li et al
(24) | 2008 | China | 805 | 835 | Blood | PCR-RFLP | Melanoma | 26 |
| Pittman et al
(25) | 2008 | UK | 4016 | 3749 | Blood | AS-PCR | Colorectal
cancer | 21 |
| Yang et al
(14) | 2008 | China | 397 | 907 | Blood | PCR-RFLP | Pancreatic
cancer | 24 |
| Gangwar et al
(26) | 2009 | India | 212 | 250 | Blood | PCR-RFLP | Bladder cancer | 29 |
| Wang et al
(27) | 2009 | China | 365 | 368 | Blood | PCR-RFLP | Bladder cancer | 26 |
| Li et al
(28) | 2010 | USA | 1023 | 1052 | Blood | PCR-RFLP | Head and neck
cancer | 26 |
| Liu et al
(29) | 2010 | China | 373 | 838 | Blood | PCR-RFLP | Colorectal
cancer | 25 |
| Lv et al
(30) | 2010 | China | 100 | 544 | Blood | TaqMan | Lymphoma | 26 |
| Srivastava et
al (31) | 2010 | India | 230 | 230 | Blood | PCR-RFLP | Gallbladder
cancer | 24 |
| Chatterjee et
al (32) | 2011 | South Africa | 445 | 1221 | Blood | PCR-RFLP | Cervical
cancer | 18 |
| Hart et al
(33) | 2011 | Norway | 442 | 440 | Blood/tissue | TaqMan | Lung cancer | 20 |
| Kesarwani et
al (34) | 2011 | India | 175 | 198 | Blood | PCR-RFLP | Prostate
cancer | 24 |
| Ma et al
(35) | 2011 | China | 218 | 285 | Blood | Mass-Array | Ovarian cancer | 18 |
| Theodoropoulos
et al (36) | 2011 | Greece | 402 | 480 | Blood | PCR-RFLP | Colorectal
cancer | 18 |
| Umar et al
(37) | 2011 | India | 259 | 259 | Blood | PCR-RFLP | Esophageal
cancer | 20 |
 | Table IIThe genotype distribution of CASP-8
−652 6N polymorphism in case and control groups. |
Table II
The genotype distribution of CASP-8
−652 6N polymorphism in case and control groups.
| Authors
(Refs.) | Case
| Control
| HWE test
|
|---|
| Total | ins | del | ins/ins | ins/del | del/del | Total | ins | del | ins/ins | ins/del | del/del | χ2 | P-value | Test |
|---|
| Son et al
(20) | 432 | 654 | 210 | 247 | 160 | 25 | 432 | 659 | 205 | 249 | 161 | 22 | 0.38 | 0.54 | HWE |
| Sun et al
(21) | 4938 | 7917 | 1959 | 3173 | 1571 | 194 | 4919 | 7373 | 2465 | 2771 | 1831 | 317 | 0.39 | 0.53 | HWE |
