Introduction
According to the National Cancer Institute, although
lung cancer mortality rates have declined due to reduced tobacco
use, lung cancer remains the leading cause of cancer-related
mortality worldwide (1). A high rate
of metastasis at diagnosis is the principal reason for poor
prognosis (2). Tumor invasion and
metastasis are complex and dynamic processes controlled by multiple
factors, including tumor cells and the tumor microenvironment
(3). Lung cancer patients are prone
to distant metastases; thus, the survival period is shortened and
life quality is affected (4).
Nevertheless, the mechanism of metastasis in lung cancer remains
unclear.
The phosphoinositide 3-kinase-protein kinase B (Akt)
signaling pathway is believed to be closely associated with
metastasis (5). Girdin, an Akt
substrate and newly discovered nuclear actin-binding protein, has a
key role in promoting cell migration and angiogenesis during
embryonic development, inflammation and tumor angiogenesis, and it
is highly expressed in several human malignant carcinomas, such as
colon, breast, glioblastoma and esophageal carcinomas (6–12). A
study by Song et al (13)
assessed Girdin expression and the correlation between its
expression and clinical-pathological parameters and survival in a
cohort of 36 consecutive patients with non-small cell lung cancer
(NSCLC), and observed a significant correlation between elevated
Girdin expression and blood vessel infiltration of the tumor.
The Janus kinase-signal transducer and activator of
transcription (STAT) signaling pathway is also closely associated
with many biological processes, particularly metastasis (14). The STAT family consists of six
members (STAT1-STAT6), of which STAT3 is one of the most common
sustained activated signaling proteins (15). A study by Dunkel et al
(16) demonstrated that Girdin is
capable of forming a positive feedback loop to increase the
activity of STAT3, thereby promoting tumor invasion and migration.
In a previous study, to explore whether Girdin is mediated by STAT3
in lung cancer, the authors of the present study depleted
endogenous STAT3 and observed that Girdin expression decreased
(17). It was also found that
interleukin (IL)-17 promotes tumor angiogenesis in NSCLC by
activating STAT3/Girdin signaling in NSCLC cell lines, which
subsequently upregulates vascular endothelial growth factor
(17). Nevertheless, few studies
have explored the expression of Girdin protein and STAT3, as well
as their relationship with lung cancer.
In the present study, the correlation between Girdin
protein and STAT3 protein in lung cancer was evaluated using
immunohistochemistry (IHC). A prognostic model based on clinical
parameters was also generated to determine whether Girdin could act
as a prognostic biomarker for lung cancer.
Patients and methods
Patient tissue samples
A total of 334 NSCLC tissue sections, 20 benign lung
disease tissue sections, 20 adjacent normal lung tissues sections,
24 fresh NSCLC tissues and 5 fresh normal lung tissue sections were
obtained with informed consent at the Harbin Medical University
Cancer Hospital (Harbin, China) between January 2005 and December
2006. All patients included in the present study had been
surgically resected and diagnosed with stage I–IIIA NSCLC. Patients
with any other types of cancer, or who missed follow-up
appointments were excluded from the study. This retrospective
analysis was approved by the Ethics Committee of Harbin Medical
University Cancer Hospital. The clinical parameters extracted from
medical records included: Age; sex; smoking history; Eastern
Cooperative Oncology Group (ECOG) performance status (18); histological type and grade; stage
(IASLC 7th TNM Staging system) (19); metastasis sites; diameter of the
carcinoma; and specimen sites.
IHC
For IHC, 4-µm-thick formaldehyde-fixed (fixed with
4% formaldehyde at room temperature for 24 h), paraffin-embedded
sections of 334 NSCLC, 20 benign lung disease and 20 adjacent
normal lung tissues were deparaffinized in xylene and then
rehydrated in serially graded alcohols. Antigens were retrieved by
boiling the samples in 10 mM sodium citrate buffer at pH 6.0 for 30
min. Subsequently, the sections were washed with phosphate-buffered
saline (pH 7.4), blocked with 3% hydrogen peroxide at room
temperature for 20 min and incubated overnight at 4°C with
anti-Girdin (ab111035; 1:100; Abcam, Cambridge, UK) and anti-STAT3
(ab119352; 1:500; Abcam) antibodies. The slides were incubated with
horseradish peroxidase-conjugated anti-rabbit immunoglobulin G
secondary antibodies (SC2040, 1:400, Santa Cruz Biotechnology,
Inc., Dallas, TX, USA) for 30 min at room temperature, followed by
signal detection with diaminobenzidine. The slides were
counterstained with hematoxylin at room temperature for 5 min. The
mean percentage of positive tumor cells was determined in at least
five fields at magnification, ×200 using a light microscope.
The slides were evaluated independently by two
experienced pathologists who reached a consensus. The percentages
of positive cells were categorized as follows: 0, 0%; 1, 0–10%; 2,
10–50%; and 3, >50%. The staining intensity was scored as
follows: 0, negative; 1, weak; 2, moderate; and 3, strong. The
scores for the percentage of positive cells and staining intensity
were multiplied to achieve a weighted score for each case. Cases
with scores ≤4 were defined as low expression and cases with scores
>4 were defined as high expression.
