Introduction
The aging population in China is becoming
increasingly evident along with the acceleration in the pace of
life. The number of individuals with neck pain has also increased,
but the pathogenesis of this health problem remains unclear. The
cartilage end-plate is an important part of the intervertebral
disc. The degeneration of this end-plate is closely associated with
intervertebral disc degeneration, which is a cell-mediated process.
The chondrocytes are the major cell type in the cartilage
end-plate. These cells have an important role in maintaining the
physiological functions of the intervertebral disc and the
integrity of the extracellular matrix (1). Apoptosis is considered an important
factor in disc degeneration (2).
Extensive clinical and animal model studies have shown that cell
structure loss and cell death are associated with intervertebral
disc degeneration (3). Therefore,
studying the pathological physiology of chondrocytes is
important.
Autophagy is a form of apoptosis. In this process,
cells engulf cytoplasmic proteins or organelles which are then
packed into vesicles, and form an autophagolysosome with the
lysosome. This process modulates metabolic materials and certain
organelles. The autophagic process has four parts:
substrate-induced porautophagosome, autophagy, fusion of the
autophagosome with lysosomes and degradation of the contents of the
autophagosome (4). Autophagy has a
significant role in various degenerative pathological processes.
For example, this mechanism is associated with cancer, microbial
infections, heart diseases and even life extension (5,6).
However, the association between autophagy and cartilage end-plate
degeneration has rarely been studied.
In this study, surgically removed cartilage
end-plates from patients with cervical spondylosis were used to
establish degenerative chondrocyte cultures through enzyme
digestion. Cultured chondrocytes obtained from the cartilage
end-plates of patients with cervical vertebral fracture or
dislocation served as the control. In addition, autophagy in the
cultured chondrocytes was observed and the significance of
variations in autophagy in the degeneration of cervical vertebral
end-plate chondrocytes was investigated.
Materials and methods
Subjects
The subjects were cervical spine surgery patients
admitted to Yijishan Hospital, Wannan Medical College (Wuhu, China)
between February 2011 and August 2012. A total of 31 cases of
cervical spondylosis (the cervical spondylosis group) were
selected, with 19 males and 12 females, aged 38 to 72 years. The
average age was 52 years. Seventeen cases of fracture and
dislocation patients (the control group) were also selected, with
11 males and 6 females, aged 23 to 36 years. The average age was 30
years. Patients with tumors, tuberculosis, diabetes, infections and
