Introduction
Endometrial carcinoma (EC) is the most common
gynecologic malignancy and is associated with a poor prognosis when
diagnosed at an advanced stage (1).
Endometrial cancer is traditionally classified into type I and type
II subtypes (2). Type I cancers
account for 80–85% of EC cases, are of endometrioid histology, more
often well differentiated and associate with favorable prognosis
(2). In contrast the type II cancers
are non-endometrioid carcinomas, poorly differentiated and
associate with poorer survival (2).
However, patients with deep myometrial invasion, poor
differentiation, serous or clear cell histology or extension of
disease to other organs or lymph nodes within the pelvic region are
at higher risk for disease recurrence (3,4).
Therefore, it is imperative to find new therapeutic targets to
elaborate the molecular mechanisms underlying progression of
endometrial carcinogenesis.
L1 cell adhesion molecule (L1CAM, CD171) is a
200–220-kDa transmembrane glycoprotein composed of 6
immunoglobulin-like domains, 5 fibronectin-type III domains, a
transmembrane stretch, and a short cytoplasmic tail (5). L1CAM was originally identified as a
neural cell adhesion molecule in the central nervous system that
plays an important role in initiating cerebellar cell migration and
neurite outgrowth (6). L1CAM
expression is also found in other cell types such as lymphoid and
myelomonocytic cells, kidney tubule epithelial cells, and
intestinal crypt cells (7–10). In addition, L1CAM expression has been
identified in a variety of tumor types and correlates with poor
prognosis and metastasis (11).
L1CAM functions mostly in proliferation, migration, invasion, and
survival through L1CAM homophilic interaction or heterophilic
interactions with other cell adhesion molecules, integrins, or
growth factor receptor, while the cellular properties are not
homogeneous among different types of cancers (12). Recently, it has been reported that
L1CAM was involved in progression of endometrial cancer (13).
Cyclophilin A (CypA) is a highly abundant protein,
accounting for up to ~0.6% of the total cytosolic protein content
(14). CypA is involved in a growing
number of biological processes, including protein folding, signal
transduction, viral infection, trafficking, receptor assembly,
immune response, and transcription regulation (15). Although several proteins have been
identified to interact with CypA (16–19), the
underlying mechanism of the CypA action and the physiological
implications of the interactions remain in most cases unknown. CypA
exhibits peptidyl-prolyl cis-trans isomerase (PPIase)
activity by catalyzing cis-trans isomerization of peptide
bonds preceding proline residues (20). CypA can in principle act as an enzyme
or a binding partner (21) in
mediating the biological processes.
In this study, we showed that L1CAM promotes EMT
with increased characteristics of CICs and paclitaxel resistance in
human endometrial cancer.
Materials and methods
HEC-1A cells line
HEC-1A cells were obtained from Peking Union Medical
College (Beijing, China). Briefly, cells were maintained in
Dulbecco's modified Eagle's medium (DMEM; Sigma, Shanghai, China)
containing 10% fetal bovine serum (FBS; Shanghai ExCell Biology,
China) and 100 mg/ml penicillin and streptomycin (Gibco, Shanghai,
China) at 37°C in a humidified atmosphere with 5%
CO2.
L1CAM expressing plasmids/empty
vectors and transfection experiments
L1CAM expressing plasmids and empty vectors
(pcDNA3.1) were obtained from Tiangen (Beijing, China).
Transfections were performed with Lipofctamine 2000 transfection
reagent (Invitrogen, Carlsbad, USA) following the manufacturers'
protocols.
Western blot analysis
It was performed as described previously (22,23).
Total protein was prepared using extraction buffer comprising
NaCl/Pi containing 0.5% Triton X-100, 1 mM EDTA, 1 mM
phenylmethyl sulfonyl fluoride, and complete protease inhibitors
(Roche). The concentration of each protein lysate was determined
using a BCA™ protein assay kit (Thermo Scientific, Rockford, IL,
USA). Equal amounts of total protein were subjected to 12%
SDS/PAGE. Then samples were transferred to nitrocellulose membranes
and blocked for 60 min at room temperature in 5% skim milk powder
(w/v) in NaCl/Pi. The membranes were immunoblotted using
primary anti-body anti-L1CAM (1:500; Abcam, Cambridge, MA, USA),
anti-E-cadherin (1:500; Abcam, Cambridge, MA, USA), anti-Vimentin
(1:500; Abcam, Cambridge, MA, USA), anti-Musashi-1 (1:500; Abcam,
Cambridge, MA, USA), anti-CD133 (1:500; Abcam, Cambridge, MA, USA),
anti-Cyclophilin A (1:500; Abcam, Cambridge, MA, USA) and
anti-β-actin (1:500; Abcam, Cambridge, MA, USA) overnight at 4°C,
anti-rabbit secondary antibodies (1:10,000; Abcam, Cambridge, MA,
USA) were used for 30 min at room temperature. The specific
proteins were visualized by Odyssey™ Infrared Imaging
System (Gene Company, Lincoln, NE, USA). β-actin expression was
used as an internal control to show equal loading of the protein
samples.
Immunofluorescence staining
It was performed as described previously (24,25).
Cells were plated on glass coverslips in six-well plates and
transfected as indicated. At 48 h after transfection, the cells
were fixed in 4% paraformaldehyde for 15 min, and then blocked with
goat serum blocking solution for 20 min at room temperature.