Western blot analysis
A total of 24 fresh NSCLC and 5 normal tissues were
washed three times with PBS solution and treated by ultrasonic
lysis with a radioimmunoprecipitation lysis buffer (P0013C;
Beyotime Institute of Biotechnology, Haimen, China) for protein
extraction. Protein were quantified by BCA. A total of 30 µg of
protein were loaded per lane and separated by 10% SDS-PAGE, after
which the proteins were transferred to a polyvinylidene difluoride
membrane. Subsequently, the membrane was blocked with 5% skim milk
for 1 h at room temperature and incubated with primary antibodies
directed against Girdin, (ab113890; 1:500; Abcam) and β-actin
(4970P; 1:1,000; CST Biological Reagents Co., Ltd., Shanghai,
China) overnight at 4°C. Appropriately diluted specific secondary
antibodies (anti-rabbit IgG; ZB2301; 1:1,000; OriGene Technologies,
Inc., Beijing, China) were added and incubated for 1 h at room
temperature. An enhanced chemiluminescence kit (Pierce; Thermo
Fisher Scientific, Inc., Waltham, MA, USA) was used to detect and
analyze immunostained protein bands using a charge-coupled camera
(LAS4000; Fujifilm, Tokyo, Japan) and Gel-Pro Analyzer software
version 4.0 (Media Cybernetics, Inc., Rockville, MD, USA).
Statistical analysis
Data were presented as the mean ± standard
deviation. Statistical analysis was performed using SPSS 18.0
software (SPSS, Inc., Chicago, IL, USA). P<0.05 was considered
to indicate a statistically significant difference. ANOVA and
Dunnett's post hoc test was performed for the comparison of Girdin
expression between fresh tumor and normal tissues. A multivariate
Cox regression model was used to analyze prognostic variables for
survival measures. Chi-squared tests were used to examine the
statistical association between clinical-pathological and IHC data.
