Introduction
Osteosarcoma is one of the most common primary bone
malignancies and is the third most frequent cancer in
adolescentsand young adults (1).
Osteosarcoma localizes to the proximal tibia or the distal femur
and has a highly malignant tendency to invade the surrounding
normal tissues and to metastasize. Usually, the treatments for
osteosarcoma are chosen as surgical resection, chemotherapy and
radiotherapy, but the prognosis remains poor because of aggressive
invasion into local tissue and extremely-high metastatic rate.
Hence, novel therapeutic approaches, such as target therapies
against specific targets, are urgently required.
MicroRNAs (miRNAs/miRs) are a class of small
noncoding and endogenous regulatory RNA molecules that are
approximately 19–25 nucleotides in length, which can bind the
3′-untranslated region (3′-UTR) of specific genes to inhibit mRNA
translation then impair the protein process, act as
posttranscriptional regulations of gene expression (2). Micro RNAs can regulate the biological
function in cancer cells, be involved in a variety of critical
cellular processes, including cell differentiation, proliferation
and metabolism. Besides, some miRNAs has been used to classify
human cancers and markers for predicting the prognosis (3,4).
MiRNA play a significant role in tumor progression,
including tumorigenesis, invasion, metastasis and angiogenesis in
malignant tissues, such as osteosarcoma. They can regulate the cell
function through tumor suppressors and oncogenes in osteosarcoma,
such P53 (5), SPRED-1 (6), and BCL-2 (7,8). He
et al (5) found that miR-34
family as direct transcriptional targets of p53 tumor suppressor,
which regulate the DNA damage, cellular stress and improper cell
proliferation. Moreover, they found that ectopic expression of
miR-34 induces cell cycle arrest across osteosarcoma cells and cell
lines. Maire et al (6) found
that miR-126 is overexpressed in human osteosarcoma specimens.
miR-126 has been reported to repress SPRED-1, which is an inhibitor
of mitogen-activated protein kinase (MAPK) signaling, and stimulate
vascular endothelial growth factor (VEGF)-induced angiogenesis, to
enhance cell migration, proliferation and survival. Therefore, the
miRNA play a significant role in osteosarcoma development.
Recent study showed that miR-187 is significantly
downregulated in many types of human malignancies, including renal
carcinoma, pancreatic cancer, thyroid cancer, gastric cancer,
esophageal cancer, prostate cancer, breast cancer, ovarian cancer,
and lung carcinomas (9–18). The biological functions were
controversial to date. In breast cancer, miR-187 could leads to a
more aggressive, invasive cancer type and had a worse prognosis
(16). In pancreatic cancer, miR-187
can predict short overall survival (OS) in patients after radical
surgery. In colorectal cancer, the miR-187 could inhibit the tumor
growth and invasion (19). To date,
the differences of miR-187 function in different cancers has not
been clear.
In this study, we studied the miR-187 expression in
osteosarcoma tissues and cell lines, then investigated its
functions on osteosarcoma cell migration and invasion after
transfected miR-187 mimic plasmids. Furthermore, we explored that
MAPK12 was the direct target for miR-187 in osteosarcoma cells and
was involved in the functional influence of miR-195 on osteosarcoma
cells migration and invasion.
Materials and methods
Patients and specimens
The human clinical samples were collected at 304
hospitals surgical specimens. The study was approved by the ethical
board of the hospital. All samples were collected in liquid
nitrogen for a quick frozen and then stored at −80°C until RNA
extraction.
Cell culture and transfections
The human osteosarcoma cancer cell line, U2OS cells
was cultured in DMEM supplemented with 10% FBS at 37°C in 5%
CO2.
miR-187 mimicsplasmids, MAPK12 siRNA, and scramble
control were ordered from Jima company and transfected into
osteosarcoma cells with Lipofectamine® 3000 following
protocols.
Cell proliferation assay
The transfected cells were incubated with 10% CCK-8
at 37°C for 2 h then measured at 450 nm absorption for examining
the effect of cell proliferation rate. Proliferation rates were
detected at days 1, 2,3, 4 and 5 after plasmids and siRNAs
transfection. The proliferation rates then were calculated with
software.
