Introduction
Genetic diseases, trauma and caries may lead to
tooth loss. Implants or dentures are used clinically to repair
missing teeth (1,2). However, there are a number of
disadvantages in these approaches (3). For example, the implanted or denture
tooth is generally not as strong as the natural tooth (3). With developments in tissue engineering,
it is becoming possible to use stem cells and tissue engineering
techniques to repair defected teeth (1,2). Dental
pulp stem cells (DPSCs) are mesenchymal stem cells (MSCs) capable
of differentiating into neural cells, adipocytes and odontoblasts
(4). Studies have demonstrated that
DPSCs may be used to repair the defected tooth; however, how DPSCs
are directionally differentiated into odontoblasts remains unclear
(5–7). Studies have demonstrated that during
the differentiation of DPSCs into odontoblasts the undifferentiated
cells are smaller than the differentiated cells (4). DPSCs have been demonstrated to
differentiate into a variety of cell types. Depending on induction
conditions DPSCs may differentiate into odontoblasts, myotubes or
adipocytes (8,9). hDPSCs isolated from dental pulp of
young permanent teeth may differentiate into osteoblasts in
vitro (10). In addition, DPSCs
also express markers for glial cells and neuronal precursors,
including glial fibrillary acidic proteins and nestin, and may
differentiate into neurons in vitro; indicating that
cultured DPSCs have the potential to differentiate into neuron-like
cells (11–13). Studies have also revealed that DPSCs
can differentiate into chondrocytes in vitro (13,14).
Runt-related transcription factor 2 (Runx2), also
known as core binding factor α1, belongs the Runt domain gene
family of transcription factors. It regulates protein expression at
transcriptional level and serves a crucial role in bone and tooth
development (15). A previous study
demonstrated that, as an osteoblast differentiation-specific
transcription factor, Runx2 may directly induce the differentiation
of mesenchymal stem cells to osteoblasts (16). Runx2 is expressed in odontogenic and
osteogenic mesenchymal cells during the embryonic period, and its
expression patterns in osteoblast differentiation and odontoblast
differentiation are different (17–19).
Therefore, it may be a key attribute that determines odontoblast
differentiation.
The Rho/Rho-associated protein kinase (ROCK)
signaling pathway has been indicated to have a role in cell
proliferation, differentiation and apoptosis, and in regulating the
differentiation of MSCs into osteoblasts and adipocytes (20). Therefore, the Rho/ROCK signaling
pathway may also regulate the expression of Runx2 to direct the
differentiation of human dental pulp stem cells (hDPSCs) into
odontoblasts (21). The aim of the
present study was to investigate the expression of Runx2 during
odontoblast and osteoblast differentiation and effect of the
Rho/ROCK signaling pathway on the expression of Runx2 and other
mineralization genes using a pathway inhibitor during odontoblast
differentiation.
Materials and methods
Human bone marrow MSCs (hBMSCs)
hBMSCs were purchased from the American Type Culture
Collection (Manassas, VA, USA) and maintained in Dulbecco's
modified Eagle's medium (DMEM) supplemented with 12% fetal bovine
serum (FBS; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA,
USA) at 37°C in a humidified atmosphere containing 5%
CO2. The present study was approved by the Ethics
Committee of Xiamen Stomatological Hospital (Xiamen, China).
Reagents and instruments
A BCA protein assay kit, enhanced chemiluminescent
ultra-sensitive luminescence solution, mouse anti-GAPDH monoclonal
antibodies (cat. no. AF0006) and horseradish peroxidase-labeled
goat anti-rabbit immunoglobulin G (H+L) antibodies (cat. no. A0208)
were purchased from Beyotime Institute of Biotechnology (Beijing,
China). Peroxidase-conjugated goat anti-mouse IgG (H+L) antibodies
(cat. no. ZB-2305) were purchase from OriGene Technologies, Inc.
(Rockville, MD, USA). Rabbit anti-dentin sialophosphoprotein (DSPP;
cat. no. ab216892), osteocalcin (OCN; cat. no. ab13420), Runx2
(cat. no. ab76956), RhoA (cat. no. ab54835) and ROCK (cat. no.
ab45171) polyclonal antibodies, anti-cluster of differentiation
(CD)29-phycoerythrin (PE) (cat. no. ab93759), CD90-PE (cat. no.