Survival curves were plotted using the Kaplan-Meier method and
differences were assessed using the log-rank test. The correlation
between Girdin and STAT3 was calculated using Spearman's rank
correlation coefficient.
Results
Patient characteristics
To evaluate the clinical significance of Girdin
expression in NSCLC, an IHC analysis of 334 NSCLC tissues samples,
20 benign lung disease tissues (pulmonary hamartoma, pulmonary
fibroma, pulmonary hemangioma and pneumonia) and 20 adjacent normal
lung tissues was performed. The mean age of the 334 NSCLC patients
enrolled in the present study was 50.87 years (range, 29–80 years).
Of these, 178/334 (53.23%) patients had lymph node metastasis and
82/334 (24.53%) exhibited distant metastasis (Table I).
 | Table I.Correlation between Girdin and STAT3
expression in non-small cell lung cancer tissues. |
Table I.
Correlation between Girdin and STAT3
expression in non-small cell lung cancer tissues.
|
| Girdin |
|
|
|---|
|
|
|
|
|
|---|
| Transcription
factor | High | Low | r | P-value |
|---|
| STAT3 |
|
|
|
|
| High | 110 | 29 | 0.696 | <0.001 |
| Low | 20 | 175 |
|
|
Expression and localization of Girdin
and STAT3 in NSCLC, benign lung disease and normal lung
tissues
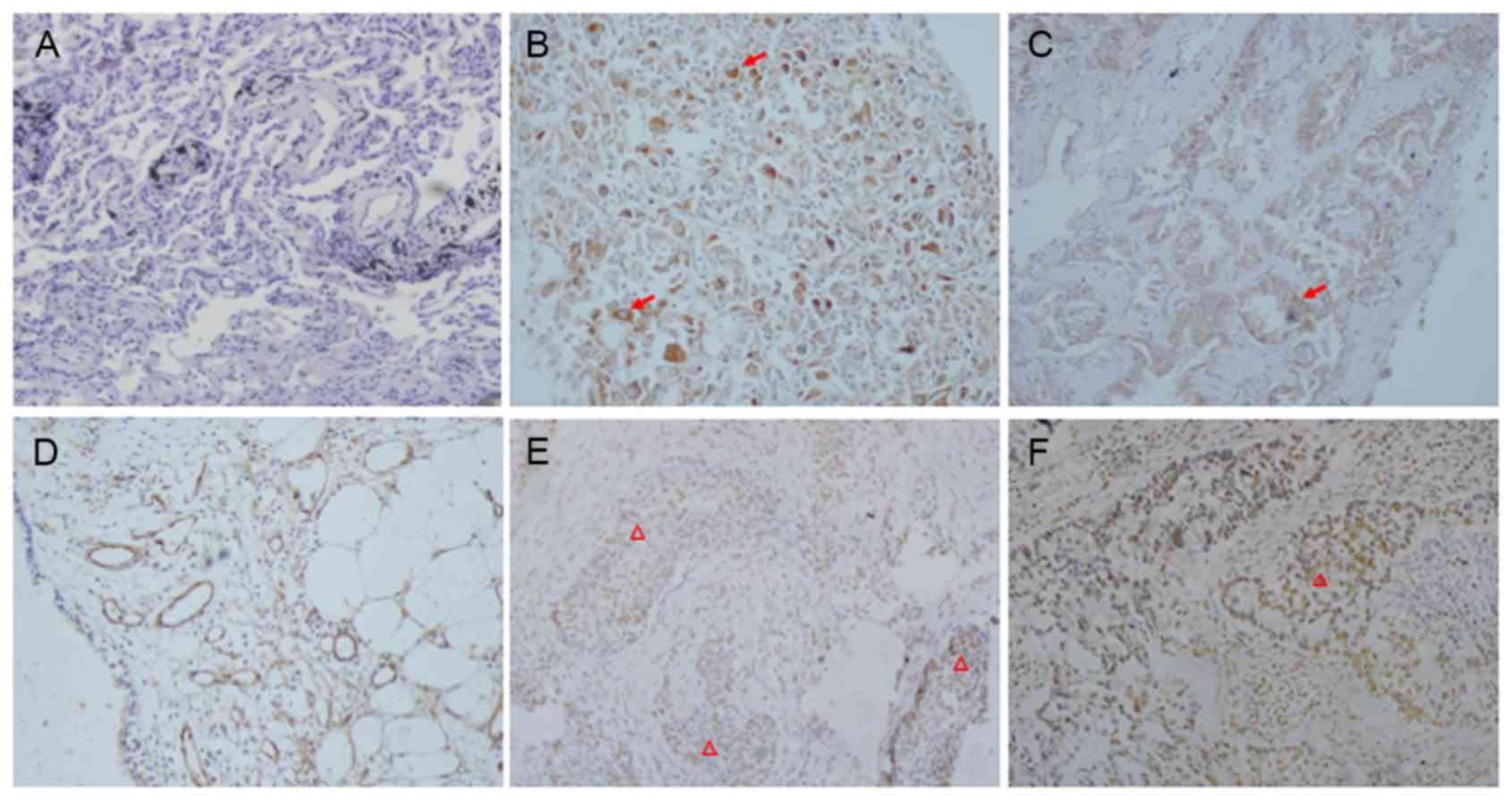
As demonstrated in Fig.
1, cytoplasmic and membrane Girdin immunoreactivity was
detected in 130/334 lung cancer samples (38.93%) and 10% (2/20, one
pulmonary hamartoma and one pulmonary hemangioma) of benign cases,
whereas it was not present in the adjacent normal tissues (0/20).
STAT3 was predominately localized in the nuclei of tumor cells.
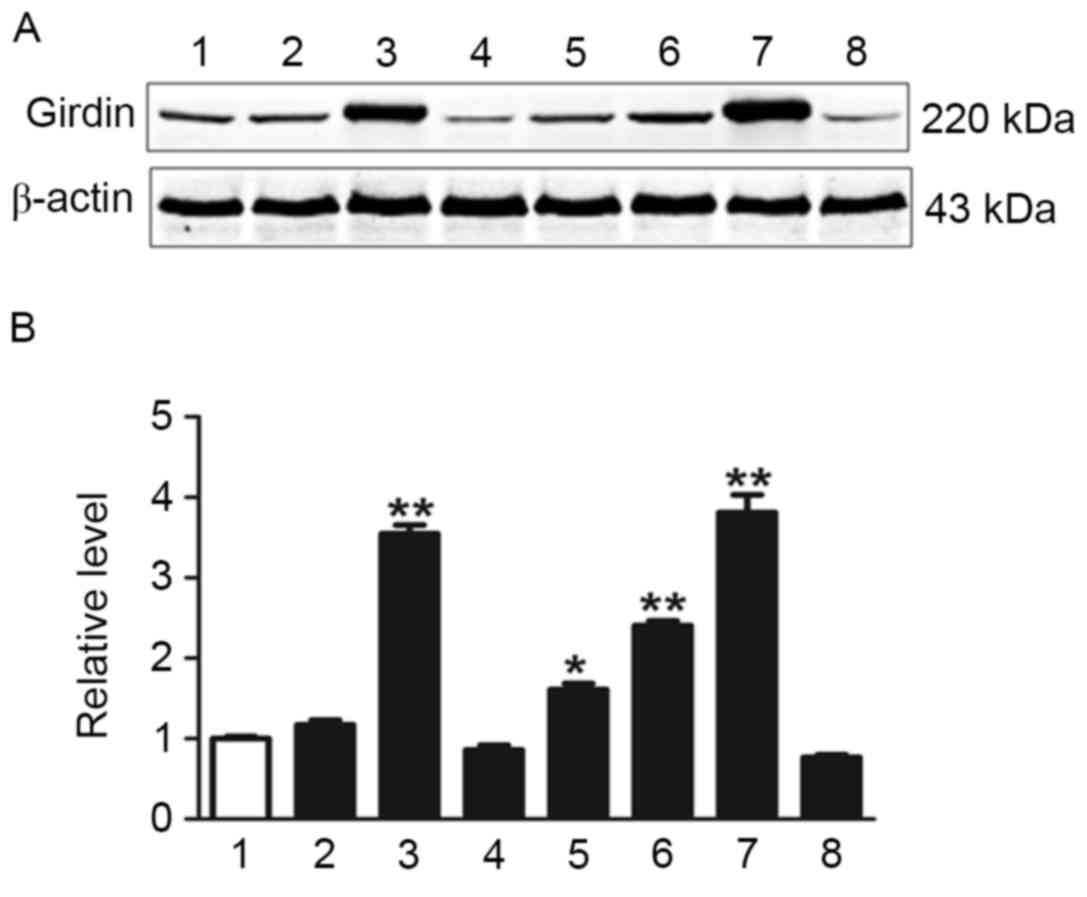
Western blotting was used to investigate Girdin expression in fresh
tumor tissues and normal tissues (Fig.