Cell migration and invasion
assays
After transfected, osteosarcoma cells were seeded
onto a Transwell and Matrigel chamber inserting into a well of a
24-well plate in serum-free medium. 10% FBS was added to bottom
chamber. The seeded cells was counted as 1×105 each
chamber. After 24 h, migrated and invasive cells on the lower
surface of the chamber were stained with 0.1% crystal violet and
counted.
RNA extraction and real-time PCR
assays
Total RNA was extracted using TRIzol from tissues
and cells according to the manufacturer's instructions.
Quantitative PCR was performed on a Bio-Rad CFX96 Real-Time PCR
System with TaqMan probes. The PCR conditions were as follows: 95°C
for 30 sec, followed by 42 cycles of 95°C for 5 sec, and 58°C for
35 sec. MAPK12 primes forward: 5′-CCACCTTCACCTTCCACCT-3′. Reverse:
5′-GCGTCTGCTCTGATGGATG-3′.
Luciferase assay
For the dual luciferase assay, U2OS cells in a
96-wells plate were transfected with miR-195 or miRNA negative
control. The cells were then cotransfected with 0.2 mg/ml of vector
with the 3′-UTR of Smad7 gene. After 48 h, luciferase activity was
measuredwith the Dual-Luciferase reporter assay system (Promega
Corp., Madison, WI, USA). Firefly luciferase activity was then
normalized to the corresponding Renilla luciferase activity.
Luciferase assays were performed in quadruplicate and repeated in 3
independent experiments.
Western blotting
Cells were collected and lysed in ice-cold RIPA
buffer (Beyotime Biotech, Jiangsu, China) according to the
manufacturer's instructions. Protein concentration was quantified
using an Enhanced BCA protein assay kit (Beyotime Biotech). The
proteins were separated by sodium dodecyl sulfate polyacrylamide
gel electrophoresis, and transferred onto polyvinylidene difluoride
membranes (Millipore Corp., Bedford, MA, USA). After being blocked
with 5% free-fat milk for 1.5 h at room temperature, membranes were
incubated with anti-MAPK12 antibody (Cell Signaling Technology,
Inc., Danvers, MA, USA; 1:1,000 dilution overnight at 4°C, followed
by incubation with horseradish peroxidase Y conjugated mouse and
rabbit secondary antibody (Beyotime Biotech) for 2 h at room
temperature.
Statistics
Student's t-test (two tailed) was performed to
analyze the data. Values of P<0.05 were considered
significantly.
Results
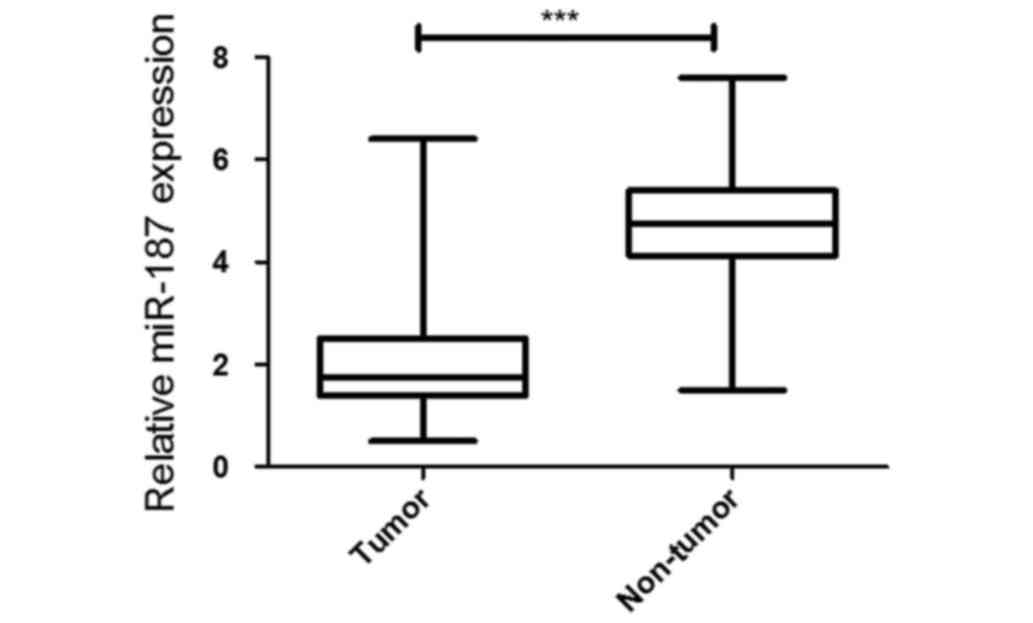
miR-187 expression is significantly
downregulated in osteosarcoma tissues and cell lines
To analyze 42 paired tumor and adjacent non-tumor
tissue samples in 304 hospital during 2011–2015, the miR-187 was
significantly decreased in the osteosarcoma by using quantitative
PCR. Meanwhile, miR-187 expression was significantly decreased in
the in cell lines compared with that of U2OS cells (Fig. 1).