Coverslips were stained with the mentioned antibody mentioned
anti-L1CAM antibodies (1:500; Abcam, Cambridge, MA, USA), After
washing three times with NaCl/Pi, cells were incubated with
appropriate secondary antibodies (Abcam, Cambridge, MA, USA) for 30
min at 37°C. 4′6-diamidino-2-phenylindole (DAPI) staining (blue)
was used to indicate nuclei. Microscopic analysis was performed
with a confocal laser-scanning microscope (Leica Microsystems,
Bensheim, Germany). Fluorescence intensities were calculated from a
few viewing areas for 300 cells per coverslip and analyzed by
ImageJ 1.37v software (http://rsb.info.nih.gov/ij/index.html).
Quantitative real-time RT-PCR
(qRT-PCR)
Quantitative real-time RT-PCR were described before
(21). The specific primer sets for
PCR were as follows: GAPDH, forward primer:
5′-GAAGGTGAAGGTCGGAGTCA-3′, and reverse primer
5′-GAAGATGGTGATGGGATTTC-3′; E-Cadherin, forward primer
5′-TCAACGATCCTGACCAGCAGTTCG-3′ and reverse primer
5′-GGTGAACCATCATCTGTGGCGATG-3′; N-cadherin, forward primer
5′-CATCCCTCCAATCAACTTGC-3′ and reverse primer
5′-ATGTGCCCTCAAATGAAACC-3′; Vimentin, forward primer
5′-GACAATGCGTCTCTGGCACGTCTT-3′ and reverse primer
5′-TCCTCCGCCTCCTGCAGGTTCTT-3′; ZEB1, forward primer
5′-TTAGTTGCTCCCTGTGCAGTT-3′ and reverse primer
5′-TAGGAGCCAGAATGGGAAAAG-3′. GAPDH was a loading control.
MTT assay
The proliferation of cells was assessed by the
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium (MTT) assay
(Sigma, St Louis, MO, USA). The MTT analysis was performed as
described previously (26–29). In brief, the cells were plated in
96-well plates in Dulbecco's modified Eagle's medium containing 10%
fetal bovine serum at a density of 8×103 cells per well
at 37°C in a 5% CO2 incubator for 12 h. Cells were
transfected with L1CAM expressing plasmid or empty vectors for 24 h
and then were treated with different doses of paclitaxel
(10−4-102). After 24 h, MTT (5
mg·ml−1) was added to the wells (20 µl per well). The
plates were incubated in a cell incubator for 4 h, then the
supernatant was removed and 150 µl of dimethyl sulfoxide was added
to each well. After incubation for 10 min, the absorbance of each
well was measured using a Synergy™ 4 (BioTek Instruments, Winooski,
VT, USA) with a wavelength of 570 nm, with the reference wavelength
set at 630 nm. Absorbance was directly proportional to the number
of survival cells.
Sphere formation assay
It was performed as described previously (30). Cells (103/ml) in
serum-free RPMI1640/1 mM Na-pyruvate were seeded on 0.5% agar
precoated 6-well plates. After 1 week, half the medium was
exchanged every third day. Single spheres were picked and
counted.
Anoikis assays
It was performed as described previously (31). Anoikis resistance was evaluated by
seeding 7.5×104 cells in ultralow attachment plates
(Corning). After 24 h of anchorage-independent culture, cells were
transfected as indicated and resuspended in 0.4% trypan blue
(Sigma, St. Louis, MO, USA) and cell viability was assessed.
miRNA microarray
It was performed as described previously (32). Total RNA from cultured cells, with
efficient recovery of small RNAs, was isolated using the mirVana
miRNA Isolation Kit (Ambion, Austin, TX, USA). cRNA for each sample
was synthesized by using 3′ IVT EXPRESS KIT (Affymetrix, Santa
Clara, CA, USA) according to the manufacturer's protocols. The
purified cRNA was fragmented by incubation in fragmentation buffer
(provided in the 3′IVT express kit) at 95°C for 35 min and chilled
on ice. The fragmented labeled cRNA was applied to MicroRNA2.0
Array (Affymetrix, Santa Clara, CA, USA) and hybridized in Genechip
hybridization oven 640 (Affymetrix, Santa Clara, CA, USA) at 45°C
for 18 h. After washing and staining in Genechip fluidics station
450 (Affymetrix, Santa Clara, CA, USA), the arrays were scanned by
using Genechip scanner 3000 (Affymetrix, Santa Clara, CA, USA). The
gene expressions levels of samples were normalized and compared by
using Partek GS 6.5 (Partek, Inc, St. Louis, MO, USA).
Average-linkage hierarchical clustering of the data was applied by
using the Cluster [Eisen et al (33), Stanford, Stanford University, CA,
USA; http://rana.lbl.gov] and the results were
displayed by using TreeView [Eisen et al (33), Stanford, Stanford University, CA,
USA; http://rana.lbl.gov].
Statistical analysis
Results were analyzed using SAS software (9.4). Data
were presented as mean ± standard error of the mean (SEM) of
separate experiments (n=3). P-values less than 0.05 were considered
to be significant.