metabolic bone diseases were excluded. All the patients underwent
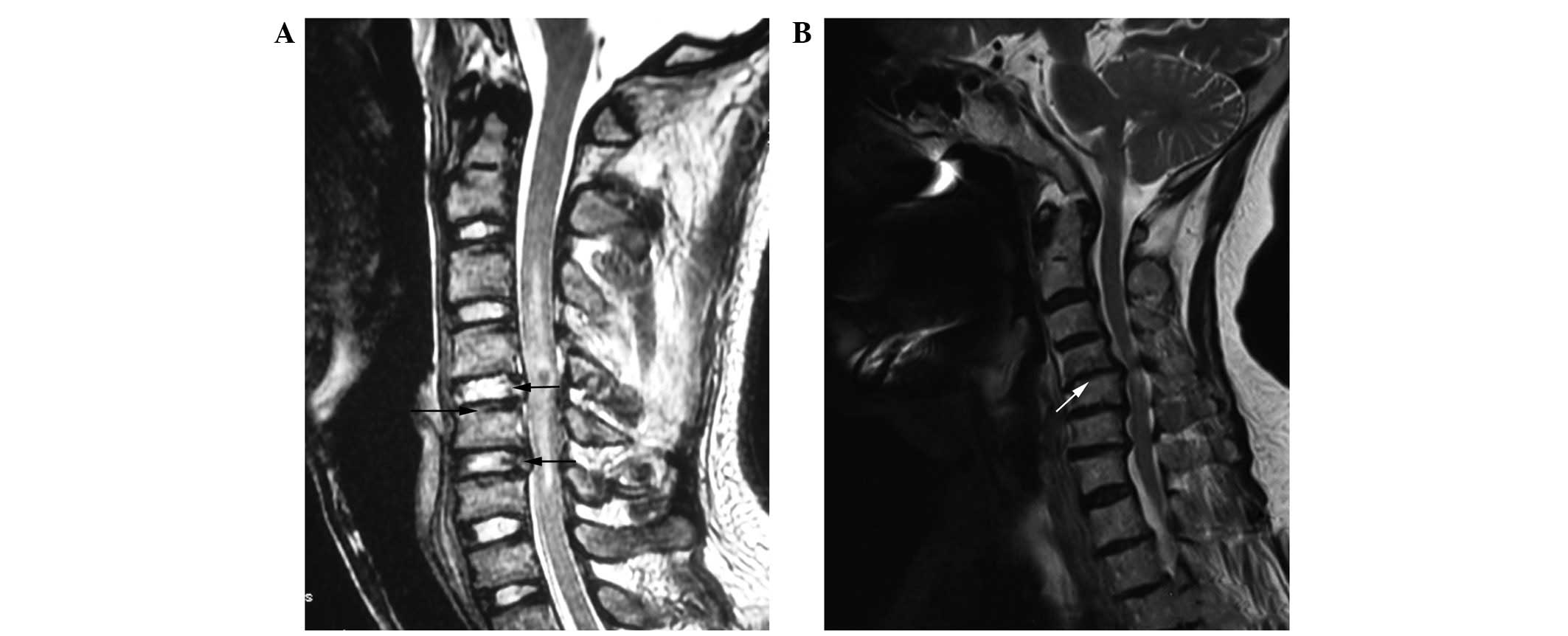
magnetic resonance imaging (MRI) examination prior to surgery. The
degeneration of pathological grade cartilage of the end-plate and
intervertebral disc was classified according to Miller (7) and Thompson et al (8). Representative MRI results are shown
in Fig. 1. The control group had 5
cases of Miller grade 0 and 12 cases of Miller grade 1 according to
MRI examination of the cartilage end-plate prior to surgery. After
surgery, 7 patients did not exhibit pathological degeneration. A
total of 10 cases of pathological disc degeneration were Thompson
grade 1. The cervical spondylosis group had 8 cases of Miller grade
2 and 23 cases of Miller grade 3 according to MRI examination of
the cartilage end-plate prior to surgery. In addition, 6 cases were
Thompson grade 3, 15 cases were Thompson grade 4 and 10 cases were
Thompson grade 5 according to MRI examination of disc degeneration.
This study was conducted in accordance with the Declaration of
Helsinki and with approval from the Ethics Committee of Wannan
Medical College. Written informed consent was obtained from all
participants.
Chondrocyte isolation and culture
The cartilage end-plate tissue samples of the
patients were excised from the disc and immediately sent to the
central laboratory (Yijishan Hospital, Wannan Medical College,
Wuhu, China). The samples were placed under a dissecting microscope
(magnification, ×4) and processed by aseptic technique in a
biological safety cabinet. The cartilage end-plate, nucleus
pulposus and annulus of the samples were separated carefully. The
chondrocytes were isolated (Type II collagenase and trypsin;
HyClone, Logan, UT, USA) and cultured as in preliminary studies
(9). The seeding density and
culture conditions of the two groups of cells were the same.
Changes in morphology and growth were observed regularly under an
inverted microscope (Olympus, Tokyo, Japan).
Toluidine blue and hematoxylin-eosin
(H&E) staining
The two groups of cells were mounted on a coverslip.
On day 9 of cell culture, ~60% fusion was observed. The cells were
then removed, seeded and stained. i) Toluidine blue staining: the
coverslip was washed thrice with phosphate buffer and fixed with 4%
paraformaldehyde for 30 min. The coverslip was washed with water
for 15 min and stained with toluidine blue for 4 h. After the
coverslip was washed with tap water, the cells were air-dried,
neutral gum was added and the cells were observed under a
microscope (Olympus, Japan). ii) H&E staining: the coverslip
was washed thrice with phosphate buffer and fixed with 4%
paraformaldehyde for 30 min. The coverslip was washed with water
for 15 min and stained with H&E. After the coverslip was washed
with tap water, the cells were air-dried, neutral gum was added and
the cells were observed under a microscope.