| Cybulski et
al (22) | 1103 | 1184 | 1022 | 317 | 550 | 236 | 965 | 1047 | 883 | 274 | 499 | 192 | 1.68 | 0.20 | HWE |
| Frank et al
(23) | 7390 | 7592 | 7188 | 1946 | 3700 | 1744 | 7693 | 7767 | 7619 | 1949 | 3869 | 1875 | 0.27 | 0.60 | HWE |
| Li et al
(24) | 805 | 871 | 739 | 243 | 385 | 177 | 835 | 854 | 816 | 207 | 440 | 188 | 2.47 | 0.12 | HWE |
| Pittman et
al (25) | 3879 | 3887 | 3871 | 995 | 1897 | 987 | 3661 | 3656 | 3666 | 892 | 1872 | 897 | 1.88 | 0.17 | HWE |
| Yang et al
(14) | 397 | 647 | 147 | 268 | 111 | 18 | 907 | 1365 | 449 | 521 | 323 | 63 | 1.76 | 0.19 | HWE |
| Gangwar et
al (26) | 212 | 326 | 98 | 121 | 84 | 7 | 250 | 367 | 133 | 133 | 101 | 16 | 0.30 | 0.58 | HWE |
| Wang et al
(27) | 365 | 591 | 139 | 238 | 115 | 12 | 365 | 545 | 185 | 205 | 135 | 25 | 0.19 | 0.67 | HWE |
| Li et al
(28) | 1023 | 1078 | 968 | 311 | 456 | 256 | 1052 | 1056 | 1048 | 257 | 542 | 253 | 1.54 | 0.21 | HWE |
| Liu et al
(29) | 370 | 582 | 158 | 233 | 116 | 21 | 838 | 1334 | 342 | 528 | 278 | 32 | 0.38 | 0.54 | HWE |
| Lv et al
(30) | 100 | 142 | 58 | 48 | 46 | 6 | 544 | 867 | 221 | 344 | 179 | 21 | 0.15 | 0.70 | HWE |
| Srivastava et
al (31) | 228 | 363 | 93 | 147 | 69 | 12 | 230 | 328 | 132 | 122 | 84 | 24 | 2.66 | 0.10 | HWE |
| Chatterjee et
al (32) | 445 | 455 | 435 | 102 | 251 | 92 | 1221 | 1255 | 1187 | 308 | 639 | 274 | 2.75 | 0.10 | HWE |
| Hart et al
(33) | 436 | 460 | 412 | 125 | 210 | 101 | 433 | 421 | 445 | 106 | 209 | 118 | 0.50 | 0.48 | HWE |
| Kesarwani et
al (34) | 170 | 244 | 96 | 86 | 72 | 12 | 198 | 301 | 95 | 109 | 83 | 6 | 4.42 | 0.04 | Non-HWE |
| Ma et al
(35) | 218 | 343 | 93 | 128 | 87 | 3 | 285 | 398 | 172 | 138 | 122 | 25 | 0.07 | 0.79 | HWE |
| Theodoropoulos
et al (36) | 402 | 407 | 397 | 103 | 201 | 98 | 480 | 494 | 466 | 120 | 254 | 106 | 1.68 | 0.19 | HWE |
| Umar et al
(37) | 259 | 381 | 137 | 139 | 103 | 17 | 259 | 369 | 149 | 138 | 93 | 28 | 3.97 | 0.05 | Non-HWE |