Results
L1CAM promotes EMT in endometrial
cancer HEC-1A cells
To investigate whether L1CAM can affect epithelial
or mesenchymal status of HEC-1A cells, we performed western blot to
test whether L1CAM expressing plasmids could express L1CAM protein
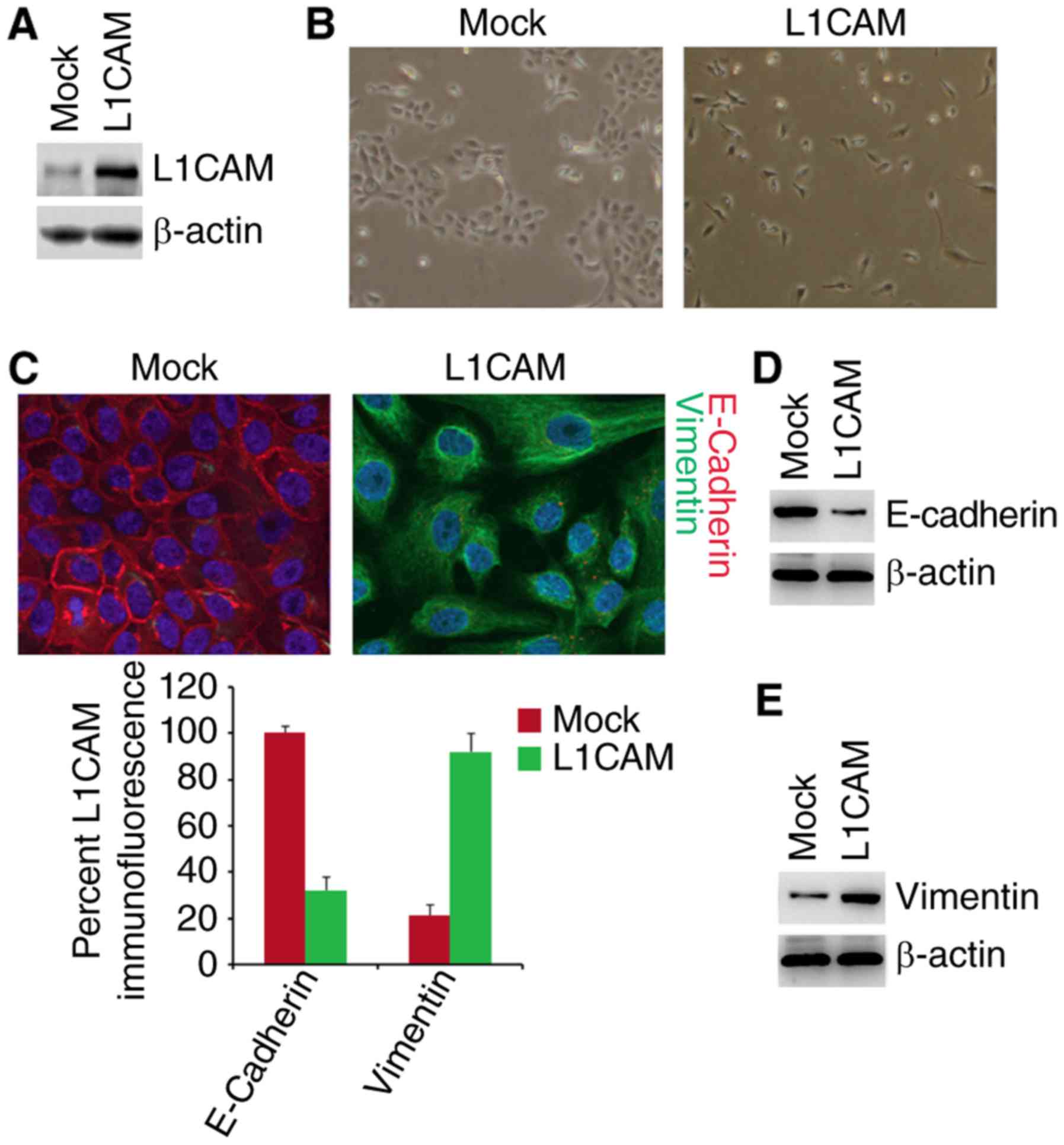
in HEC-1A cells. The results of western blot showed that L1CAM
expressing plasmids can significantly up-regulate L1CAM protein
expression in the cells (Fig. 1A).
To determine whether L1CAM can promote EMT, we transfected HEC-1A
cells with L1CAM expressing plasmids and then observed that its
overexpression promoted evident changes in the cells morphology
(EMT, epithelial to mesenchymal transiton) (Fig. 1B). To confirm that the changes of
morphology are induced by EMT, we performed immunoflurescence
analysis to detect epithelial and mesenchymal markers of HEC-1A
cells transfected with L1CAM expressing plasmids and empty vectors.
We found that that the E-Cadherin protein (epithelial marker) was
inhibited and Vimentin protein (mesenchymal marker) were induced by
L1CAM in HEC-1A cells (Fig. 1C). To
further analyze whether L1CAM could affect E-Cadherin and Vimentin
protein, we used western blotting to detect their expression in the
cells transfected with L1CAM expressing plasmids and empty vectors.
The results demonstrated that E-Cadherin was downregulated and
Vimentin was upregulated by L1CAM (Fig.