Monodansylcadaverine (MDC) fluorescence
staining
The two groups of cells were passaged and cultured.
When the logarithmic growth phase of the cells reached a density of
70–80%, the digested cells were mixed and blown into a single-cell
suspension. Cells (10 μl) were collected and counted at
5×104/ml cells per well, with an additional 2 ml for
each well. The cells were cultured for 24 h after adhesion and
washed twice for 5 min with phosphate-buffered saline (PBS). The
cells were treated with MDC (0.05 mmol/l; Sigma, St. Louis, MO,
USA) diluted with PBS at 37°C for 15 min and then washed with PBS.
The cells were observed under a fluorescence microscope (Leica,
Wetzlar, Germany) with an emission filter of 356 and 545 nm.
Laser scanning confocal microscopy
Logarithmic phase cells were trypsinized and the
precipitate was collected following centrifugation. The cells were
drawn up using a pipette and mixed with a RPMI-1640 medium without
antibiotics, producing a final count of 2×105 cells per
well. Immunofluorescence was obtained following cell adhesion for
24 h. The specific steps were as follows. Fixing: The cells were
washed thrice with PBS for 5 min each time, then fixed with 4%
paraformaldehyde for 15 min. Blocking: The cells were washed thrice
with PBS for 5 min each time. Fetal bovine serum albumin (5%) was
added to the blocking solution at 37°C for 2 h. The cells were
washed thrice with PBS for 5 min each time, anti-LC3B antibody (5
μg/ml, Sigma) was added at 4°C and the cells were incubated
overnight. The cells were then washed thrice with PBS for 5 min
each time. The diluted fluorescent secondary antibody [Alexa
Fluor® 555 donkey anti-rabbit IgG (H+L); 10 μg/ml; Santa
Cruz Biotechnology, Inc., Santa Cruz, CA, USA] was added to the
cells at 37°C for 1.5 h. The cells were washed thrice with PBS for
5 min each time. MDC (50 μM) was added to the cells as a dye at 4°C
for 30 min and kept in the dark. A confocal laser scanner
(Molecular Devices, Silicon Valley, CA, USA) was used to observe
and capture photos of the cells the following day.
Reverse transcription polymerase chain
reaction (RT-PCR)
One-step TRIzol (Invitrogen, Carlsbad, CA, USA) was
used to extract total RNA from cells. The purity and concentration
were measured using an ultraviolet spectrophotometer (Olympus,
Japan). A total of 3 μg RNA was extracted to synthesize cDNA.
Aggrecan, type II collagen and β-actin were amplified using 3 μl
cDNA as a template. β-actin was used as the internal reference. The
gene primer sequences and the amplified fragment sizes are shown in
Table I. The reaction conditions
were as follows: predenaturation at 94°C for 5 min, denaturation at
94°C for 1 min and annealing at 56.1°C for 30 sec. Type II collagen
was maintained at 60°C for 30 sec. β-actin was maintained at 51.9°C
for 30 sec, extended at 72°C for 40 sec and amplified for 35
cycles. The reaction was terminated at 72°C after 5 min. The
products were stored at 4°C. The PCR products were separated
through 1.5% agarose gel electrophoresis. The Tanon Gel Image
system 1D (Tanon, Shanghai, China) was used for semi-quantitative
analysis. The relative band intensity percentages of the target
gene and its internal reference were considered as the relative
expression levels of the target gene mRNA for statistical
analysis.
 | Table IGene primer sequences and the
amplified fragment lengths. |
Table I
Gene primer sequences and the
amplified fragment lengths.