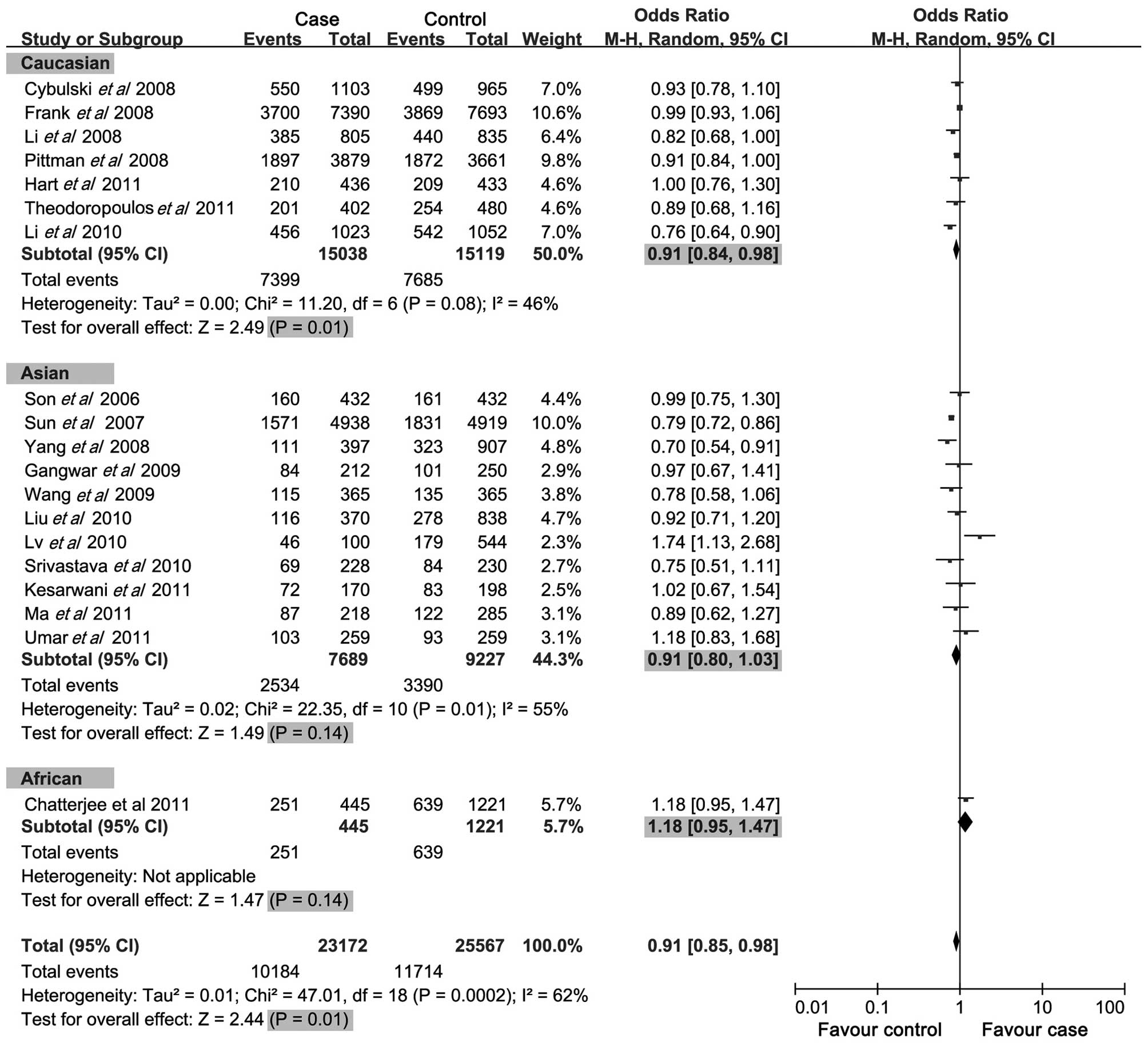
Main results and subgroup analysis
A summary of the meta-analysis findings of the
association between CASP-8 −652 6N ins/del polymorphism and cancer
risk is provided in Table III. The
meta-analysis results showed that the del allele, del allele
carrier and ins/del genotypes of −652 6N ins/del in CASP-8 gene
were negatively associated with cancer risk (OR=0.91, 95%
CI=0.84–0.98, P=0.01; OR=0.88, 95% CI=0.80–0.96, P=0.005; OR=0.91,
95% CI=0.85–0.98, P<0.001; respectively) (Figs. 2–4), while no significant correlation was
observed between the del/del genotypes of −652 6N ins/del and
cancer risk (OR=0.89, 95% CI=0.79–1.01, P=0.08). In the subgroup
analysis by ethnicity, we found that the del allele of −652 6N
ins/del was a protective factor for cancer risk in the Caucasian
and Asian populations (OR=0.96, 95% CI=0.93–1.00, P=0.05; OR=0.86,
95% CI=0.75–1.00, P=0.05; respectively), although not in the
African population (OR=1.01, 95% CI=0.87–1.18, P=0.891). For the
del allele carrier of −652 6N ins/del polymorphism, negative
associations with cancer risk were found in the Caucasian
population (OR=0.89, 95% CI=0.83–0.97, P=0.005), but not in the
Asian and African populations (OR=0.86, 95% CI=0.73–1.01, P=0.06;
OR=1.13, 95% CI=0.88–1.47, P=0.33; respectively). Notably, no
associations were found between the del/del genotype (variant
homozygote) of the −652 6N ins/del polymorphism and cancer risk in
the three populations (OR=0.89, 95% CI=0.79–1.10, P=0.08). However,
with regards to the ins/del genotype (heterozygote) of the −652 6N
ins/del polymorphism, protective associations with cancer risk were
found in the Caucasian population (OR=0.91, 95% CI=0.84–0.98,
P=0.01), whereas no correlation was found in the Asian and African
populations (OR=0.91, 95% CI=0.80–1.03, P=0.14; OR=1.18, 95%
CI=0.95–1.47, P=0.14; respectively).