1D and E). We also performed real-time PCR to detect epithelial
and mesenchymal markers. As anticipated, we found that epithelial
marker (E-cadherin) was downregulated and mesenchymal markers (such
as N-Cadherin, Vimentin, and ZEB1) was upregulated by L1CAM in
HEC-1A cells (Fig. 1F).
L1CAM promotes formation of CICs in
HEC-1A cells
EMT can contribute to increased formation of CICs in
cancer cells (34–37). To determine whether L1CAM could
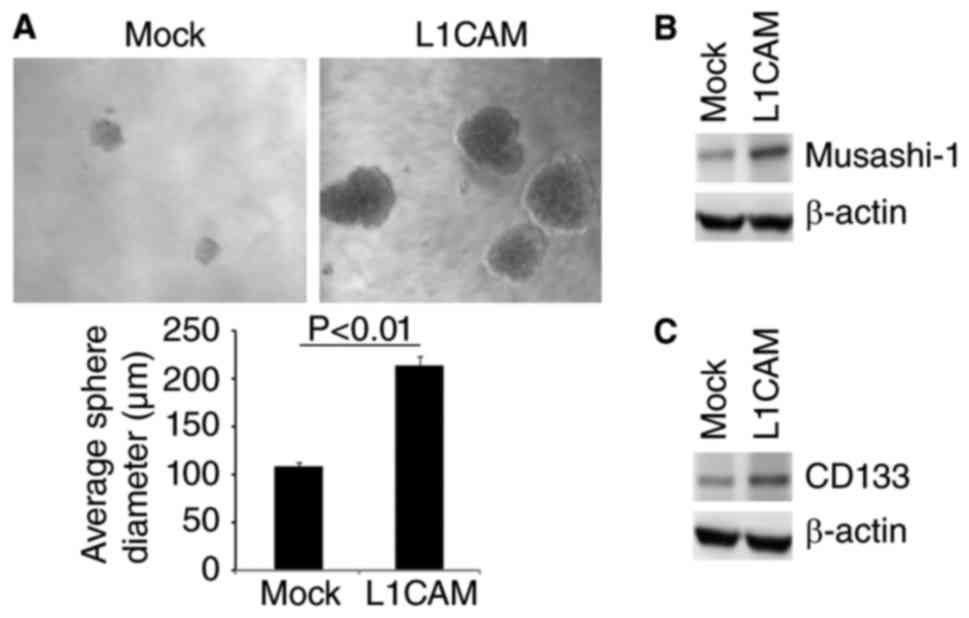
affect characteristics of CICs, we performed sphere forming assay
to evaluate the formation of CICs in HEC-1A cells. The results of
sphere forming assay showed that formation of spheres were
increased by L1CAM in HEC-1A cells (Fig.
2A). Moreover, we performed western blot to detect whether
L1CAM could regulate CICs markers-Musashi-1 and CD133 expression in
the cells. We found that Musashi-1 and CD133 protein were evidently
upregulated by L1CAM in HEC-1A cells (Fig. 2B and C).
L1CAM promotes paclitaxel resistance
in human endometrial cancer HEC-1A cells
To further identify whether L1CAM can affect
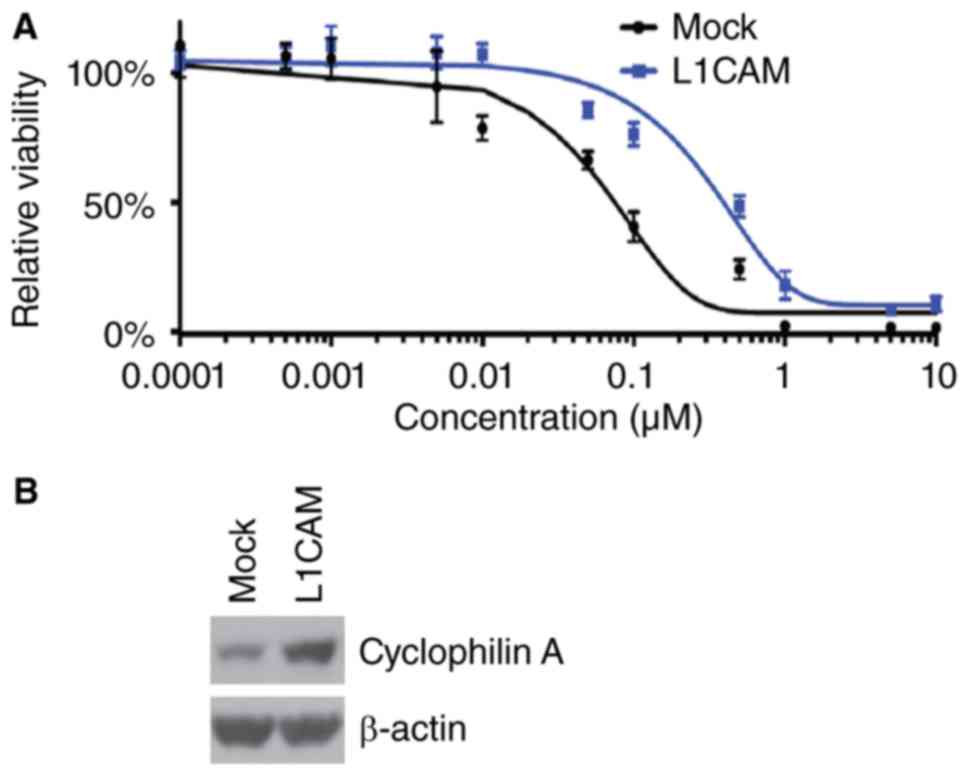
paclitaxel efficacy in HEC-1A cells, we performed MTT assay in
HEC-1A cells treated as indicated (Fig.