ab93758), CD105-PE (cat. no. ab11414), CD146-PE (cat. no. ab75769),
CD34-PE (cat. no. ab81289) and CD45-PE (cat. no. ab10558)
antibodies were purchased from were purchased from Abcam
(Cambridge, MA, USA). Hank's balanced salt solution (HBSS) and
radioimmunoprecipitation assay (RIPA) lysis buffer were from
Shanghai Solarbio Science & Technology Co., Ltd. (Shanghai,
China). Collagenase I and neutral collagenase II were obtained from
Gibco (Thermo Fisher Scientific, Inc.) and a TRIzol kit and
one-step reverse transcription (RT)-polymerase chain reaction (PCR)
kit were obtained from Invitrogen (HiFiScript cDNA synthesis kit;
cat no. CW2569M; CWBIO, Beijing, China). C3 exoenzyme was purchased
from Sigma-Aldrich (Merck KGaA, Darmstadt, Germany). The flow
cytometer FACScalibur was supplied by BD Biosciences (San Jose, CA,
USA). Evolution™ 220 UV-Visible spectrophotometer was
from Thermo Fisher Scientific, Inc. and electrophoresis apparatus,
electrophoresis transfer apparatus and the gel imaging system
(ChemiDocTM XRS) were obtained from Bio-Rad Laboratories, Inc.
(Hercules, CA, USA). The fluorescence quantitative PCR instrument
LightCycler96 was purchased from Roche Diagnostics (Basel,
Switzerland) and the PCR thermal cycler PTC-200 was purchased from
(Bio-Rad Laboratories, Inc.).
hDPSC isolation
hDPSCs were isolated as described previously
(22,23). Impacted third molars were collected
from 29 healthy young (aged 19–29) male volunteers; the pulps were
extracted under sterile conditions. The volunteers who possessed
healthy teeth tissue, and no periodontal or periapical diseases
were enrolled in the present study at Xiamen Stomatological
Hospital (Xiamen, China) between July and December 2015. Patients
who had previously had extraction were excluded. All volunteers
provided their prior written, informed consent. The pulps were
washed with HBSS, and digested with 3% collagenase I and 4% neutral
collagenase II for 1 h at 37°C, and filtered through a 70 µm mesh
to generate a single cell suspension. The suspension was
centrifuged at 500 × g at 25°C for 10 min, resuspended in DMEM
supplemented 15% FBS, inoculated into wells in 96-well plates and
cultured at 37°C in an atmosphere containing 5% CO2. The
culture medium was refreshed every 3 days and subcultured when the
confluency reached 80–90%.
hDPSC characterization
hDPSCs at the third passage were digested with 0.25%
EDTA-trypsin at 37°C for 2 min when the confluency reached 80–90%
to obtain a single cell suspension. The cells were adjusted to a
density of 1×106 cells/ml and incubated with CD29-PE,
CD90-PE, CD105-PE, CD146-PE, CD34-PE and CD45-PE antibodies (all
1:100) for 1 h at room temperature and analyzed by performing flow
cytometry using a flow cytometer. The cells were blocked in 1%
bovine serum albumin (BSA; CWBIO) for 30 min at 37°C and washed
with PBS prior to the analysis. The data were analysed using BD
CellQuest™ software (version 5.1; BD Biosciences).
Induction and characterization of
odontoblasts and osteoblasts
hDPSCs and hBMSCs at the third passage were
inoculated to the wells of 6-well plates, combined with 100 µl
mineralization induction solution (10 nmol/l dexamethasone, 10
mmol/l β-glycerophosphate and 50 µg/ml vitamin C) and α-minimum
essential medium (Gibco; Thermo Fisher Scientific, Inc.)
supplemented with 100 ml/l FBS, and cultured at 37°C. Cells were
harvested at day 0, 7, 14 and 21 for analysis of DSPP, OCN and
Runx2 levels using western blot analysis.
Treatment with C3 exoenzyme
hDPSCs (1×106) at the third passage were
inoculated into the wells of 6-well plates and C3 exoenzyme at a
final concentration of 50 µg/ml was added. Following 48 h of
incubation at 37°C, cells were harvested and assessed for the
expression of RhoA, ROCK, Runx2, alkaline phosphatase (ALP),
collagen type I (Col I), Msh homeobox 2 (MSX2), distal-less
homeobox 2 (DLX2), DSPP, dentin matrix protein-1 (DMP-1), bone
sialoprotein (BSP) and OCN at the protein or mRNA level. Cells
cultured in DMEM and not treated with C3 exoenzyme were used as
control.
RT-quantitative PCR (RT-qPCR)
Total RNA was extracted hDPSCs and hBMSCs using a
TRIzol kit, according to the manufacturer's protocols. The purity
and quantity of RNA were determined using a UV spectrophotometer.