 | Table IIIMeta-analysis of the association
between the −652 6N ins>del polymorphism in CASP-8 and cancer
risk. |
Table III
Meta-analysis of the association
between the −652 6N ins>del polymorphism in CASP-8 and cancer
risk.
| Subgroup | Case no./N | Control no./N | OR (95% CI) | P-value | Effect model |
|---|
| del allele | 18220/46344 | 20678/51134 | 0.91
(0.84–0.98) | 0.01 | Random |
| Caucasian | 14597/30076 | 14943/30238 | 0.96
(0.93–1.00) | 0.05 |
| Asian | 3188/15378 | 4548/18454 | 0.86
(0.75–1.00) | 0.05 |
| African | 435/890 | 1187/2442 | 1.01
(0.87–1.18) | 0.89 |
| del allele
carrier | 14202/23172 | 16196/25567 | 0.87
(0.80–0.96) | 0.005 | Random |
| Caucasian | 10998/15038 | 11314/15119 | 0.89
(0.83–0.97) | 0.005 |
| Asian | 2861/7689 | 3969/9227 | 0.86
(0.73–1.01) | 0.06 |
| African | 343/445 | 913/1221 | 1.13
(0.88–1.47) | 0.33 |
| del/del | 4018/23172 | 4482/25567 | 0.89
(0.79–1.01) | 0.08 | Random |
| Caucasian | 3599/15308 | 3629/15119 | 1.00
(0.95–1.05) | 0.90 |
| Asian | 327/7689 | 579/9227 | 0.73