MiR-187 expression was as potential
marker for diagnosis and prognosis to the osteosarcoma
The relation between miR-187 expression level and
clinical parameters was further analyzed in 42 osteosarcoma
patients. The statistical analysis showed that lower miR-187
expression level was significantly correlated with pathological
grading and distant metastasis. (Table
I). These results suggested that the miR-187 downregulation
maybe involved in the invasion and metastasis of osteosarcoma
(Table I).
 | Table I.Correlation of Mir-187 expression with
clinical pathological parameters of patients with Osteosarcoma. |
Table I.
Correlation of Mir-187 expression with
clinical pathological parameters of patients with Osteosarcoma.
|
|
| miR-187
expression |
|
|---|
|
|
|
|
|
|---|
|
| No. of patients | (Low) | (High) | P-value |
|---|
| Age (years) |
|
|
| 0.2165 |
|
>20 | 20 | 12 | 8 |
|
| ≤20 | 22 | 9 | 13 |
|
| Sex |
|
|
| 0.0213 |
| Male | 29 | 18 | 11 |
|
|
Female | 13 | 3 | 10 |
|
| Clinical stage |
|
|
| 0.0023 |
| I–II | 22 | 6 | 16 |
|
|
III–IV | 20 | 15 | 5 |
|
| Histologic grade |
|
|