2). Girdin expression was significantly higher in several NSCLC
tissue samples compared with normal tissues as determined by
one-way ANOVA (P<0.05; Fig.
2).
The potential correlation between the expression of
Girdin and STAT3 in NSCLC was assessed. Spearman's rank correlation
analysis revealed that Girdin expression was closely correlated
with STAT3 expression in the NSCLC patient cohort (r=0.696;
P<0.001) (Table I).
Relationship between Girdin and STAT3
overexpression and clinical-pathological parameters
The correlations between Girdin and STAT3 expression
and the clinicopathological characteristics of NSCLC are
demonstrated in Table II. Lung
tumor expression of Girdin was not dependent on age, sex, smoking
history, family history, histology, ECOG performance status,
histopathological subtype, degree of differentiation, tumor size,
metastatic site or T stage. Lung tumor expression of STAT3 in the
lung cancer cases was not dependent on age, sex, smoking history,
family history, histology, ECOG performance status,
histopathological subtype, degree of differentiation or specimen
sites. Elevated expression of Girdin was associated with positive
lymph node metastasis status (P=0.001), positive distant metastasis
status (P<0.001), later TNM stage (P<0.001) and more tumor
sites (P=0.034). Elevated expression of STAT3 was correlated with
later TNM stage (P=0.007), positive lymph node metastasis status
(P<0.001), positive distant metastasis (P=0.011), later T stage
(P=0.004) and larger tumor diameter (P=0.002).
 | Table II.Association between Girdin, STAT3 and
clinicopathological factors in non-small cell lung cancer
(n=334). |
Table II.
Association between Girdin, STAT3 and
clinicopathological factors in non-small cell lung cancer
(n=334).
|
| Girdin
expression |
| STAT3 expression |
|
|---|
|
|
|
|
|
|
|---|
| Parameter | Low | High | P-value | Low | High | P-value |
|---|
| Age, years |
|
| 0.625 |
|
| 0.822 |
|
<55 | 68 | 40 |
| 64 | 44 |
|
| ≥55 | 136 | 90 |
| 131 | 95 |
|
| Gender |
|
| 0.508 |
|
| 0.223 |
| Male | 142 | 86 |
| 128 | 100 |
|
|
Female | 62 | 44 |
| 67 | 39 |
|
| Family history |
|
| 0.448 |
|