The RT reaction was performed with 200 ng RNA in a total volume of
10 µl containing 2 µl (2 µg) RNA, 2 µl dNTP (500 µg/ml) and 2 µl
MgCl2 (50 mM), and the resulting complementary DNA was
quantified using a one-step PCR kit, according to the
manufacturer's protocols, and the primers listed in Table I in a total volume of 25 µl. Human
GADPH (Hs03929097_g1), was used as an internal control. The cycling
conditions performed were the following: Pre-denaturation at 95°C
for 5 min, followed by 40 cycles of 30 sec at 95°C, 30 sec at 60°C
and 30 sec at 72°C. qPCR was performed using 30 cycles of
denaturation at 95°C for 30 sec, annealing at 51°C for 20 sec and
extension at 75°C for 30 sec. Experiments were performed in
triplicate and the mean value was calculated for each case. The
data were assessed using RQ Manager software (version 1.2.1.;
Applied Biosystems; Thermo Fisher Scientific, Inc.). Relative
expression was calculated by using the 2−ΔΔCq method and
obtaining the fold change value, according to previously described
protocol (17).
 | Table I.Reverse transcription-quantitative
polymerase chain reaction primers. |
Table I.
Reverse transcription-quantitative
polymerase chain reaction primers.
| Gene of
interest | Direction | Primer sequence
(5′-3′) | Expected product
size (bp) |
|---|
| Alkaline
phosphatase | Forward |
CTCGTTGACCACCTGGAAGAGCTTCAAACCG | 168 |
|
| Reverse |
GGTCCGTCACGTTGTTCCTGTTCAGC |
|
| Collagen type
I | Forward |
AGGGCTCCAACGAGATCGAGATCCG | 223 |
|
| Reverse |
TACAGGAAGCAGACAGGGCCAACGTCG |
|
| Msh homeobox 2 | Forward |
CATCCCAGCTTCTAGCCTTC | 103 |
|
| Reverse |
GCAGCCATTTTCAGCTTTTC |
|
| Distal-less
homeobox 2 | Forward |
GGCTCCTACCAGTCCCACG | 133 |
|
| Reverse |
CGTTGTTGACCGGAGACGAA |
|
| Dentin
sialophosphoprotein | Forward |
GTACCCAAACAACACAGGACA | 114 |
|
| Reverse |
AATCAGGGATTAGAAACCGAAG |
|
| Dentin matrix
protein-1 | Forward |
CATGACACCTTTGGAGACGAA | 145 |
|
| Reverse |
CTTGGGCACTGTTTTCTTGAG |
|
| Bone
sialoprotein | Forward |
GCAGTTAGCAATACTACAGGCA | 111 |
|
| Reverse |
GACCATTTATCAAGCAGTACACC |
|
| Osteocalcin | Forward |
CATGAGAGCCCTCACA | 315 |
|
| Reverse |
AGAGCGACACCCTAGAC |
|
| GAPDH | Forward |
AGCCACATCGCTCAGACA | 314 |
|
| Reverse |
TGGACTCCACGACGTACT |
|
Western blot analysis
Following different treatments, hDPSCs and hBMSCs
were lysed with RIPA lysis buffer that contained protease and
phosphatase inhibitors. The supernatants were collected following
centrifugation at 7,000 × g at 4°C for 20 min. The protein was
quantified using the BCA method, 20 µg protein was loaded per lane
and separated using 12% SDS-PAGE, transferred to a PVDF membrane.
The membranes were blocked with 2% BSA (Beyotime Institute of
Biotechnology) at room temperature for 1 h, then the target
proteins were detected by incubating the membrane with primary
antibodies against DSPP, osteocalcin, Runx2, RhoA, ROCK and GAPDH
(all 1:100) at 4°C overnight, and horseradish peroxidase-labeled
goat anti-rabbit secondary and peroxidase-conjugated goat
anti-mouse antibodies (both 1:2,000) at 37°C for 1 h. Visualization
was achieved with a chemiluminescence kit. The intensity of blot
signals was quantitated using Quantity one analysis software
(version 4.62; Bio-Rad Laboratories, Inc.).
Statistical analysis
All data are expressed as the mean ± standard error
of the mean obtained from at least three independent experiments.
Statistical comparisons between groups were assessed by using the
Student's t-test. P<0.05 was considered to indicate a
statistically significant difference. Data were analyzed using SPSS
software (version 10.1; SPSS, Inc., Chicago, IL, USA).
Results
hDPSCs are spindle-shaped, and
positive for MSC and endothelial cell markers
As indicated in Fig.
1, the primary hDPSCs were spindle-shaped and fibrous-like
after 3 days of culture in vitro. The culture reached 80%
confluence in 7–8 days. As indicated in Fig. 2, flow cytometry analysis revealed
that >95% of the cells were positive for MSC markers CD29, CD90,
CD105, and for endothelial cell marker CD146, whereas <5% were
positive for hematopoietic stem cell markers CD34 and CD45. These
results suggested that the purity of the hDPSCs was high enough for
the subsequent experiments.