(0.53–1.01) | 0.06 |
| African | 92/445 | 274/1221 | 0.90
(0.69–1.18) | 0.44 |
| ins/del | 10184/23172 | 11741/25567 | 0.91
(0.85–0.98) | <0.001 | Random |
| Caucasian | 7399/125038 | 7685/15119 | 0.91
(0.84–0.98) | 0.01 |
| Asian | 2534/7689 | 3390/9227 | 0.91
(0.80–1.03) | 0.14 |
| African | 251/445 | 639/1221 | 1.18
(0.95–1.47) | 0.14 |
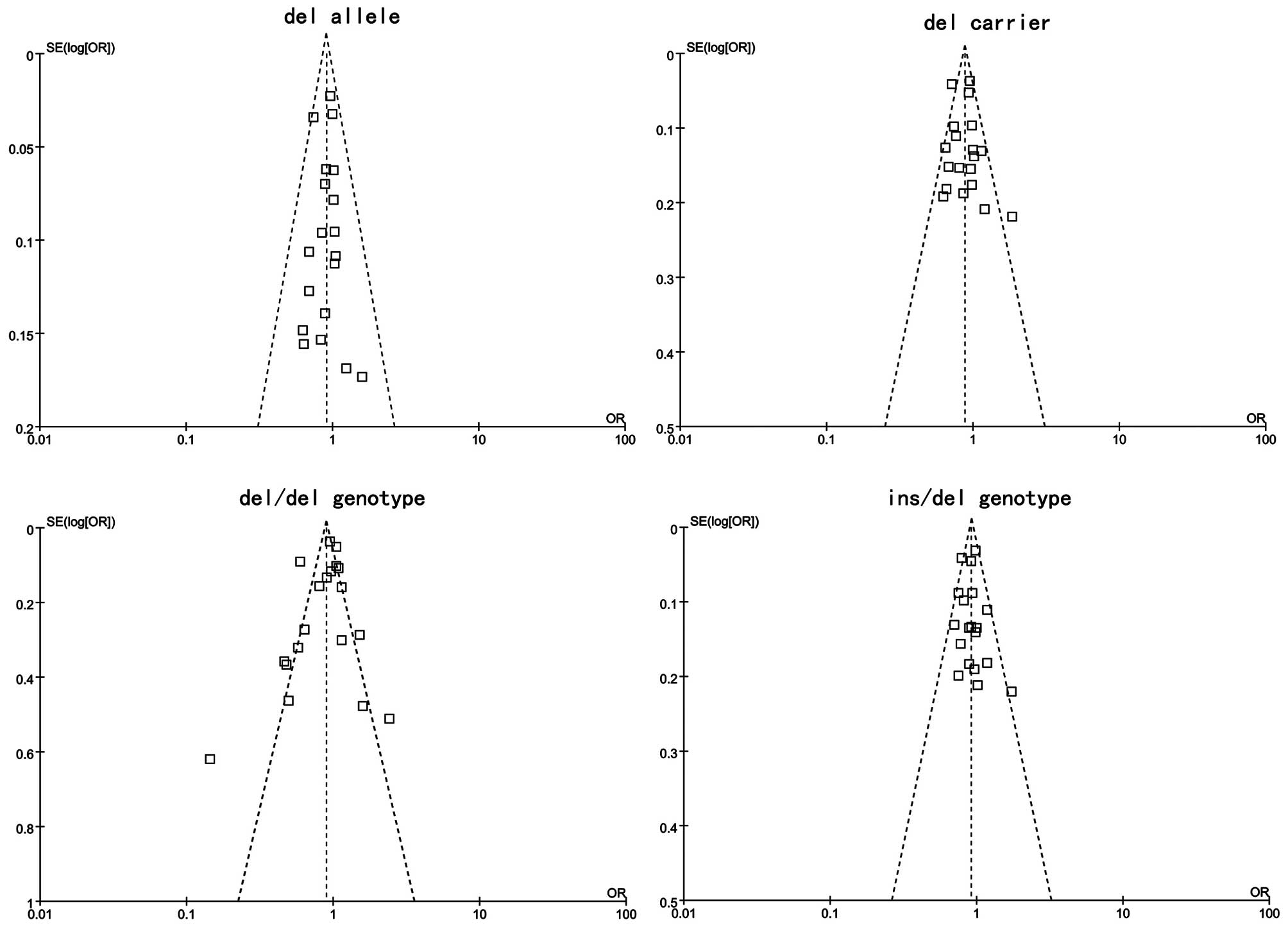
Publication bias
Publication bias of the literature was accessed by
Begger’s funnel plot and Egger’s linear regression test. Egger’s
linear regression test was used to measure the asymmetry of the
funnel plot. The graphical funnel plots of included studies
appeared to be symmetrical (Fig.
5). Egger’s test also showed that there was no statistical
significance for all evaluations of publication bias (all
P>0.05). Findings of Egger’s publication bias test are shown in
Table IV.
 | Table IVEvaluation of publication bias by
Egger’s linear regression test. |
Table IV
Evaluation of publication bias by
Egger’s linear regression test.