| Gene | Forward primer | Reverse primer | Product length
(bp) |
|---|
| Type II collagen |
5′-CTCCTGGAGCATCTGGAGAC-3′ |
5′-ACCACGATCACCCTTGATCT-3′ | 153 |
| Aggrecan |
5′-AGTATCATCAGTCCCAGAATCTAGCA-3′ |
5′-AATGCAGAGGTGGTTTCACTCA-3′ | 132 |
| β-Actin |
5′-GCATCCCCCAAAGTTCACAA-3′ |
5′-AGGACTGGGCCATTCTCCTT-3′ | 153 |
Western blotting
Radioimmunoprecipitation assay buffer and
phenylmethanesulfonyl fluoride were used to extract protein from
the two groups of cells. The total protein was measured using the
bicinchoninic acid method. Sodium dodecyl sulfate-polyacrylamide
gel electrophoresis was performed on the protein. Human anti-LC3B,
and anti-β-actin were diluted (1:1,000) in TBS-Tween 20 containing
1% BSA. The membranes were incubated for 2 h at 37°C with the
primary antibody, washed three times in TBS-Tween 20 and for 1 h
with the secondary antibody goat anti-rabbit IgG-HRP and goat
anti-mouse IgG-HRP (1:5,000). Subsequent detection was performed
using the ECL western blotting system (Amersham Biosciences,
Chalfont St Giles, UK) according to the manufacturer’s
instructions. Specificity of the antibody was assessed by omitting
the first antibody in western blotting experiments.
Statistical analysis
Experimental data are expressed as mean ± SD. The
two groups were compared using two independent sample Student’s
t-tests. The results were determined using SPSS 18.0 software
(SPSS, Inc., Chicago, IL, USA). P<0.05 was considered to
indicate a statistically significant result.
Results
Morphological changes of the
chondrocytes
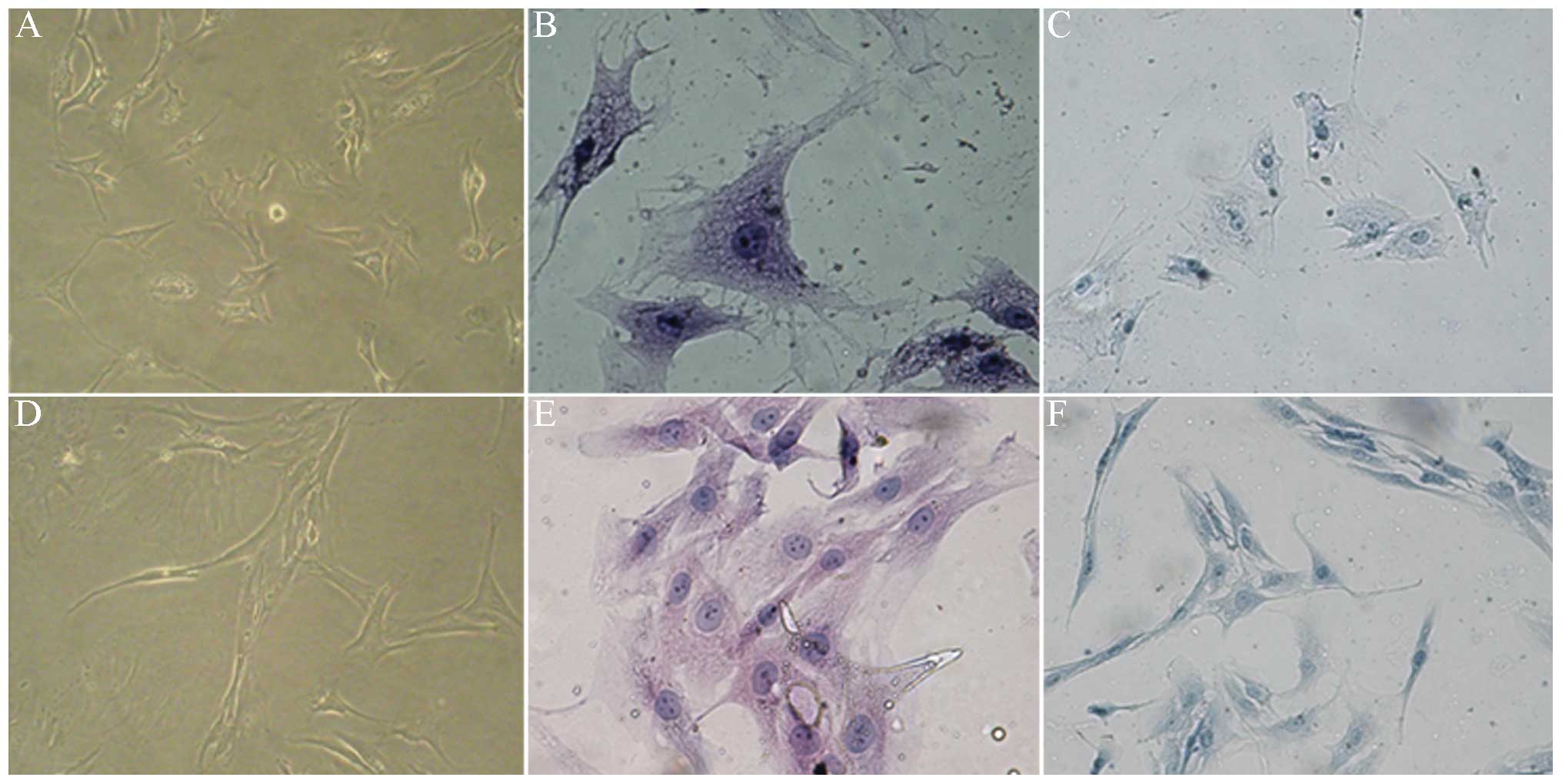
Under an inverted microscope, it was observed that