DSPP protein expression increases
throughout the differentiation of hDPSCs into odontoblasts
The differentiation of hDPSCs into odontoblast was
investigated by determining the protein expression levels of DSPP.
Results indicated that, following treatment with mineralization
induction solution, the protein expression levels of DSPP in hDPSCs
were increased in a time-dependent manner, and at 7, 14 and 21
days, expression levels were significantly increased compared with
0 days (P<0.01; Fig. 3). These
findings suggested that odontoblast differentiation may have been
induced in the hDPSCs.
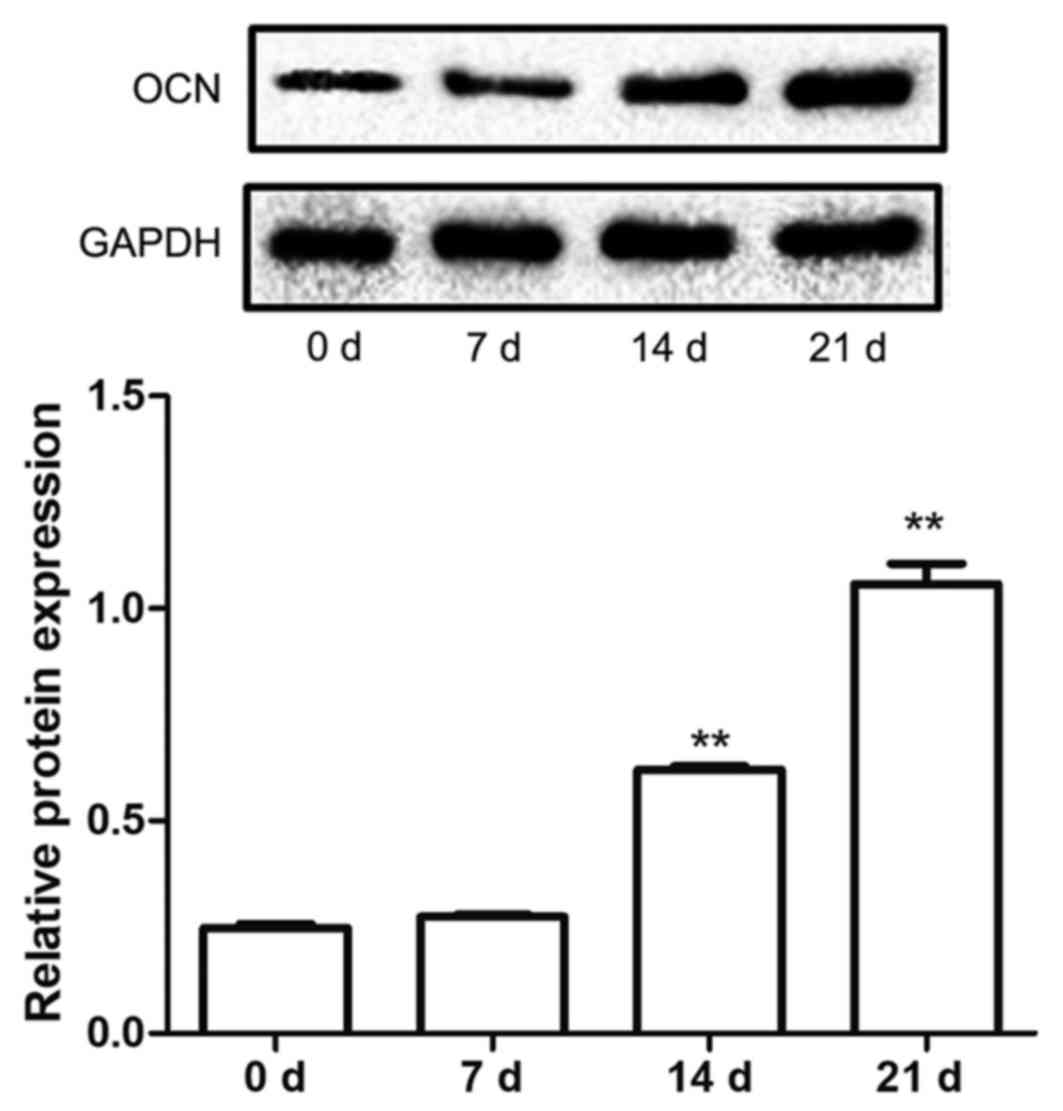
OCN protein expression increases
throughout the differentiation of hBMSCs into osteoblast
The protein expression levels of OCN in hBMSCs
cultured in mineralization solution were also examined in the
present study. The data demonstrated that the protein expression
levels of OCN were significantly upregulated at 14 and 21 days
after induction compared with 0 days (P<0.01; Fig. 4), which indicated that mineralization
may have induced hBMSCs to differentiate into osteoblasts.