| 0.0002 |
| I–II | 20 | 4 | 16 |
|
|
III–IV | 22 | 17 | 5 |
|
| Metastasis |
|
|
| 0.0042 |
| Yes | 10 | 9 | 1 |
|
| No | 32 | 12 | 20 |
|
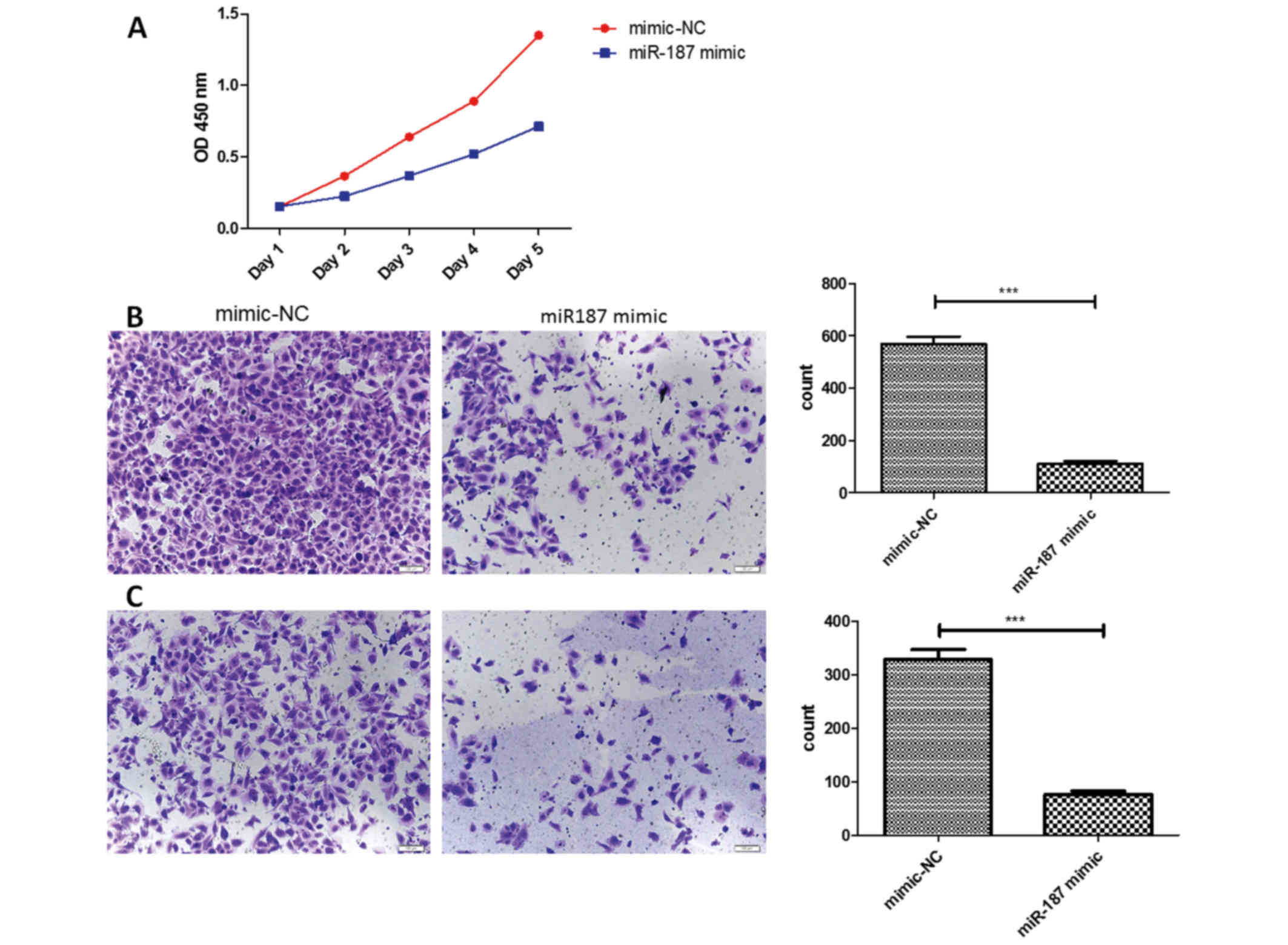
miR-187 could suppress the
osteosarcoma cells proliferation, migration and invasion
The miR-187 was found significantly decreased in the
osteosarcoma cells, then we investigated the miR-187 biological
function in osteosarcoma cells. After transfected in miR-187 mimics
plasmids, we found the difference of biological behavior with the
corresponding negative control. In CCK-8 assay, we discovered the
proliferation ability of osteosarcoma cells was affected. We found
that the proliferation rate was lower in the miR-187 mimics groups,
especially in the 4th day. Besides, the Transwell assay showed that
the miR-187 mimics groups had a less cells in the migrated sphere,
suggested the miR-187 could suppress the migration in the
osteosarcoma cells. In the Matrigel assay, the miR-187 mimics group
also showed the less cells migrated, suggested the miR-187 could
suppress the invasiveness in the osteosarcoma cells (Fig. 2).