| 0.094 |
| Yes | 40 | 30 |
| 47 | 23 |
|
| No | 164 | 100 |
| 148 | 116 |
|
| Smoking status |
|
| 0.473 |
|
| 0.616 |
|
Non-smoker | 118 | 70 |
| 112 | 76 |
|
|
Smoker | 86 | 60 |
| 83 | 63 |
|
| ECOG status |
|
| 0.799 |
|
| 0.158 |
|
0–1 | 190 | 122 |
| 179 | 133 |
|
| ≥2 | 14 | 8 |
| 16 | 6 |
|
| Histology
grade |
|
| 0.104 |
|
| 0.633 |
|
Well-differentiated | 2 | 2 |
| 2 | 2 |
|
|
Moderately differentiated | 62 | 26 |
| 55 | 33 |
|
| Poorly
differentiated | 140 | 102 |
| 138 | 104 |
|
| Histological
type |
|
| 0.496 |
|
| 0.764 |
|
Adenocarcinoma | 106 | 64 |
| 96 | 74 |
|
|
Squamous cell carcinoma | 80 | 58 |
| 83 | 55 |
|
|
Other | 18 | 8 |
| 16 | 10 |
|
| TNM stage |
|
| <0.001 |
|
| 0.007 |
|
I–IIIA | 150 | 56 |
| 132 | 74 |
|
|
IIIB-IV | 54 | 74 |
| 63 | 65 |
|
| Tumor stage |
|
| 0.693 |
|
| 0.004 |
| T1 | 44 | 26 |
| 48 | 22 |
|
| T2 | 114 | 72 |
| 108 | 78 |
|
| T3 | 26 | 22 |
| 18 | 30 |
|
| T4 | 20 | 10 |
| 21 | 9 |
|
| Lymph node
metastasis |
|
| 0.001 |
|
| <0.001 |
|
Yes | 94 | 84 |
| 88 | 90 |
|
| No | 110 | 46 |
| 107 | 49 |
|
| Distant
metastasis |
|
| <0.001 |
|
| 0.011 |
|
Yes | 26 | 56 |
| 38 | 44 |
|
| No | 178 | 74 |
| 157 | 95 |
|
| Diameter of tumor,
cm |
|
| 0.434 |
|
| 0.002 |
| ≤3 | 58 | 32 |
| 64 | 26 |
|
|
3–7 | 134 | 86 |
| 123 | 97 |
|
|
>7 | 12 | 12 |
| 8 | 16 |
|
| Sites of
specimen |
|
| 0.034 |
|
| 0.797 |
| Primary
tumor site | 190 | 112 |
| 177 | 125 |
|
|
Metastasis tumor site | 14 | 18 |
| 18 | 14 |
|
Elevated Girdin and STAT3 expression
is associated with poor prognosis in NSCLC
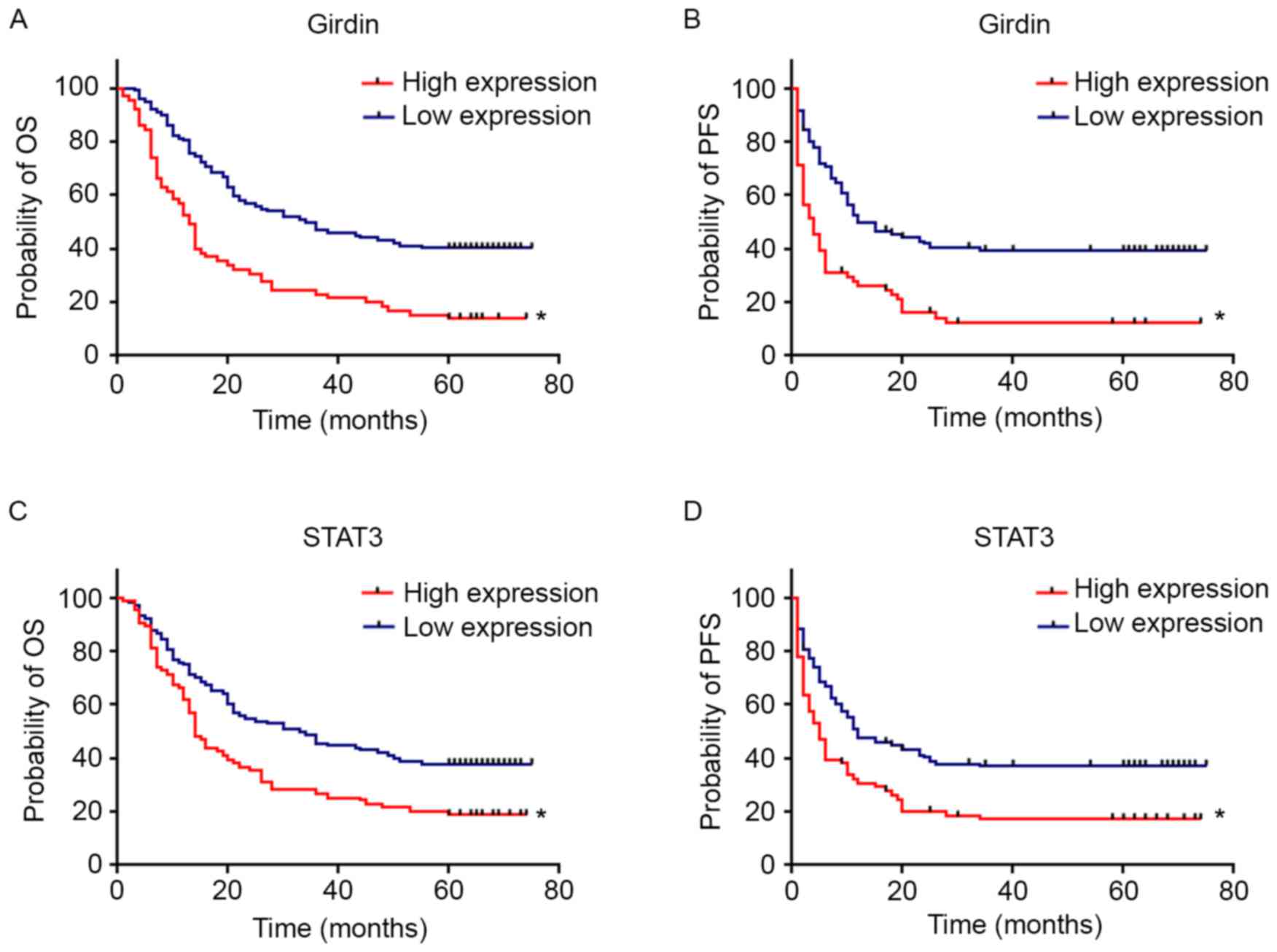
The Kaplan-Meier survival curves of Girdin and STAT3
for overall survival (OS) and progression-free survival (PFS) are
demonstrated in Fig. 3. Patients
with elevated Girdin expression were observed to have significantly
shorter OS (P<0.001) and PFS (P<0.001) compared with those
with lower expression. Patients with increased STAT3 expression
were observed to have significant shorter OS (P<0.001) and PFS
(P<0.001) rates compared with patients with low-level
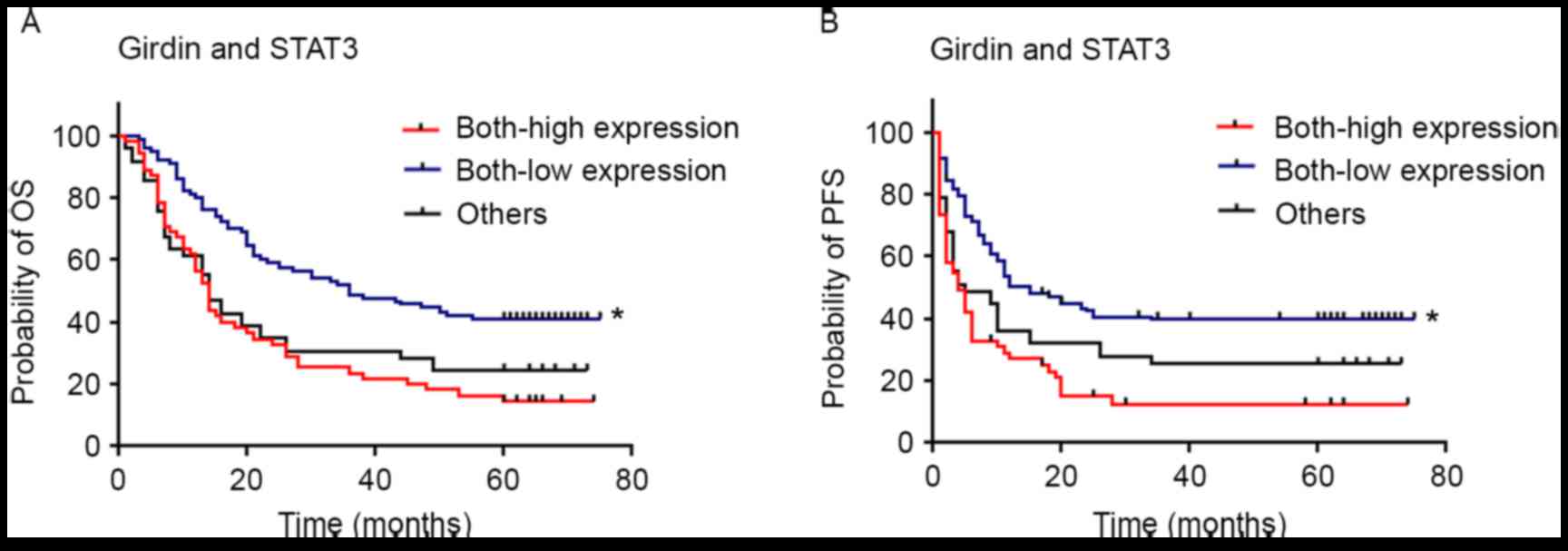
expression. Furthermore, patients with low expression of both