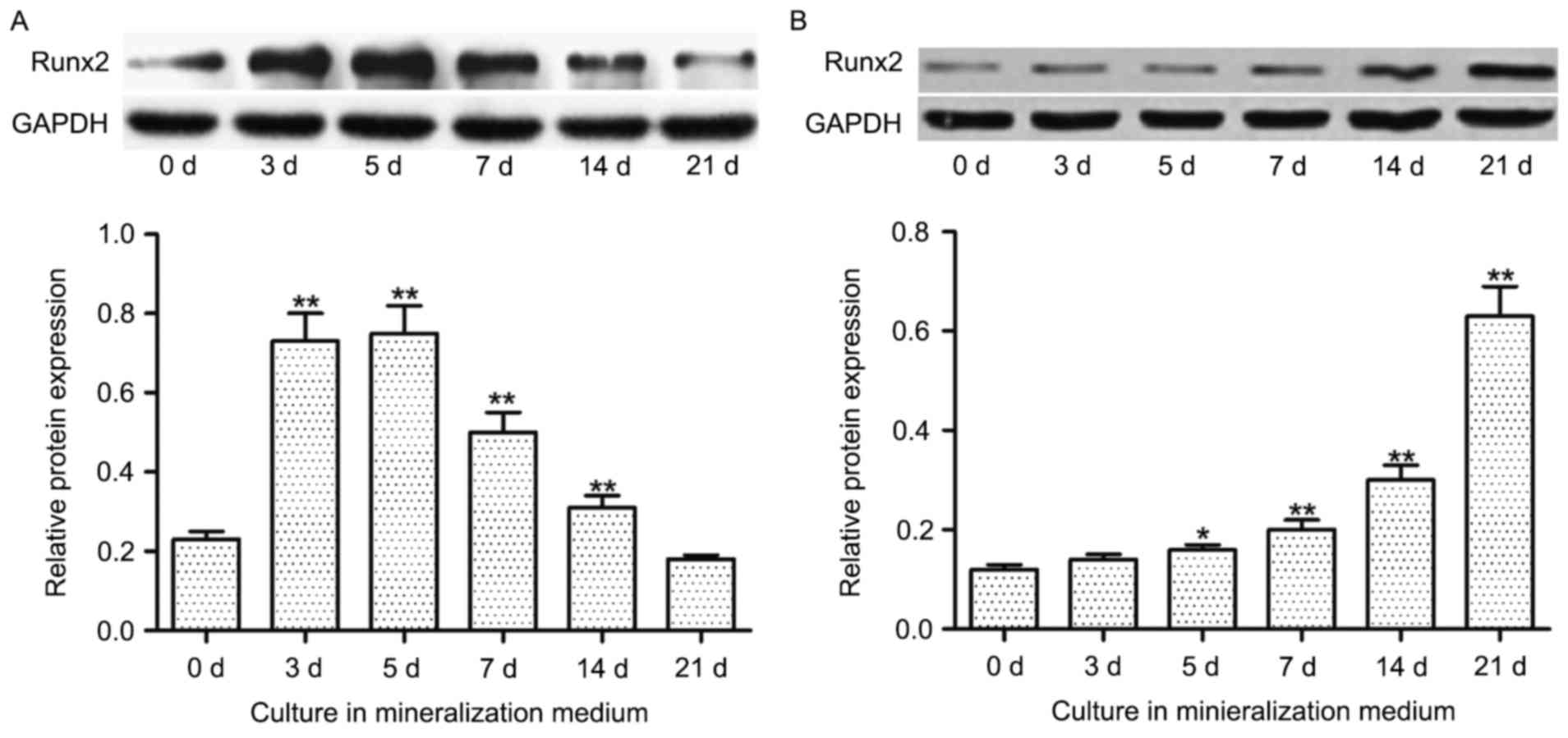
Runx2 protein expression decreases in
hDPSCs and increases in hBMSCs during odontoblast and osteoblast
differentiation, respectively
In hDPSCs, increased incubation time in
mineralization medium resulted in increased protein expression
levels of Runx2 until day 5, after which the expression levels
declined. Furthermore, significantly increased expression levels of
Runx2 protein were indicated at 3, 5, 7 and 14 days in culture
compared with 0 days in culture (P<0.01; Fig. 5A). Conversely, the expression of
Runx2 protein in hBMSCs was increased in a time-dependent manner,
with a significant increase in expression indicated at 5
(P<0.05), 7 (P<0.01), 14 (P<0.01) and 21 (P<0.01) days
compared with 0 days (Fig. 5B).