| SNP | Coefficient | SE | t | P-value | 95% CI |
|---|
| del allele | −0.298 | 0.932 | −0.320 | 0.753 | (−2.265,
1.669) |
| del carrier | 0.375 | 0.834 | 0.450 | 0.658 | (−1.384,
2.135) |
| del/del
genotype | −0.745 | 0.645 | −1.160 | 0.264 | (−2.105,
0.615) |
| ins/del
genotype | 0.192 | 0.664 | 0.290 | 0.776 | (−1.208,
1.592) |
Discussion
CASP-8, located on chromosome 2q33–q34, encoded by
the CASP-8 gene, is a caspase protein that plays a key role in the
execution-phase of cell apoptosis (28). When induced by Fas and various
apoptotic stimuli, this protein is involved in apoptosis (29). Caspase-8 is known to activate
during death receptor-initiated apoptosis, inducing apoptosis and
maintaining immune homeostasis and immune surveillance, while the
single genetic variants in CASP-8 and their function in human
cancer susceptibility remain to be elucidated (21). The −652 6N ins/del (rs3834129), a
common SNP in the CASP-8 gene, is strongly associated with the
CASP-8 expression. Investigators have reported a correlation
between the −652 6N ins/del polymorphism and susceptibility to
various types of cancer. Sun et al observed that the CASP-8
−652 6N ins/del allele was associated with a reduced risk of
developing different types of human cancer, including lung,
esophageal, colorectal, cervical and breast cancer, as well as
gastric cancer, indicating that this variant allele may confer
protection against multiple cancers (21). Frank et al showed that the
CASP-8 −652 6N ins/del variant has no significant effect on breast
cancer risk in Europeans (23). In
their study, Li et al observed that the CASP-8 −652 6N
ins/del variant genotypes (ins/del, ins/del+del/del) were
associated with significantly lower cutaneous melanoma risk than
were the ins/ins genotypes (24).
In our study, we examined the association of the −652 6N ins/del
polymorphism in the CASP-8 gene with the risk for cancer by
meta-analysis. A negative association was observed between the del
allele, del allele carrier and ins/del genotype of the −652 6N
ins/del polymorphism in CASP-8 gene and cancer risk. In the
stratified analysis by ethnicity, Caucasians who harbored the
ins/del genotypes or del allele or del allele carrier were found to
exhibit a significantly lower risk for cancer. In addition, a
negative association was also found between the del allele of −652
6N ins/del in CASP-8 gene and cancer risk in the Asian
population.
Limitations of this study should be acknowledged.
First, although the funnel plot and Egger’s test did not show any
publication bias, selection bias may have occurred because only
studies published in English or Chinese were included. Second, the
control subjects of the present study might not be representative
of the general population, necessitating well-designed
population-based studies with large sample sizes and detailed
exposure information to validate our findings. Third, there was
significant between-study heterogeneity from studies of the −652 6N
ins/del polymorphism, while the geno-type distribution also showed
deviation from HWE in some studies. Fourth, our meta-analysis was
based on unadjusted OR estimates as not all published studies
presented adjusted ORs, or when they did, the ORs were not adjusted
by the same potential confounders, such as age, gender, ethnicity
and exposures. In addition, our analysis did not consider the
possibility of gene-gene or SNP-SNP interactions or the possibility
of linkage disequilibrium between polymorphisms. Therefore, our
conclusions should be interpreted with caution.
In conclusion, findings of this study have shown a
common insertion-deletion variation in the promoter region of the
CASP-8 gene as a low penetrance susceptibility locus for certain
common types of human cancers. The del allele, del allele carrier
and ins/del genotype of the −652 6N ins/del polymorphism in CASP-8
gene may serve as protective factors for cancer risk. However,
these findings should be validated by large-scale, prospective
studies investigating more diverse ethnic groups and more detailed
environmental exposure data.
Acknowledgements
We would like to thank J.L. Liu
(MedChina medical information service Co., Ltd.) for his valuable
contribution and kindly revising the manuscript.
References
|
1
|
He X, Chang S, Zhang J, Zhao Q, Xiang H,
Kusonmano K, Yang L, Sun ZS, Yang H and Wang J: MethyCancer: the
database of human DNA methylation and cancer. Nucleic Acids Res.