Girdin and STAT3 were observed to have significantly longer OS
(P<0.001) and PFS (P<0.001) compared with individuals with
high/high expression and others (low/high, high/low) expression
(Fig. 4).
Elevated Girdin expression is
independently associated with OS and PFS in NSCLC
To identify prognostic variables of NSCLC, a
multivariate analysis was performed. It was identified that TNM
stage (P=0.002 and P=0.001), lymph node metastasis (P=0.009 and
P=0.004), distant metastasis (P=0.048 and P=0.001) and Girdin
expression (P=0.004 and P=0.001) were prognostic factors in NSCLC
for OS and PFS, respectively (Table
III).
 | Table III.Multivariate survival analysis of OS
and PFS in patients with non-small cell lung cancer (n=334). |
Table III.
Multivariate survival analysis of OS
and PFS in patients with non-small cell lung cancer (n=334).
|
| OS | PFS |
|---|
|
|
|
|
|---|
| Parameter | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| TNM stage |
|
|
|
|
|
|
|
I–IIIA | 1.000 |
|
| 1.000 |
|
|
|
IIIB-IV | 0.490 | 0.316–0.757 | 0.002 | 0.480 | 0.311–0.739 | 0.001 |
| Tumor stage |
|
|
|
|
|
|
| T1 | 1.000 |
|
| 1.000 |
|
|
| T2 | 0.454 | 0.251–0.821 | 0.009 | 0.449 | 0.249–0.810 | 0.008 |
| T3 | 0.901 | 0.544–1.491 | 0.684 | 0.912 | 0.551–1.511 | 0.722 |
| T4 | 1.303 | 0.736–2.307 | 0.363 | 1.780 | 1.009–3.141 | 0.047 |
| Lymph node
metastasis |
|
|
|
|
|
|
|
Yes | 1.000 |
|
| 1.000 |
|
|
| No | 1.462 | 1.101–1.942 | 0.009 | 1.505 | 1.139–1.989 | 0.004 |
| Distant
metastasis |
|
|
|
|
|
|
|
Yes | 1.000 |
|
| 1.000 |
|
|
| No | 1.561 | 1.005–2.427 | 0.048 | 2.116 | 1.355–3.311 | 0.001 |
| Girdin
expression |
|
|
|
|
|
|
|
Low | 1.000 |
|
| 1.000 |
|
|
|
High | 1.894 | 1.228–2.921 | 0.004 | 2.127 | 1.366–3.311 | 0.001 |
| STAT3
expression |
|
|
|
|
|
|
|
Low | 1.000 |
|
| 1.000 |
|
|
|
High | 0.796 | 0.527–1.202 | 0.277 | 0.686 | 0.445–1.058 | 0.088 |
Discussion
Currently, the expression status of Girdin protein
and its prognostic value for lung cancer are unclear. In the
present study, it was identified that Girdin expression is
significantly associated with TNM stage and tumor metastasis in
human lung cancer.