C3 exoenzyme incubation decreased
Runx2, Rho and ROCK protein expression in hDPSCs
To examine whether the Rho/ROCK signaling pathway is
involved in regulating Runx2 expression, hDPSCs were treated with
the pathway inhibitor, C3 exoenzyme. Results indicated that
compared with the control, hDPSCs treated with the inhibitor
exhibited significantly decreased protein expression levels of
RhoA, ROCK and Runx2 (P<0.01; Fig.
6A). Furthermore, following mineralization induction, Runx2
protein expression levels in the inhibitor-treated hDPSCs were
significantly decreased at 7 and 14 days compared with that in the
control (P<0.01; Fig. 6B).
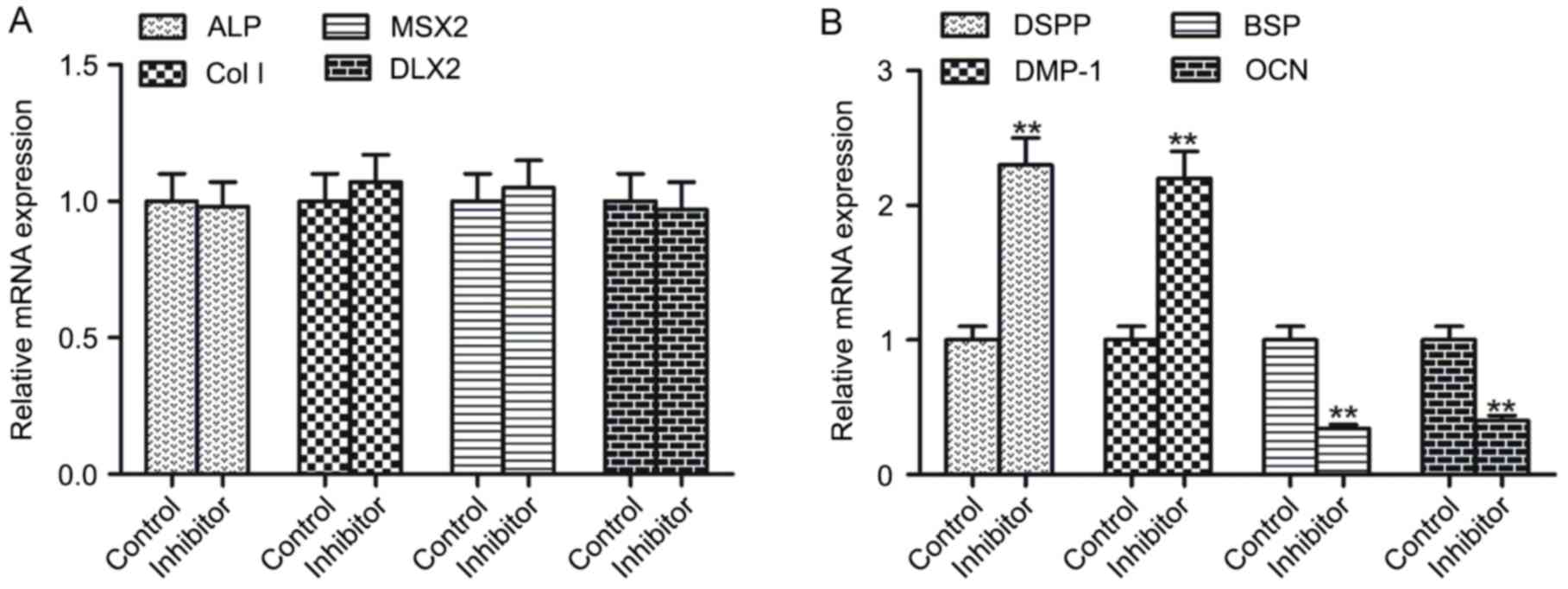
C3 exoenzyme incubation alters the
mRNA expression of late mineralization genes
The present study evaluated the impact of the
Rho/ROCK signaling pathway on the mRNA expression levels of early
and late mineralization genes. Compared with the control, the mRNA
expression levels of early mineralization genes ALP, Col I, MSX2
and DLX2 were not significantly different in inhibitor-treated
hDPSCs (Fig. 7A). However, the mRNA
expression levels of late mineralization genes, DSPP and DMP-1,
were significantly upregulated in inhibitor-treated hDPSCs, whereas
BSP and OCN were significantly downregulated compared with the
controls (P<0.01; Fig. 7B).