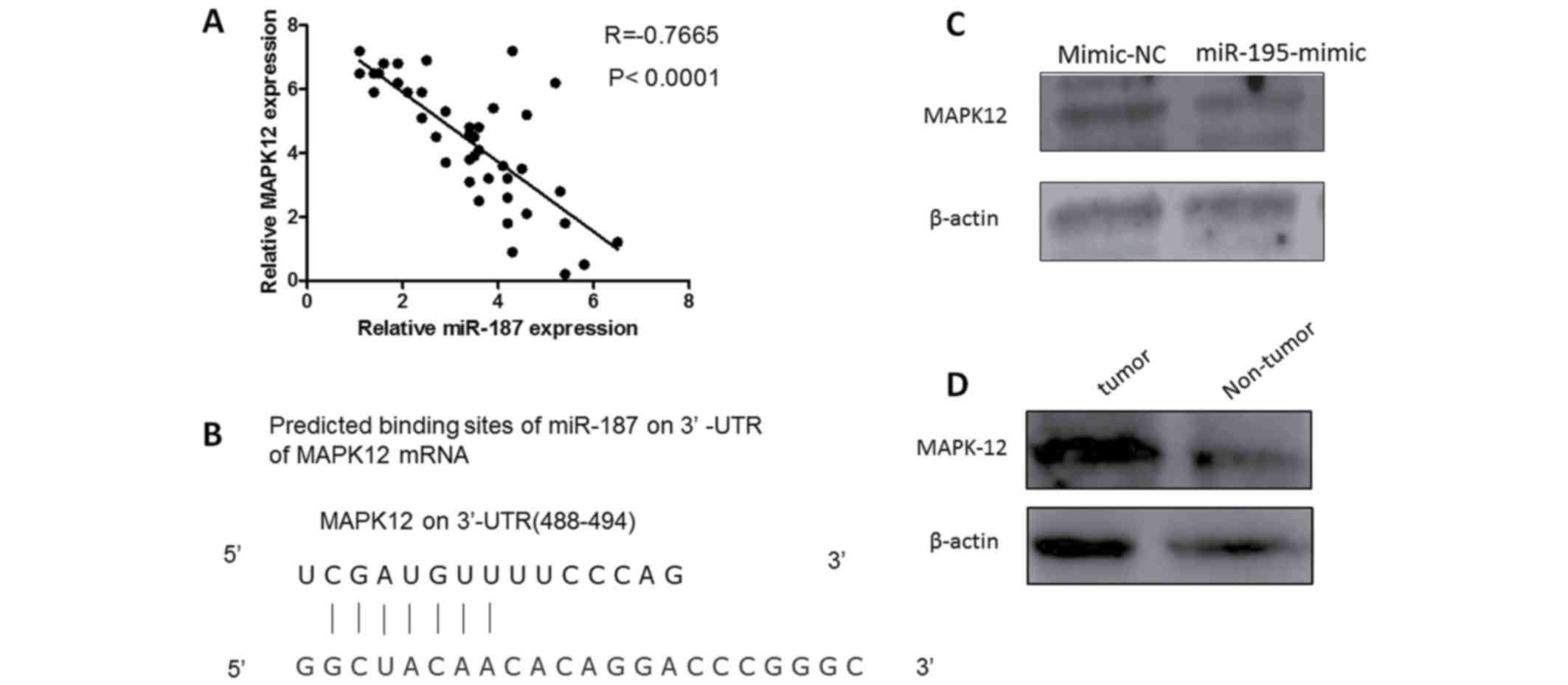
MAPK12 was the direct target of
miR-187
To discover the mechanism of MiR-87 affecting the
biological behavior in osteosarcoma cells, we constructed
luciferase reporter vector containing MAPK12 3′-UTR. Luciferase
activity assay demonstrated that miR-187 significantly inhibited
the luciferase activity of MAPK12-3′-UTR reporter comparing with
control group (Fig. 3).
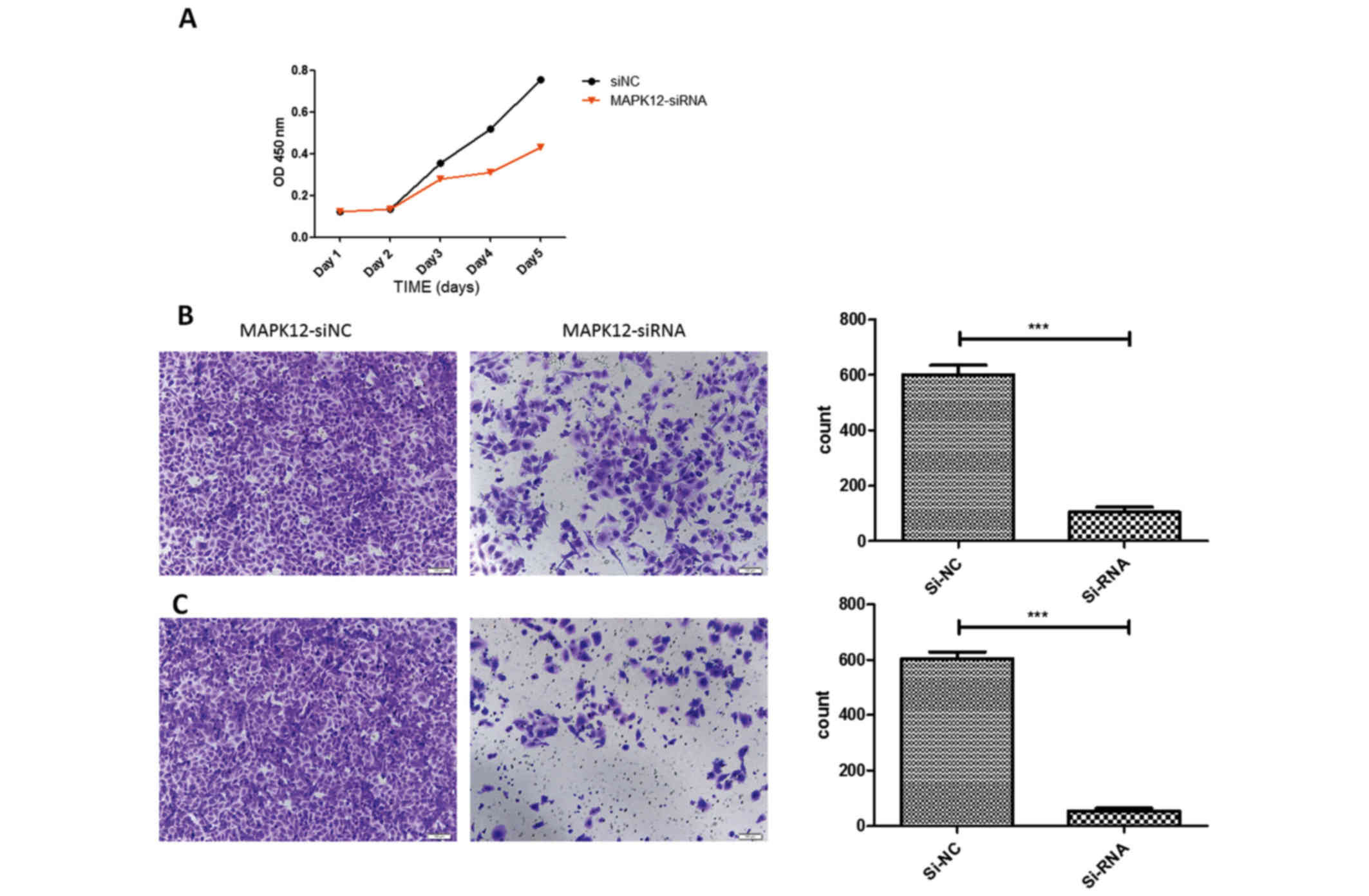
Knockdown of MAPK12 can inhibit
osteosarcoma cell proliferation, migration and invasion
To investigate the role of MAPK12 in osteosarcoma
progression, we knocked down MAPK12 expression by siRNAs in two
osteosarcoma cell lines. Knockdown of MAPK12 attenuated
osteosarcoma cell migration significantly as demonstrated by CCK-8,
Transwell and Matrigel assay, suggested MAPK12 may play an
important role in the osteosarcoma cells proliferation, migration
and invasion. The aberrant downregulation of miR-187 in
osteosarcoma may give rise to upregulation of MAPK12 and further to
promote osteosarcoma cancer progression (Fig. 4).
Discussion
In this study, we explored the miR-187 expression in
osteosarcoma tissues and cells, and results showed that the miR-187
was decreased in the both osteosarcoma tissues and cells. After
transfected the miR-187 mimics plasmids, we found that the
osteosarcoma cells proliferation, migration and invasion ability
was inhibited. The Luciferase activity assay demonstrated that
miR-187 targeted the MAPK12 for regulating the cell biological
function. Knockdown MAPK12 could inhibit the proliferation and
migration ability of osteosarcoma cells.