the majority of the control cells were polygonal. The cells from
the patients with cervical spondylosis gradually stretched and the
majority of these cells were shuttle type. The growth rate of the
cells in the cervical spondylosis group was significantly slower
than that of the cells in the control group. H&E staining
demonstrated that the cytoplasms contained particulate matter in
the control group; the nuclei were large and round, and two to
three nucleoli per cell were observed. The spindle was the primary
morphology of the cell nucleus in the cervical spondylosis group.
The nucleolus was unclear and the gap between cells increased. The
cytoplasm and cells surrounded by chondrocytes were stained and
metachromatic in the two groups (Fig.
2). This result confirmed that the cells used in this
experiment were end-plate chondrocytes.
MDC fluorescence staining
Under a dark field fluorescence microscope, the
autophagosome absorbed MDC and appeared as a green granular
structure. The control and cervical spondylosis groups both
contained fluorescent granular autophagosomes (Fig. 3).
Laser scanning confocal microscopy
Laser scanning confocal microscopy showed that the
LC3 proteins of the autophagosome were primarily concentrated in
the intracellular and perinuclear regions (Fig. 4).
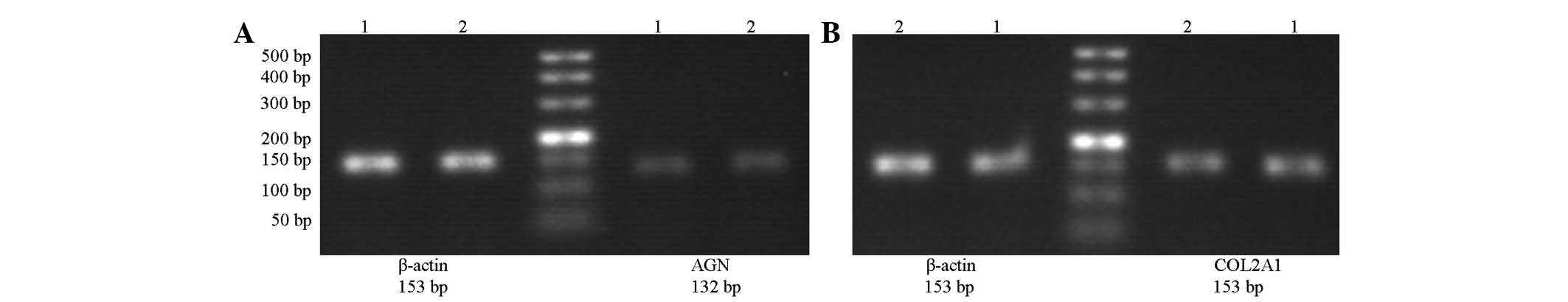
RT-PCR
The mRNA expression levels of the aggrecan gene
(0.715±0.194) and the type II collagen gene (0.628±0.254) in the
cervical spondylosis group were much lower than those in the
control group (0.913±0.254 and 0.845±0.186, respectively). The
difference between the two groups was statistically significant
(both P<0.05; Fig. 5).