Discussion
Research has demonstrated that DPSCs may be isolated
and cultured in vitro (24).
DPSCs form dentin-like complex structures on the fracture surfaces
in the teeth of immunodeficient nude mice or dogs where DPSC-seeded
stents were implanted (25), which
suggests that, under certain conditions, DPSCs may differentiate to
form the pulp-dentin complex and may be used as seed cells for pulp
tissue regeneration and dentin repair. To elucidate the molecular
regulation of odontoblast differentiation in hDPSCs, the expression
of genes involved in the differentiation and impact of the Rho/ROCK
signaling pathway were investigated in the present study. To
accompany these, hDPSCs were isolated with high purity according to
microscopy and flow cytometry analysis. Furthermore, the present
results also demonstrated that the isolated hDPSCs may
differentiate toward odontoblasts in vitro as the expression
of DSPP, an odontoblast-specific protein (19), was significantly increased over time
when the hDPSCs were cultured in mineralization induction medium.
Additionally, results indicated that hBMSCs may be induced by the
mineralization medium to differentiate into osteoblasts, as the
protein expression levels of osteoblast-specific protein, OCN, were
significantly upregulated following 14 and 21 days in culture
compared with 0 days. OCN is vitamin K-dependent small molecule
protein, and has high affinity to calcium and hydroxyapatite
(26). Therefore, OCN is regarded as
late stage marker for bone formation (26).
Runx2, a member of the Runt family, is expressed in
osteogenic and odontogenic mesenchymal cells during the embryonic
period, and has an important role in bone and tooth development
(23,27). It is capable of binding specific
sites on promoter sequences to regulate the transcription of
downstream genes and, subsequently, the development of tissues and
organs (23,27). Studies have revealed that Runx2 may
regulate the differentiation of BMSCs into osteoblasts, which has
an important role in bone tissue repair, and it is continuously and
highly expressed during osteogenic differentiation (17,19).
During the differentiation of hBMSCs to odontoblasts, Runx2
expression increases in the early stage and declines in the late
stage (17,19), which suggests that the expression
pattern of Runx2 may be a key factor that impacts the
differentiation direction to odontoblast or to osteoblast. The
present study indicated that, following mineralization induction,
Runx2 protein expression patterns in hDPSCs and hBMSCs were
different. In hDPSCs, Runx2 protein expression levels increased in
the early differentiation period, peaked on day 5 and then declined
in late differentiation period. However, in hBMSCs, Runx2 protein
expression levels increased over the full duration of the
experimental period and in the late mineralization stage. These
results are consistent with earlier studies (24,25),
which implies that regulating Runx2 expression may influence the
direction of differentiation in these stem cells.
The Rho/ROCK signaling pathway is associated with
cell proliferation and differentiation, apoptosis, the cell cycle,
cell polarity, the cytoskeleton and vasoconstriction (23,27,28). The
signaling pathway also regulates the differentiation of MSCs to
adipocytes, osteoblasts and chondrocytes (11,13). The
upstream protein Rho GTPase belongs to the Ras superfamily,
including RhoA, RhoB and RhoC subfamilies (28,29). Rho
GTPase exists in all eukaryotic organisms and has roles in cell
migration, movement, proliferation and differentiation (28,29). Rho
GTPase has a number of downstream effector proteins, including
fatty kinase and serine/threonine kinases (30). Among them, ROCK has been intensively
studied. Activated RhoA may bind directly to the C-terminus of ROCK
to activate it (31). Activated ROCK
phosphorylates myosin and its regulatory proteins to regulate
changes of and contract the cytoskeleton (31). Therefore, RhoA/ROCK signaling is
important for cytoskeleton reorganization, cell migration,
movement, contraction and proliferation (32). Furthermore, the RhoA/ROCK signaling
influences cell polarity and morphology by regulating cytoskeletal
polymerization, thereby affecting the final differentiation of a
cell (32). -Studies have indicated
that the regulation of the RhoA/ROCK signaling pathway induces the
differentiation of cells toward osteoblasts (33,34). For
example, through activating the ROCK signaling pathway, BMSCs were
prompted to differentiate into osteoblasts with increased Runx2 and
osteocalcin expression observed (35). However, an alternative study
indicated that inhibition of the ROCK signaling pathway decreased
LIPUS-induced differentiation of MSCs to osteoblasts and
downregulated Runx2 expression (36). These studies demonstrate that the
RhoA/ROCK signaling pathway may have regulatory roles on Runx2
expression in osteoblasts and suggests that the pathway may also
have a role in regulating Runx2 expression in odontoblast
differentiation. In the present study, the RhoA/ROCK signaling
pathway inhibitor, C3 exoenzyme (37), was used to analyze the impact of the
signaling pathway in hDPSCs. Results demonstrated that C3 exoenzyme
treatment significantly downregulated the protein expression levels
of RhoA, ROCK and Runx2 in hDPSCs. Furthermore, the protein
expression levels of Runx2 were also significantly decreased
following culture for 7 and 14 days in mineralization medium. These
findings suggest that the RhoA/ROCK signaling pathway may have a
regulatory role on Runx2 expression in hDPSCs during dentin
differentiation.