3A). The results showed that overexpressing L1CAM could promote
paclitaxel resistance (Fig. 3A). In
addition, we performed western blot to analyze cyclophilin A
protein expression in L1CAM expressing plasmids and empty vectors
transfected HEC-1A cells. We found that cyclophilin A protein can
be increased by L1CAM (Fig. 3B).
L1CAM regulates paclitaxel
resistance-associated microRNAs expression in HEC-1A cells
Oncogenes can promote endometrial cancer progression
by regulating microRNA expression and microRNA involved in
endometrial cancer pathogenesis can function as oncogene or tumor
suppressor gene (38–40). Thus, we reasoned that L1CAM could
function as an oncogene by regulating miRNAs expression. We
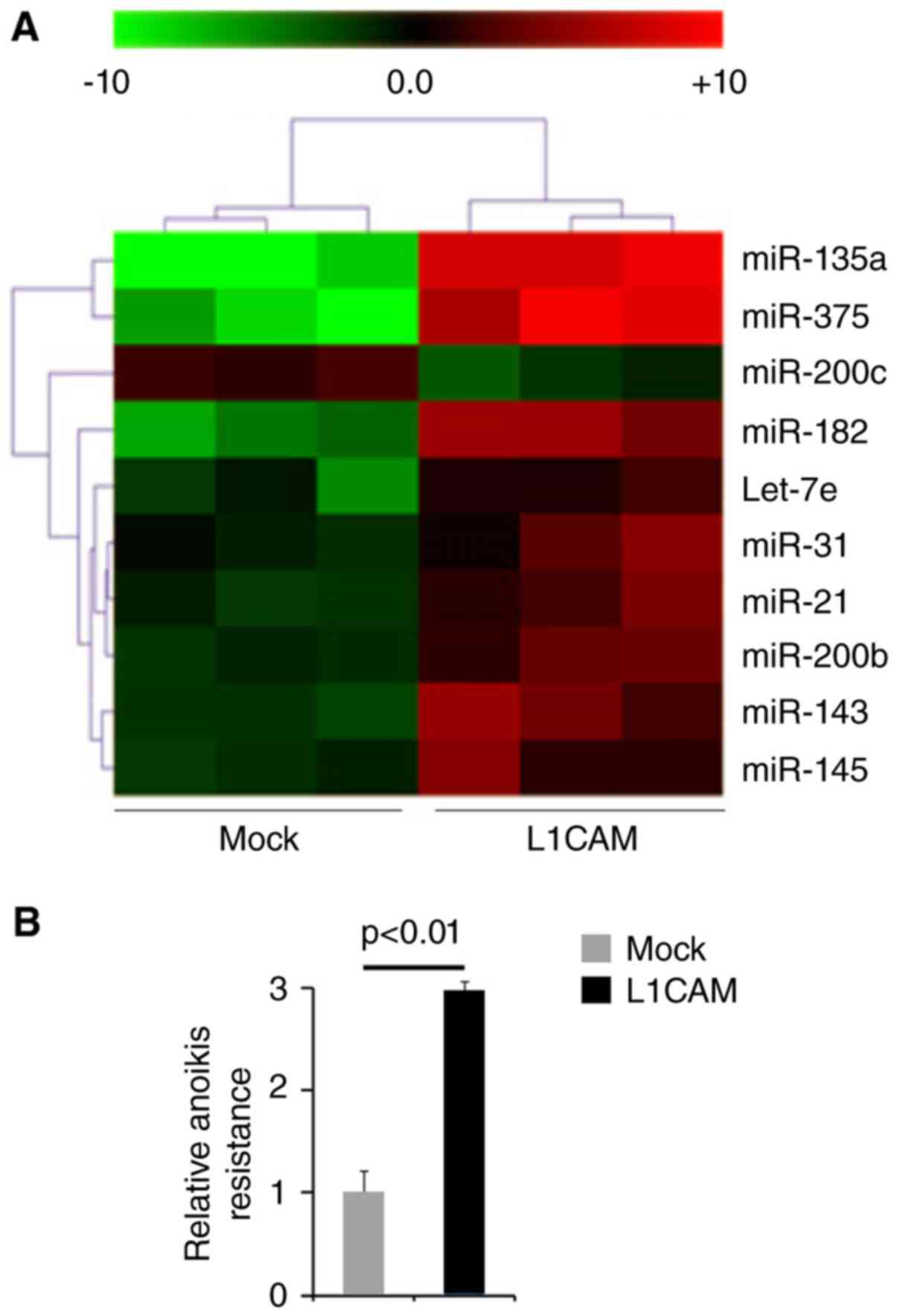
performed microarrays to detect miRNA expression. RNAs isolated
from L1CAM expressing plasmids or empty vectors transfected HEC-1A
cells were hybridized to a custom miRNA microarray platform. We
found that miR-135a, miR-375, miR-200c, miR-182, let-7e, miR-31,
miR-21, miR-200b, miR-143 and miR-145 were changed more than 10
folds in the cells (Fig. 4A).