Western blotting
The expression of LC3-II in the cervical spondylosis
group was downregulated compared with that in the control group.
LC3-I was upregulated, whereas the ratio LC3-II/LC3-I was also
decreased. The expression of β-actin did not differ between the two
groups (Fig. 6).
Discussion
The cartilage end-plate constitutes the upper and
lower bounds of the intervertebral disc. This disc is the largest
avascular structure in the body with a relative lack of nutrient
supply. The supply of nutrients and the discharge of metabolites in
the disc are conducted through the end-plate. Therefore, the
cartilage end-plate has an important role in the nutrition path of
the disc, since material exchange occurs between the disc and its
external environment. The delivery of nutrients to the cartilage
end-plate is primarily affected by the shape, size and charge of
the solute, as well as the composition of the cartilage end-plate.
The cartilage end-plate is composed of cells and stromata. The
major cell types in the cartilage end-plate are chondrocytes and
fibroblasts. The matrices are primarily composed of aggrecan and
type II collagen. The maintenance of normal decomposition and
anabolism by a matrix is essential for its biological function. The
cartilage end-plate prevents small molecules called proteoglycans
from being lost in the disc. Following the degeneration of the
cartilage end-plate, this protective effect is weakened, thus the
loss of disc proteoglycan increases (10–12).
As the nutrition path of the cartilage end-plate breaks, the matrix
syntheses of type II collagen and aggrecan, as well as the amount
of water in the cartilage end-plate, are reduced. Consequently,
disorders of the cartilage end-plate structure result and the
normal function of the cartilage end-plate is lost, eventually
leading to disc degeneration (13). In the present study, chondrocyte
phenotypes were distinguished by toluidine blue staining, H&E
staining and RT-PCR. The results show that the expression levels of
type II collagen and aggrecan in the cervical spondylosis group
end-plates were significantly lower than those in the control
group. Furthermore, the results are in accordance with previous
studies (9,14).
Autophagic cell death has been observed from as
early as the 1960s; however, cause for concern has not been
identified until recently. As an innate host defense mechanism,
autophagy is involved in the removal of damaged organelles
(15). This mechanism is an
important physiological process for maintaining cellular
homeostasis, such as in the growth and development of an organism,
cell differentiation and proliferation, remodeling, and degradation
of aging or damaged intracellular organelles, hamartoma and excess
proteins (16). Autophagy is
primarily involved in clearing damaged organelles and in reusing
macromolecular substances of cells. Autophagy also has an important
role in maintaining cell homeostasis and in promoting cell
survival. In addition, autophagy serves as the main degradation
pathway for macromolecular proteins and organelles. Autophagy has a
protective effect on various tissue cells (17). Lapatinib induces autophagy and is
able to impede the growth of breast cancer (18). The regulation of autophagy also
prevents diabetes, which induces damage to the endoplasmic
reticulum (19). Autophagy is
closely associated with human osteoarthritis and the arthritis
model in mice, and may have a protective effect against
osteoarthritis (20). In addition,
autophagy has an extremely important role in preventing damage to
the mitochondrion and the endoplasmic reticulum, and may
effectively remove damaged organelles, thus delaying or preventing
apoptosis initiation, reversing cell injury or degeneration, or
even death (21). If autophagy is
enhanced in the degeneration of cartilage cells, then various
metabolic waste materials in cartilage cells are promptly removed,
thus allowing the cells to develop a suitable environment. The
degeneration of articular cartilage may be delayed effectively or
even blocked. Apoptosis has an important role in disc degeneration
and numerous other diseases. Autophagy has a significant role in
blocking apoptosis, however, the specific mechanisms of autophagy
in disc degeneration remain unclear.
Autofluorescent MDC labels the autophagic structures
of cells and may be used as an autophagy tracer (11). In the present study, autophagic
bodies in the control and cervical spondylosis groups were observed
through MDC staining. LC3, which is the human homolog of the
autophagy-related gene Atg8 in mammalian cells, is also commonly
used in detecting autophagy (22).