Several studies have investigated the association
between aberrant miR-187 expression and the risk of diverse
cancers. Emerging evidence has shown that aberrant expression of
miRNAs contributes to tumorigenesis and potentiallyservesas
biomarkers for prediction and prognosis in various cancers
including osteosarcoma. Recently, miR-187 was found downregulated
in cancer tissues, such as thyroid cancer, gastric cancer,
esophageal cancer, prostate cancer, breast cancer, ovarian cancer,
and lung carcinomas (11,14–16,19,20). In
this study, we further confirmed that miR-187 can be downregulated
in the osteosarcoma. Osteosarcoma is a severe disease that occurs
in a very young age and with a bad prognosis. Despite of
conventional therapy, many patients still suffers with a very poor
prognosis. Therefore, new therapy and early diagnosis methods
should be developed for an in-time diagnosis. In several studies,
the miR-187 expression was a marker for some malignant tumor, such
as miR-187 playing oncogenic roles in oral carcinogenesis and
Plasma miR-187 could be validated as a marker of OSCC for
diagnostic uses. In this study, we found that miR-187 expression
level was significantly correlated with pathological grading and
distant metastasis, suggested that the miR-187 may be the suitable
marker for predicting the prognosis in the future.
Moreover, we found that miR-187 could suppress the
ostrosacoma cells proliferation. The results suggested miR-187
maybe act as a tumor suppressor in ostrosacoma. Downregulation or
deletion of miR-187 may initiate the oncology activity. In this
study, we found that the proliferation was inhibited in mimic-187
group, so we redid the cell counts to make sure there being same
cell population in both control group and mimic-187 group before
planting cells in the upper chamber of Transwell and Matrigel
assays. We did this part to avoid the influence of the
proliferation. The results showed that miR-187 could suppress the
ostrosacoma cells migration and invasion. However, some studies
showed that the miR-187 also promote the migration and
proliferation in other cancer cells, suggested that the miR-187 may
affect the biological behavior in specific cancers.
To date, some studies showed that CD276, BCL6 was
the target of miR-187 (19). In this
study, we also found that the MAPK12 was a target of miR-187.
MAPK12 has been shown to regulate cancer cell metastases in some
types of cancer. MAPK11 and MAPK12 are two of the four p38 MAPKs
that play important roles in the cellular responses evoked by
extracellular stimuli, and MAPK12 plays a role in repressing cell
proliferation through the downregulation of cyclin D1 in response
to hypoxia in adrenal cells. In this study, downregulating the
MAPK-12 could suppress the proliferation, migration and
invasiveness in ostrosacoma cells, which verified that MAPK12
promotes the biological behavior in the ostrosacoma cells. And we
also confirmed that MAPK12 was a direct target of miR-187, which
expanded the potential target network of miRNA-187. Although we
identified miR-195 maybe act as a direct regulator of MAPK12
expression, we could not conclude that MAPK12 was only regulated by
miR-187, there was the possibility of modification of MAPK12
protein levels through miRNAs other than miR-187, which may be
found in future studies. Besides, as discussed above, miR-187 also
regulate the cell cycle to regulate cancer cell growth. These data
reflect the complexity of the regulation of tumorigenesis by a
network of molecules in cancers with micro RNAs.
In conclusion, we found that miR-187 was decreased
in the ostrosacoma tissues and cells, which had clinical
relationships with advantage stages and metastasis. The miR-187
suppressed the proliferation, migration and invasion by targeting
the MAPK12.
References
|
1
|
He JP, Hao Y, Wang XL, Yang XJ, Shao JF,
Guo FJ and Feng JX: Review of the molecular pathogenesis of
osteosarcoma. Asian Pac J Cancer Prev. 15:5967–5976. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Flynt AS and Lai EC: Biological principles
of microRNA-mediated regulation: Shared themes amid diversity. Nat
Rev Genet. 9:831–842. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Friedman RC, Farh KK, Burge CB and Bartel
DP: Most mammalian mRNAs are conserved targets of microRNAs. Genome
Res. 19:92–105. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Griffiths-Jones S, Saini HK, van Dongen S
and Enright AJ: miRBase: Tools for microRNA genomics. Nucleic Acids
Res. 36(Database issue): D154–D158. 2008.PubMed/NCBI
|
|
5
|
He L, He X, Lim LP, de Stanchina E, Xuan
Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, et al: A microRNA
component of the p53 tumour suppressor network. Nature.