L1CAM promotes anoikis resistance in
human endometrial cancer HEC-1A cells
To study the roles of L1CAM on metastasis, we used
anoikis assays to detect its role regulating anoikis resistance.
Cells transfected with L1CAM expressing plasmids showed about 200%
increased resistance to anoikis-mediated cell death (Fig. 4B).
Discussion
The expression of L1CAM is a strong predictor of
poor outcome in endometrial cancer (41). EMT plays an important role in
invasion and metastasis of endometrial cancer and enables cancer
cells to obtain malignant characters and traits of CICs (42). Critical molecular features of this
process are the deregulation of E-cadherin and vimentin expression
(43). Consistent with previous
report that L1CAM was inversely associated with E-cadherin
expression (44), we found that
overexpressing L1CAM induced EMT and inhibited E-cadherin
expression in endometrial cancer HEC-1A cells.
CICs have been proposed as the major power of EMT
and responsible for poor survival (45). Musashi-1 and CD133 have been proposed
as markers of CICs for endometrial cancer (46). In line with previous report that
L1CAM is required for maintaining CICs and targeting L1CAM may
represent a novel therapeutic strategy (47), we showed that its overexpression
evidently promoted formation of CICs traits and upregulated
Musashi-1 and CD133 protein. All the results indicated that L1CAM
might be a therapeutic target for eradicating CICs in endometrial
cancer.
Chemotherapy is a common therapeutic strategy for
cancer, but it fails to eradicate cancer cells, because of primary
resistance or acquired drug resistance. Elucidating the mechanisms
of drug resistance for cancer will yield vital information about
how to improve cancer chemotherapy and circumvent the resistance.
In line with previous report that L1CAM can confer chemoresistance
in malignant tumor (48,49), we showed that over-expressing L1CAM
could promote paclitaxel resistance in endometrial cancer HEC-1A
cells. Cyclophilin A expression was increased in
paclitaxel-resistant endometrial cancer cells, as well as silencing
Cyclophilin A reversed paclitaxel resistance (50). We showed that Cyclophilin A was
upregulated by L1CAM. microRNAs have recently been identified as
key genes implicated in mechanisms of chemoresistance. Upregulation
of miR-135a and miR-375 can contribute to paclitaxel resistance.
Up-regulating miR-200c in ovarian cancer reduced tumor burden and
improved paclitaxel sensitivity. We showed that L1CAM significantly
upregulatedmiR-135a and miR-375 expression and downregulated
miR-200c expression. The results indicated that L1CAM may induce
paclitaxel resistance by regulating microRNAs.
Glossary
Abbreviations
Abbreviations:
|
CICs
|
cancer initiating cells
|
|
EMT
|
epithelial to mesenchymal
transition
|
|
EC
|
endometrial cancer
|
References
|
1
|
Koval OA, Sakaeva GR, Fomin AS, Nushtaeva
AA, Semenov DV, Kuligina EV, Gulyaeva LF, Gerasimov AV and Richter
VA: Sensitivity of endometrial cancer cells from primary human
tumor samples to new potential anticancer peptide lactaptin. J
Cancer Res Ther. 11:345–351. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Di Cristofano A and Ellenson LH:
Endometrial carcinoma. Annu Rev Pathol. 2:57–85. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kadar N, Homesley HD and Malfetano JH:
Prognostic factors in surgical stage III and IV carcinoma of the
endometrium. Obstet Gynecol. 84:983–986. 1994.PubMed/NCBI
|
|
4
|
Morrow CP, Bundy BN, Kurman RJ, Creasman
WT, Heller P, Homesley HD and Graham JE: Relationship between
surgical-pathological risk factors and outcome in clinical stage I
and II carcinoma of the endometrium: A gynecologic oncology group
study. Gynecol Oncol. 40:55–65. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Haspel J and Grumet M: The L1CAM
extracellular region: A multi-domain protein with modular and
cooperative binding modes. Front Biosci. 8:s1210–s1225. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Rathjen FG and Schachner M:
Immunocytological and biochemical characterization of a new
neuronal cell surface component (L1 antigen) which is involved in
cell adhesion. EMBO J. 3:1–10. 1984.PubMed/NCBI
|
|
7
|
Pancook JD, Reisfeld RA, Varki N, Vitiello
A, Fox RI and Montgomery AM: Expression and regulation of the
neural cell adhesion molecule L1 on human cells of myelomonocytic
and lymphoid origin. J Immunol. 158:4413–4421. 1997.PubMed/NCBI
|
|
8
|
Debiec H, Christensen EI and Ronco PM: The
cell adhesion molecule L1 is developmentally regulated in the renal
epithelium and is involved in kidney branching morphogenesis. J
Cell Biol. 143:2067–2079. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Thor G, Probstmeier R and Schachner M:
Characterization of the cell adhesion molecules L1, N-CAM and J1 in
the mouse intestine. EMBO J. 6:2581–2586. 1987.PubMed/NCBI
|
|
10
|
Ebeling O, Duczmal A, Aigner S, Geiger C,
Schöllhammer S, Kemshead JT, Möller P, Schwartz-Albiez R and
Altevogt P: L1 adhesion molecule on human lymphocytes and
monocytes: Expression and involvement in binding to alpha v beta 3
integrin. Eur J Immunol. 26:2508–2516. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Altevogt P, Doberstein K and Fogel M:
L1CAM in human cancer. Int J Cancer. 138:1565–1576. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Colombo F and Meldolesi J: L1-CAM and
N-CAM: From adhesion proteins to pharmacological targets. Trends
Pharmacol Sci. 36:769–781. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Notaro S, Reimer D, Duggan-Peer M, Fiegl
H, Wiedermair A, Rössler J, Altevogt P, Marth C and Zeimet AG:
Evaluating L1CAM expression in human endometrial cancer using
qRT-PCR. Oncotarget. 7:40221–40232. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ryffel B, Woerly G, Greiner B, Haendler B,
Mihatsch MJ and Foxwell BM: Distribution of the cyclosporine
binding protein cyclophilin in human tissues. Immunology.