The LC3 protein rapidly moves to an autophagic environment
(23). In the present study, the
LC3 proteins of the autophagosome were visible in the cytoplasm and
in the perinuclear space under a laser scanning confocal
microscope. These proteins appeared as dot-like structures, thus
confirming that autophagy occurs in the chondrocytes of the
intervertebral disc. Autophagy occurs in the degenerative discs of
rats (24), as well as in human
cervical vertebrae, as shown in the current study. LC3 has two
forms: LC3-I and LC3-II. The latter is located in the membrane of
the autophagosome and is considered as a marker of autophagy. As
autophagy increases, LC3-II is upregulated and LC3-I is
downregulated, thus the LC3-II/LC3-I ratio is proportional to
autophagy (25). Western blotting
results showed that compared with the control group, LC3-I in the
cervical spondylosis group was upregulated, whereas LC3-II was
downregulated and the LC3-II/LC3-I ratio decreased. The results are
consistent with a previous study (26), and imply that autophagy has a
negative correlation with the degeneration of chondrocytes.
Autophagy occurs extensively in the degradation and
recirculation of system eukaryotic cells. This mechanism is an
important physiological process that allows primary lysosomes to
process endogenous substrates, and organisms to maintain the
metabolic balance of proteins and the stability of the
intracellular environment. In the present study, it was observed
that autophagy has an important role in human cervical disc
degeneration. The regulation of autophagy may prevent disc
degeneration in cartilage end-plate cells. Information concerning
autophagy has been increasing as numerous studies are conducted.
However, few data on autophagy in chondrocytes, as well as its
effect on the growth and degeneration of such cells, are available.
In the present study, autophagy was demonstrated to be closely
associated with the degeneration of chondrocytes at the molecular
level. However, the specific role of autophagy in disc degeneration
requires further study. The regulation activity of autophagy in
chondrocytes may provide a new direction for the treatment of
spinal degenerative diseases.
Acknowledgements
This study was supported by the Chinese National
Natural Sciences Fund project (81272048), the special Fund of
Public Service Sectors of The Ministry of Health (201002018) and by
the Chinese Anhui Province Natural Science Foundation
(1308085MH152).
References
|
1
|
Roberts S and Johnson WE: Analysis of
aging and degeneration of the human intervertebral disc. Spine
(Phila Pa 1976). 24:500–501. 1999.PubMed/NCBI
|
|
2
|
Jing L-B, Zhang X-L, Xu H-Z, et al:
Protective effect of autophagy on nucleus pulposus cells under
starvation. Chin J Pathophysiol. 28:1302–1307. 2012.(In
Chinese).
|
|
3
|
Haschtmann D, Stoyanov JV, Gédet P and
Ferguson SJ: Vertebral endplate trauma induces disc cell apoptosis
and promotes organ degeneration in vitro. Eur Spine J. 17:289–299.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Reggiori F and Klionsky DJ: Autophagy in
the eukaryotic cell. Eukaryot cell. 1:11–21. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chhangani D and Mishra A: Mahogunin ring
finger-1 (MGRN1) suppresses chaperone-associated misfolded protein
aggregation and toxicity. Sci Rep. 3:19722013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liu H, Cao W, Li Y, Feng M, Wu X, Yu K and
Liao M: Subgroup J avian leukosis virus infection inhibits
autophagy in DF-1 cells. Virol J. 10:1962013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Miller G: The spine. Berquist TH: MRI of
the Musculoskeletal System. 2nd edition. Raven Press; New York, NY:
pp. 238–240. 1990
|
|
8
|
Thompson JP, Pearce RH, Schechter MT,
Adams ME, Tsang IK and Bishop PB: Preliminary evaluation of a
scheme for grading the gross morphology of the human intervertehral
disc. Spine (Phila Pa 1976). 15:411–415. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Xu HG, Peng HX, Cheng JF and Lü K:
Establishment and significance of an in vitro model of degeneration
of human cervical endplate chondrocytes. Zhonghua Yi Xue Za Zhi.