Girdin, which is phosphorylated following epidermal
growth factor stimulation and is a novel Akt substrate, is
essential for cell metastasis (20).
It is an important factor for the leading edge of cell pseudopods
involved in cell movement (21). A
study by Garcia-Marcos et al (22) reported that the survival rate of
patients with colon cancer and Girdin-positive expression was
reduced compared with Girdin-negative expression. Girdin expression
also predicted mortality risk, independent of microsatellite
stability status. The authors concluded that Girdin may serve as a
convenient metastasis biomarker for colon cancer (22). In the present study, it was
demonstrated that patients with elevated Girdin expression had
poorer OS and PFS compared with those with lower expression levels.
These results are consistent with previous investigations of Girdin
in other cancer types. In breast cancer tissues and cell lines,
Girdin was highly expressed, and the co-expression of Girdin and
tumor necrosis factor receptor 4 led to an increased rate of lymph
node metastasis (23). A study by
Nishimae et al (24) reported
that the expression of Girdin in invasive breast cancer was
strongly associated with lymph node metastasis. In esophageal
squamous cell carcinoma (ESCC), Girdin was demonstrated to be
involved in the motility of ESCC cells, and the expression of
Girdin protein was inversely correlated with ESCC patient survival
(12). In the present study, it was
identified that the expression rate of Girdin in NSCLC was 38.93%,
which differed from 72.2% (26/36) in a study by Song et al
(13) of 36 NSCLC patients
undergoing surgery. This difference may be because 334 patients
with different stages were recruited to the present study, whereas
only patients with early-stage disease were enrolled in the study
by Song et al (13). The
present study also demonstrated that tissues with stronger
expression of Girdin were obtained from metastasis sites, which may
be because Girdin facilitates cell invasion and metastasis.
STAT3 is a STAT family member activated by tyrosine
phosphorylation in response to various factors, such as epidermal
growth factor and IL-6 (25,26). It was reported by Dunkel et al
(16) that STAT3 protein upregulates
Girdin expression, and that Girdin enhances STAT3 activation in a
positive feedback loop during wound healing and tumor metastasis.
STAT3 was also demonstrated to be essential for Girdin expression
under stimulated tension force under physiological conditions, as
well as for osteoblast proliferation and migration during
quiescence (27). These findings
suggest that STAT3/Girdin pathway activation has a critical role in
proliferation and migration. In the present study, it was revealed
that Girdin overexpression was correlated with STAT3 in patient
tissues. The results indicated that patients with high-level
expression of both Girdin and STAT3 had lower OS and PFS rates
compared with low/high, high/low and low/low expression, which
indicates that STAT3/Girdin may serve an essential role in
malignant behavior in NSCLC.
In conclusion, the present data indicated that
Girdin may be a biomarker for metastasis in patients with NSCLC.
Combined Girdin and STAT3 expression could predict poor prognosis
in patients with NSCLC.
Acknowledgements
The present study was supported by the Education
Department Foundation of Heilongjiang Province (grant no.
12521313).
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Marx V: Tracking metastasis and tricking
cancer. Nature. 494:133–136. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cairns RA, Khokha R and Hill RP: Molecular
mechanisms of tumor invasion and metastasis: An integrated view.
Curr Mol Med. 3:659–671. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cetin K, Ettinger DS, Hei YJ and O'Malley
CD: Survival by histologic subtype in stage IV nonsmall cell lung
cancer based on data from the surveillance, Epidemiology and end
results program. Clin Epidemiol. 3:139–148. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yang Y, Ahn YH, Chen Y, Tan X, Guo L,
Gibbons DL, Ungewiss C, Peng DH, Liu X, Lin SH, et al: ZEB1
sensitizes lung adenocarcinoma to metastasis suppression by PI3K
antagonism. J Clin Invest. 124:2696–2708. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Enomoto A, Murakami H, Asai N, Morone N,
Watanabe T, Kawai K, Murakumo Y, Usukura J, Kaibuchi K and
Takahashi M: Akt/PKB regulates actin organization and cell motility
via Girdin/APE. Dev Cell. 9:389–402. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ohara K, Enomoto A, Kato T, Hashimoto T,
Isotani-Sakakibara M, Asai N, Ishida-Takagishi M, Weng L, Nakayama
M, Watanabe T, et al: Involvement of Girdin in the determination of
cell polarity during cell migration. PLoS One. 7:e366812012.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ghosh P, Tie J, Muranyi A, Singh S,
Brunhoeber P, Leith K, Bowermaster R, Liao Z, Zhu Y, LaFleur B, et
al: Girdin (GIV) expression as a prognostic marker of recurrence in
mismatch repair-proficient stage II colon cancer. Clin Cancer Res.