36:D836–D841. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Salvesen GS and Abrams JM: Caspase
activation - stepping on the gas or releasing the brakes? Lessons
from humans and flies. Oncogene. 23:2774–2784. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Shi Y: Caspase activation, inhibition, and
reactivation: a mechanistic view. Protein Sci. 13:1979–1987. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kumar S: Caspase function in programmed
cell death. Cell Death Differ. 14:32–43. 2007. View Article : Google Scholar
|
|
5
|
Friedlander RM: Apoptosis and caspases in
neurodegenerative diseases. N Engl J Med. 348:1365–1375. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kumar S and Dorstyn L: Analysing caspase
activation and caspase activity in apoptotic cells. Methods Mol
Biol. 559:3–17. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Siegel RM: Caspases at the crossroads of
immune-cell life and death. Nat Rev Immunol. 6:308–317. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Degterev A, Boyce M and Yuan J: A decade
of caspases. Oncogene. 22:8543–8567. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bodmer JL, Holler N, Reynard S,
Vinciguerra P, Schneider P, Juo P, Blenis J and Tschopp J: TRAIL
receptor-2 signals apoptosis through FADD and caspase-8. Nat Cell
Biol. 2:241–243. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ashkenazi A and Dixit VM: Apoptosis
control by death and decoy receptors. Curr Opin Cell Biol.
11:255–260. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Danial NN and Korsmeyer SJ: Cell death:
critical control points. Cell. 116:205–219. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang X, Miao X, Sun T, Tan W, Qu S, Xiong
P, Zhou Y and Lin D: Functional polymorphisms in cell death pathway
genes FAS and FASL contribute to risk of lung cancer. J Med Genet.
42:479–484. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
MacPherson G, Healey CS, Teare MD,
Balasubramanian SP, Reed MW, Pharoah PD, Ponder BA, Meuth M,
Bhattacharyya NP and Cox A: Association of a common variant of the
CASP8 gene with reduced risk of breast cancer. J Natl Cancer Inst.
24:1866–1869. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yang M, Sun T, Wang L, Yu D, Zhang X, Miao
X, Liu J, Zhao D, Li H, Tan W and Lin D: Functional variants in
cell death pathway genes and risk of pancreatic cancer. Clin Cancer
Res. 14:3230–3236. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Von Elm E, Altman DG, Egger M, Pocock SJ,
Gøtzsche PC and Vandenbroucke JP; STROBE Initiative: The
Strengthening the Reporting of Observational Studies in
Epidemiology (STROBE) statement: guidelines for reporting
observational studies. Epidemiology. 18:800–804. 2007.
|
|
16
|
Zhang L, Liu JL, Zhang YJ and Wang H:
Association between HLA-B*27 polymorphisms and ankylosing
spondylitis in Han populations: a meta-analysis. Clin Exp
Rheumatol. 29:285–292. 2011.
|
|
17
|
Higgins JP and Thompson SG: Quantifying
heterogeneity in a meta-analysis. Stat Med. 21:1539–1558. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zintzaras E and Ioannidis JP:
Heterogeneity testing in meta-analysis of genome searches. Genet
Epidemiol. 28:123–137. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Peters JL, Sutton AJ, Jones DR, Abrams KR
and Rushton L: Comparison of two methods to detect publication bias
in meta-analysis. JAMA. 295:676–680. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Son JW, Kang HK, Chae MH, Choi JE, Park
JM, Lee WK, Kim CH, Kim DS, Kam S, Kang YM and Park JY:
Polymorphisms in the caspase-8 gene and the risk of lung cancer.
Cancer Genet Cytogenet. 169:121–127. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sun T, Gao Y, Tan W, Ma S, Shi Y, Yao J,
Guo Y, Yang M, Zhang X, Zhang Q, et al: A six-nucleotide
insertion-deletion polymorphism in the CASP8 promoter is associated
with susceptibility to multiple cancers. Nat Genet. 39:605–613.
2007. View
Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cybulski C, Wokołorczyk D, Gliniewicz B,
Sikorski A, Górski B, Jakubowska A, Huzarski T, Byrski T, Debniak
T, Gronwald J, et al: A six-nucleotide deletion in the CASP8
promoter is not associated with a susceptibility to breast and
prostate cancers in the Polish population. Breast Cancer Res Treat.