72:399–404. 1991.PubMed/NCBI
|
|
15
|
Nigro P, Pompilio G and Capogrossi MC:
Cyclophilin A: A key player for human disease. Cell Death Dis.
4:e8882013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bonfils C, Bec N, Larroque C, Del Rio M,
Gongora C, Pugnière M and Martineau P: Cyclophilin A as negative
regulator of apoptosis by sequestering cytochrome c. Biochem
Biophys Res Commun. 393:325–330. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bosco DA, Eisenmesser EZ, Pochapsky S,
Sundquist WI and Kern D: Catalysis of cis/trans isomerization in
native HIV-1 capsid by human cyclophilin A. Proc Natl Acad Sci USA.
99:pp. 5247–5252. 2002; View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Brazin KN, Mallis RJ, Fulton DB and
Andreotti AH: Regulation of the tyrosine kinase Itk by the
peptidyl-prolyl isomerase cyclophilin A. Proc Natl Acad Sci USA.
99:pp. 1899–1904. 2002; View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Howard BR, Vajdos FF, Li S, Sundquist WI
and Hill CP: Structural insights into the catalytic mechanism of
cyclophilin A. Nat Struct Biol. 10:475–481. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Göthel SF and Marahiel MA: Peptidyl-prolyl
cis-trans isomerases, a superfamily of ubiquitous folding
catalysts. Cell Mol Life Sci. 55:423–436. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Fischer G, Tradler T and Zarnt T: The mode
of action of peptidyl prolyl cis/trans isomerases in vivo: Binding
vs. FEBS Lett. 426:17–20. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wu L, Wang Q, Yao J, Jiang H, Xiao C and
Wu F: MicroRNA let-7g and let-7i inhibit hepatoma cell growth
concurrently via downregulation of the anti-apoptotic protein
B-cell lymphoma-extra large. Oncol Lett. 9:213–218. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xiang Y, Lu DL, Li JP, Yu CX, Zheng DL,
Huang X, Wang ZY, Hu P, Liao XH and Zhang TC: Myocardin inhibits
estrogen receptor alpha-mediated proliferation of human breast
cancer MCF-7 cells via regulating MicroRNA expression. IUBMB Life.
68:477–487. 2016. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ren ZG, Dong SX, Han P and Qi J: miR-203
promotes proliferation, migration and invasion by degrading SIK1 in
pancreatic cancer. Oncol Rep. 35:1365–1374. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liao XH, Lu DL, Wang N, Liu LY, Wang Y, Li
YQ, Yan TB, Sun XG, Hu P and Zhang TC: Estrogen receptor α mediates
proliferation of breast cancer MCF-7 cells via a
p21/PCNA/E2F1-dependent pathway. FEBS J. 281:927–942. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liao XH, Li YQ, Wang N, Zheng L, Xing WJ,
Zhao DW, Yan TB, Wang Y, Liu LY, Sun XG, et al: Re-expression and
epigenetic modification of maspin induced apoptosis in MCF-7 cells
mediated by myocardin. Cell Signal. 26:1335–1346. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Liao XH, Wang Y, Wang N, Yan TB, Xing WJ,
Zheng L, Zhao DW, Li YQ, Liu LY, Sun XG, et al: Human chorionic
gonadotropin decreases human breast cancer cell proliferation and
promotes differentiation. IUBMB Life. 66:352–360. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Liao XH, Xiang Y, Yu CX, Li JP, Li H, Nie
Q, Hu P, Zhou J and Zhang TC: STAT3 is required for
MiR-17-5p-mediated sensitization to chemotherapy-induced apoptosis
in breast cancer cells. Oncotarget. 8:15763–15774. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Liao XH, Li JY, Dong XM, Wang X, Xiang Y,
Li H, Yu CX, Li JP, Yuan BY, Zhou J and Zhang TC: ERα inhibited
myocardin-induced differentiation in uterine fibroids. Exp Cell
Res. 350:73–82. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Song Z, Liu Z, Sun J, Sun FL, Li CZ, Sun
JZ and Xu LY: The MRTF-A/B function as oncogenes in pancreatic
cancer. Oncol Rep. 35:127–138. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhang WL, Lv W, Sun SZ, Wu XZ and Zhang
JH: miR-206 inhibits metastasis-relevant traits by degrading MRTF-A
in anaplastic thyroid cancer. Int J Oncol. 47:133–142. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Xin J, Zhang XK, Xin DY, Li XF, Sun DK, Ma
YY and Tian LQ: FUS1 acts as a tumor-suppressor gene by
upregulating miR-197 in human glioblastoma. Oncol Rep. 34:868–876.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Eisen MB, Spellman PT, Brown PO and
Botstein D: Cluster analysis and display of genome-wide expression
patterns. Proc Natl Acad Sci U S A. 95:pp. 14863–148681998;
|
|
34
|
Kurrey NK, Jalgaonkar SP, Joglekar AV,