447:1130–1134. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Maire G, Martin JW, Yoshimoto M,
Chilton-MacNeill S, Zielenska M and Squire JA: Analysis of
miRNA-gene expression-genomic profiles reveals complex mechanisms
of microRNA deregulation in osteosarcoma. Cancer Genet.
204:138–146. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Aqeilan RI, Calin GA and Croce CM: miR-15a
and miR-16-1 in cancer: Discovery, function and future
perspectives. Cell Death Differ. 17:215–220. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Xia L, Zhang D, Du R, Pan Y, Zhao L, Sun
S, Hong L, Liu J and Fan D: miR-15b and miR-16 modulate multidrug
resistance by targeting BCL2 in human gastric cancer cells. Int J
Cancer. 123:372–379. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gu L, Li H, Chen L, Ma X, Gao Y, Li X,
Zhang Y, Fan Y and Zhang X: MicroRNAs as prognostic molecular
signatures in renal cell carcinoma: A systematic review and
meta-analysis. Oncotarget. 6:32545–32560. 2015.PubMed/NCBI
|
|
10
|
Bloomston M, Frankel WL, Petrocca F,
Volinia S, Alder H, Hagan JP, Liu CG, Bhatt D, Taccioli C and Croce
CM: MicroRNA expression patterns to differentiate pancreatic
adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA.
297:1901–1908. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Nikiforova MN, Tseng GC, Steward D, Diorio
D and Nikiforov YE: MicroRNA expression profiling of thyroid
tumors: Biological significance and diagnostic utility. J Clin
Endocrinol Metab. 93:1600–1608. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lo SS, Hung PS, Chen JH, Tu HF, Fang WL,
Chen CY, Chen WT, Gong NR and Wu CW: Overexpression of miR-370 and
downregulation of its novel target TGFβ-RII contribute to the
progression of gastric carcinoma. Oncogene. 31:226–237. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wijnhoven BP, Hussey DJ, Watson DI, Tsykin
A, Smith CM and Michael MZ; South Australian Oesophageal Research
Group, : MicroRNA profiling of Barrett's oesophagus and oesophageal
adenocarcinoma. Br J Surg. 97:853–861. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Casanova-Salas I, Masiá E, Armiñán A,
Calatrava A, Mancarella C, Rubio-Briones J, Scotlandi K, Vicent MJ
and López-Guerrero JA: MiR-187 targets the androgen-regulated gene
ALDH1A3 in prostate cancer. PLoS One. 10:e01255762015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Blenkiron C, Goldstein LD, Thorne NP,
Spiteri I, Chin SF, Dunning MJ, Barbosa-Morais NL, Teschendorff AE,
Green AR, Ellis IO, et al: MicroRNA expression profiling of human
breast cancer identifies new markers of tumor subtype. Genome Biol.
8:R2142007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mulrane L, Madden SF, Brennan DJ, Gremel
G, McGee SF, McNally S, Martin F, Crown JP, Jirström K, Higgins DG,
et al: miR-187 is an independent prognostic factor in breast cancer
and confers increased invasive potential in vitro. Clin Cancer Res.
18:6702–6713. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chao A, Lin CY, Lee YS, Tsai CL, Wei PC,
Hsueh S, Wu TI, Tsai CN, Wang CJ, Chao AS, et al: Regulation of
ovarian cancer progression by microRNA-187 through targeting
Disabled homolog-2. Oncogene. 31:764–775. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li X, Shi Y, Yin Z, Xue X and Zhou B: An
eight-miRNA signature as a potential biomarker for predicting
survival in lung adenocarcinoma. J Transl Med. 12:1592014.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang ZS, Zhong M, Bian YH, Mu YF, Qin SL,
Yu MH and Qin J: MicroRNA-187 inhibits tumor growth and invasion by
directly targeting CD276 in colorectal cancer. Oncotarget.
7:44266–44276. 2016.PubMed/NCBI
|
|
20
|
Schultz NA, Andersen KK, Roslind A,
Willenbrock H, Wøjdemann M and Johansen JS: Prognostic microRNAs in
cancer tissue from patients operated for pancreatic cancer-five
microRNAs in a prognostic index. World J Surg. 36:2699–2707. 2012.
View Article : Google Scholar : PubMed/NCBI
|