91:2912–2916. 2011.(In Chinese).
|
|
10
|
Le Maitre CL, Freemont AJ and Hoyland JA:
The role of interleukin-1 in the pathogenesis of human
intervertebral disc degeneration. Arthritis Res Ther. 7:R732–R745.
2005.
|
|
11
|
Biederbick A, Kern HF and Elsässer HP:
Monodansylcadaverine (MDC) is a specific in vivo marker for
autophagic vacuoles. Eur J cell Biol. 66:3–14. 1995.PubMed/NCBI
|
|
12
|
Shen C, Yan J, Jiang LS and Dai LY:
Autophagy in rat annulus fibrosus cells: evidence and possible
implications. Arthritis Res Ther. 13:R1322011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liang QQ, Cui XJ, Xi ZJ, et al: Prolonged
upright posture induces degenerative changes in intervertebral
discs of rat cervical spine. Spine (Phila Pa 1976). 36:E14–E19.
2011.PubMed/NCBI
|
|
14
|
Sowa G, Vadalà G, Studer R, et al:
Characterization of intervertebral disc aging: longitudinal
analysis of a rabbit model by magnetic resonance imaging,
histology, and gene expression. Spine (Phila Pa 1976).
33:1821–1828. 2008. View Article : Google Scholar
|
|
15
|
Shin BH, Lim Y, Oh HJ, et al:
Pharmacological activation of Sirt1 ameliorates
polyglutamine-induced toxicity through the regulation of autophagy.
PLoS One. 8:e649532013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Caramés B, Taniguchi N, Otsuki S, Blanco
FJ and Lotz M: Autophagy is a protective mechanism in normal
cartilage, and its aging-related loss is linked with cell death and
osteoarthritis. Arthritis Rheum. 62:791–801. 2010.PubMed/NCBI
|
|
17
|
Sun K, Xie X, Liu Y, et al: Autophagy
lessens ischemic liver injury by reducing oxidative damage. Cell
Biosci. 3:262013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhu X, Wu L, Qiao H, et al: Autophagy
stimulates apoptosis in HER2-overexpressing breast cancers treated
by lapatinib. J Cell Biochem. 114:2643–2653. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bachar-Wikstrom E, Wikstrom JD, Ariav Y,
Tirosh B, Kaiser N, Cerasi E and Leibowitz G: Stimulation of
autophagy improves endoplasmic reticulum stress-induced diabetes.
Diabetes. 62:1227–1237. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Caramés B, Hasegawa A, Taniguchi N, Miyaki
S, Blanco FJ and Lotz M: Autophagy activation by rapamycin reduces
severity of experimental osteoarthritis. Ann Rheum Dis. 71:575–581.
2012.PubMed/NCBI
|
|
21
|
Mori K: Tripartite management of unfolded
proteins in the endoplasmic reticulum. Cell. 101:451–454. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kim HS, Montana V, Jang HJ, Parpura V and
Kim JA: Epigallocatechin gallate (EGCG) stimulates autophagy in
vascular endothelial cells: A potential role for reducing lipid
accumulation. J Biol Chem. 288:22693–22705. 2013. View Article : Google Scholar
|
|
23
|
Sanjuan MA, Dillon CP, Tait SW, et al:
Toll-like receptor signalling in macrophages links the autophagy
pathway to phagocytosis. Nature. 450:1253–1257. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ye W, Xu K, Huang D, Liang A, Peng Y, Zhu
W and Li C: Age-related increases of macroautophagy and
chaperone-mediated autophagy in rat nucleus pulposus. Connect
Tissue Res. 52:472–478. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhu W-R, Ye W, Xu K, Lou Z-K, Ren J-K,
Chen J-H and Li C-H: Starvation-induced changes of LC-3 and
Beclin-1 expression in annulus fibrosus cells. Chin J Exp Sur.
28:983–985. 2011.(In Chinese).
|
|
26
|
Ye W, Chu Z, Xu K, et al: Expression and
significance of Beclin-1 and microtubule-associated protein 1 light
chain 3 in nucleus pulpous of rats during the aging process. Chin J
Exp Surg. 27:643–645. 2010.
|