Ghanate AD, Chaskar PD, Doiphode RY and Bapat SA: Snail and slug
mediate radioresistance and chemoresistance by antagonizing
p53-mediated apoptosis and acquiring a stem-like phenotype in
ovarian cancer cells. Stem Cells. 27:2059–2068. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan
A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, et al: The
epithelial-mesenchymal transition generates cells with properties
of stem cells. Cell. 133:704–715. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Morel AP, Lièvre M, Thomas C, Hinkal G,
Ansieau S and Puisieux A: Generation of breast cancer stem cells
through epithelial-mesenchymal transition. PLoS One. 3:e28882008.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Santisteban M, Reiman JM, Asiedu MK,
Behrens MD, Nassar A, Kalli KR, Haluska P, Ingle JN, Hartmann LC,
Manjili MH, et al: Immune-induced epithelial to mesenchymal
transition in vivo generates breast cancer stem cells. Cancer Res.
69:2887–2895. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Myatt SS, Wang J, Monteiro LJ, Christian
M, Ho KK, Fusi L, Dina RE, Brosens JJ, Ghaem-Maghami S and Lam EW:
Definition of microRNAs that repress expression of the tumor
suppressor gene FOXO1 in endometrial cancer. Cancer Res.
70:367–377. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Huang YW, Liu JC, Deatherage DE, Luo J,
Mutch DG, Goodfellow PJ, Miller DS and Huang TH: Epigenetic
repression of microRNA-129-2 leads to overexpression of SOX4
oncogene in endometrial cancer. Cancer Res. 69:9038–9046. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Dong P, Kaneuchi M, Watari H, Hamada J,
Sudo S, Ju J and Sakuragi N: MicroRNA-194 inhibits epithelial to
mesenchymal transition of endometrial cancer cells by targeting
oncogene BMI-1. Mol Cancer. 10:992011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Dellinger TH, Smith DD, Ouyang C, Warden
CD, Williams JC and Han ES: L1CAM is an independent predictor of
poor survival in endometrial cancer-An analysis of the cancer
genome atlas (TCGA). Gynecol Oncol. 141:336–340. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Friel AM, Sergent PA, Patnaude C, Szotek
PP, Oliva E, Scadden DT, Seiden MV, Foster R and Rueda BR:
Functional analyses of the cancer stem cell-like properties of
human endometrial tumor initiating cells. Cell Cycle. 7:242–249.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Tanaka Y, Terai Y, Kawaguchi H, Fujiwara
S, Yoo S, Tsunetoh S, Takai M, Kanemura M, Tanabe A and Ohmichi M:
Prognostic impact of EMT
(epithelial-mesenchymal-transition)-related protein expression in
endometrial cancer. Cancer Biol Ther. 14:13–19. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Huszar M, Pfeifer M, Schirmer U, Kiefel H,
Konecny GE, Ben-Arie A, Edler L, Münch M, Müller-Holzner E,
Jerabek-Klestil S, et al: Up-regulation of L1CAM is linked to loss
of hormone receptors and E-cadherin in aggressive subtypes of
endometrial carcinomas. J Pathol. 220:551–561. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Singh A and Settleman J: EMT, cancer stem
cells and drug resistance: An emerging axis of evil in the war on
cancer. Oncogene. 29:4741–4751. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Götte M, Wolf M, Staebler A, Buchweitz O,
Kelsch R, Schüring AN and Kiesel L: Increased expression of the
adult stem cell marker Musashi-1 in endometriosis and endometrial
carcinoma. J Pathol. 215:317–329. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Bao S, Wu Q, Li Z, Sathornsumetee S, Wang
H, McLendon RE, Hjelmeland AB and Rich JN: Targeting cancer stem
cells through L1CAM suppresses glioma growth. Cancer Res.
68:6043–6048. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Held-Feindt J, Schmelz S, Hattermann K,
Mentlein R, Mehdorn HM and Sebens S: The neural adhesion molecule
L1CAM confers chemoresistance in human glioblastomas. Neurochem
Int. 61:1183–1191. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Yoon H, Min JK, Lee DG, Kim DG, Koh SS and
Hong HJ: L1 cell adhesion molecule and epidermal growth factor
receptor activation confer cisplatin resistance in intrahepatic
cholangiocarcinoma cells. Cancer Lett. 316:70–76. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Li Z, Min W and Gou J: Knockdown of
cyclophilin A reverses paclitaxel resistance in human endometrial
cancer cells via suppression of MAPK kinase pathways. Cancer
Chemother Pharmacol. 72:1001–1011. 2013. View Article : Google Scholar : PubMed/NCBI
|