22:3488–3498. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Liu C, Zhang Y, Xu H, Zhang R, Li H, Lu P
and Jin F: Girdin protein: A new potential distant metastasis
predictor of breast cancer. Med Oncol. 29:1554–1560. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Peng WT, Hu X, Yao L, Jiang YZ and Shao
ZM: Elevated expression of Girdin in the nucleus indicates worse
prognosis for patients with estrogen receptor-positive breast
cancer. Ann Surg Oncol. 21 Suppl 4:S648–S656. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gu F, Wang L, He J, Liu X, Zhang H, Li W,
Fu L and Ma Y: Girdin, an actin-binding protein, is critical for
migration, adhesion, and invasion of human glioblastoma cells. J
Neurochem. 131:457–469. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Shibata T, Matsuo Y, Shamoto T, Hirokawa
T, Tsuboi K, Takahashi H, Ishiguro H, Kimura M, Takeyama H and
Inagaki H: Girdin, a regulator of cell motility, is a potential
prognostic marker for esophageal squamous cell carcinoma. Oncol
Rep. 29:2127–2132. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Song JY, Jiang P, Li N, Wang FH and Luo J:
Clinical significance of Girdin expression detected by
immunohistochemistry in non-small cell lung cancer. Oncol Lett.
7:337–341. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Buettner R, Mora LB and Jove R: Activated
STAT signaling in human tumors provides novel molecular targets for
therapeutic intervention. Clin Cancer Res. 8:945–954.
2002.PubMed/NCBI
|
|
15
|
Lui GY, Kovacevic Z, V Menezes S,
Kalinowski DS, Merlot AM, Sahni S and Richardson DR: Novel
thiosemicarbazones regulate the signal transducer and activator of
transcription3 (STAT3) pathway: Inhibition of constitutive and
interleukin 6-induced activation by iron depletion. Mol Pharmacol.
87:543–560. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Dunkel Y, Ong A, Notani D, Mittal Y, Lam
M, Mi X and Ghosh P: STAT3 protein up-regulates Gα-interacting
vesicle-associated protein (GIV)/Girdin expression, and GIV
enhances STAT3 activation in a positive feedback loop during wound
healing and tumor invasion/metastasis. J Biol Chem.
287:41667–41683. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Pan B, Shen J, Cao J, Zhou Y, Shang L, Jin
S, Cao S, Che D, Liu F and Yu Y: Interleukin-17 promotes
angiogenesis by stimulating VEGF production of cancer cells viathe
STAT3/GIV signaling pathway in non-small-cell lung cancer. Sci Rep.
5:160532015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Oken MM, Creech RH, Tormey DC, Horton J,
Davis TE, McFadden ET and Carbone PP: Toxicity and response
criteria of the eastern cooperative oncology group. Am J Clin
Oncol. 6:649–655. 1982. View Article : Google Scholar
|
|
19
|
Goldstraw P, Crowley J, Chansky K, Giroux
DJ, Groome PA, Rami-Porta R, Postmus PE, Rusch V and Sobin L;
International Association for the Study of Lung Cancer
International Staging Committee, ; Participating Institutions, :
The IASLC lung cancer staging project: Proposals for the revision
of the TNM stage groupings in the forthcoming (seventh) edition of
the TNM Classification of malignant tumours. J Thorac Oncol.
2:706–714. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Omori K, Asai M, Kuga D, Ushida K, Izuchi
T, Mii S, Enomoto A, Asai N, Nagino M and Takahashi M: Girdin is
phosphorylated on tyrosine 1798 when associated with structures
required for migration. Biochem Biophys Res Commun. 485:934–940.
2015. View Article : Google Scholar
|
|
21
|
Garcia-Marcos M, Ghosh P and Farquhar MG:
GIV/Girdin transmits signals from multiple receptors by triggering
trimeric G protein activation. J Biol Chem. 290:6697–6704. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Garcia-Marcos M, Jung BH, Ear J, Cabrera
B, Carethers JM and Ghosh P: Expression of GIV/Girdin, a
metastasis-related protein, predicts patient survival in colon
cancer. FASEB J. 25:590–599. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang A, Wang J, Sun L, Jin J, Ren H, Yang
F, Diao K, Wei M and Mi X: Expression of tumor necrosis factor
receptor-assicated factor 4 correlates with expression of Girdin
and promotes nuclear translocation of Girdin in breast cancer. Mol
Med Rep. 11:3635–3641. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Nishimae K, Tsunoda N, Yokoyama Y, Kokuryo
T, Iwakoshi A, Takahashi M and Nagino M: The impact of Girdin
expression on recurrence-free survival in patients with
luminal-type breast cancer. Breast Cancer. 22:445–451. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yue P, Zhang X, Paladino D, Sengupta B,
Ahmad S, Holloway RW, Ingersoll SB and Turkson J: Hyperactive EGF
receptor, Jaks and Stat3 signaling promote enhanced colony-forming
ability, motility and migration of cisplatin-resistant ovarian
cancer cells. Oncogene. 31:2309–2322. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sun F, Zhang ZW, Tan EM, Lim ZLR, Li Y,
Wang XC, Chua SE, Li J, Cheung E and Yong EL: Icaritin suppresses
development of neuroendocrine differentiation of prostate cancer
through inhibition of IL-6/STAT3 and Aurora kinase a pathways in
TRAMP mice. Carcinogenesis. 37:701–711. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hu JT, Li Y, Yu B, Gao GJ, Zhou T and Li
S: Girdin/GIV is upregulated by cyclic tension, propagates
mechanical signal transduction, and is required for the cellular
proliferation and migration of MG-63 cells. Biochem Biophys Res
Commun. 464:493–499. 2015. View Article : Google Scholar : PubMed/NCBI
|