112:367–368. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Frank B, Rigas SH, Bermejo JL, Wiestler M,
Wagner K, Hemminki K, Reed MW, Sutter C, Wappenschmidt B,
Balasubramanian SP, et al: The CASP8 −652 6N del promoter
polymorphism and breast cancer risk: a multicenter study. Breast
Cancer Res Treat. 111:139–144. 2008.
|
|
24
|
Li C, Zhao H, Hu Z, Liu Z, Wang LE,
Gershenwald JE, Prieto VG, Lee JE, Duvic M, Grimm EA and Wei Q:
Genetic variants and haplotypes of the caspase-8 and caspase-10
genes contribute to susceptibility to cutaneous melanoma. Hum
Mutat. 29:1443–1451. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Pittman AM, Broderick P, Sullivan K,
Fielding S, Webb E, Penegar S, Tomlinson I and Houlston RS: CASP8
variants D302H and −652 6N ins/del do not influence the risk of
colorectal cancer in the United Kingdom population. Br J Cancer.
98:1434–1436. 2008.
|
|
26
|
Gangwar R, Mandhani A and Mittal RD:
Caspase 9 and caspase 8 gene polymorphisms and susceptibility to
bladder cancer in north Indian population. Ann Surg Oncol.
16:2028–2034. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang M and Zhang Z, Tian Y, Shao J and
Zhang Z: A six-nucleotide insertion-deletion polymorphism in the
CASP8 promoter associated with risk and progression of bladder
cancer. Clin Cancer Res. 15:2567–2572. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li C, Lu J, Liu Z, Wang LE, Zhao H,
El-Naggar AK, Sturgis EM and Wei Q: The six-nucleotide
deletion/insertion variant in the CASP8 promoter region is
inversely associated with risk of squamous cell carcinoma of the
head and neck. Cancer Prev Res (Phila). 3:246–253. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Liu B, Zhang Y, Jin M, Ni Q, Liang X, Ma
X, Yao K, Li Q and Chen K: Association of selected polymorphisms of
CCND1, p21, and caspase8 with colorectal cancer risk. Mol Carcinog.
49:75–84. 2010.PubMed/NCBI
|
|
30
|
Lv Z: Genetic polymorphisms in CASP8, Fas
and Fas ligand and the risk of peripheral T-cell lymphoma and the
relationship between the genetic polymorphisms and the clinical
characteristics and prognosis. Peking Union Medical College
Observational Studies. Epidemiology. 18:800–804. 2010.
|
|
31
|
Srivastava K, Srivastava A and Mittal B:
Caspase-8 polymorphisms and risk of gallbladder cancer in a
northern Indian population. Mol Carcinog. 49:684–692.
2010.PubMed/NCBI
|
|
32
|
Chatterjee K, Williamson AL, Hoffman M and
Dandara C: CASP8 promoter polymorphism is associated with high-risk
HPV types and abnormal cytology but not with cervical cancer. J Med
Virol. 83:630–636. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hart K, Landvik NE, Lind H, Skaug V,
Haugen A and Zienolddiny S: A combination of functional
polymorphisms in the CASP8, MMP1, IL10 and SEPS1 genes affects risk
of non-small cell lung cancer. Lung Cancer. 71:123–129. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kesarwani P, Mandal RK, Maheshwari R and
Mittal RD: Influence of caspases 8 and 9 gene promoter polymorphism
on prostate cancer susceptibility and early development of hormone
refractory prostate cancer. BJU Int. 107:471–476. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ma X, Zhang J, Liu S, Huang Y, Chen B and
Wang D: Polymorphisms in the CASP8 gene and the risk of epithelial
ovarian cancer. Gynecol Oncol. 122:554–559. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Theodoropoulos GE, Gazouli M, Vaiopoulou
A, Leandrou M, Nikouli S, Vassou E, Kouraklis G and Nikiteas N:
Poly morphisms of caspase 8 and caspase 9 gene and colorectal
cancer susceptibility and prognosis. Int J Colorectal Dis.
26:1113–1118. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Umar M, Upadhyay R, Kumar S, Ghoshal UC
and Mittal B: CASP8 −652 6N del and CASP8 IVS12-19G>A gene
polymorphisms and susceptibility/prognosis of ESCC: a case control
study in northern Indian population. J Surg Oncol. 103:716–723.
2011.
|