To provide further insights on how the RhoA/ROCK
signaling pathway regulates the differentiation of odontoblasts,
the impact of the pathway on the expression of odontoblast
phenotype proteins was explored. Results indicated that the pathway
inhibitor did not affect the transcription of early mineralization
genes, suggesting that the pathway is not involved in the early
stage of odontoblast differentiation. However, inhibition of the
pathway led to significant upregulation of DSPP and DMP-1, and
significant downregulation of BSP and OCN late mineralization
genes. DSPP and DMP-1 are only expressed in odontoblasts and their
promoters contain Runx2 binding sites (38,39). The
expression of DSPP and DMP-1 has been demonstrated to be regulated
by Runx2 to direct the differentiation toward odontoblasts
(40,41). Furthermore, OCN and BSP are
osteoblast-specific markers, and their promoters contain Runx2
binding sites (38,39). Upregulating the expression of OCN and
BSP was previously indicated to significantly promote the
differentiation of MSCs into osteoblasts (40,41). The
present data indicates that the inhibition of the Rho/ROCK
signaling pathway would not impact odontoblast differentiation in
the early stage of odontoblast differentiation, but promote the
differentiation in the late stage. This suggests that the inhibitor
only functions in the late stage.
In conclusion, the present work indicated that the
expression of Runx2 initially increases and then decreases during
differentiation of hDPSCs into odontoblasts, whereas in hBMSCs,
Runx2 is consistently and highly expressed during osteoblast
differentiation. Furthermore, the present findings suggested that
inhibition of the RhoA/ROCK signaling pathway with C3 exoenzyme
significantly downregulated the expression of Runx2, but had no
significant impact on odontoblast differentiation in the early
stage. However, differentiation in the late stage appeared to be
promoted, suggesting that it may be possible to direct the
differentiation of hDPSCs into odontoblasts by inhibiting the
RhoA/ROCK signaling pathway to downregulate the expression of
Runx2.
References
|
1
|
Corbella S, Taschieri S, Samaranayake L,
Tsesis I, Nemcovsky C and Del Fabbro M: Implant treatment choice
after extraction of a vertically fractured tooth. A proposal for a
clinical classification of bony defects based on a systematic
review of literature. Clin Oral Implants Res. 25:946–956. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ortega-Martinez J, Pérez-Pascual T,
Mareque-Bueno S, Hernández-Alfaro F and Ferrés-Padró E: Immediate
implants following tooth extraction. A systematic review. Med Oral
Patol Oral Cir Bucal. 17:e251–e261. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gonda T, Ikebe K, Ono T and Nokubi T:
Effect of magnetic attachment with stress breaker on lateral stress
to abutment tooth under overdenture. J Oral Rehabil. 31:1001–1006.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhao Y, Wang L, Jin Y and Shi S: Fas
ligand regulates the immunomodulatory properties of dental pulp
stem cells. J Dent Res. 91:948–854. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ledesma-Martinez E, Mendoza-Núñez VM and
Santiago-Osorio E: Mesenchymal stem cells derived from dental pulp:
A review. Stem Cells Int. 2016:47095722016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Nuti N, Corallo C, Chan BM, Ferrari M and
Gerami-Naini B: Multipotent differentiation of human dental pulp
stem cells: A literature review. Stem Cell Rev. 12:511–523. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kabir R, Gupta M, Aggarwal A, Sharma D,
Sarin A and Kola MZ: Imperative role of dental pulp stem cells in
regenerative therapies: A systematic review. Niger J Surg. 20:1–8.
2014.PubMed/NCBI
|
|
8
|
Shi S, Robey PG and Gronthos S: Comparison
of human dental pulp and bone marrow stromal stem cells by cDNA
microarray analysis. Bone. 29:532–539. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Téclès O, Laurent P, Zygouritsas S, Burger
AS, Camps J, Dejou J and About I: Activation of human dental pulp
progenitor/stem cells in response to odontoblast injury. Arch Oral
Biol. 50:103–108. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang J, Dong H, Wang W and Gu JD:
Reverse-transcriptional gene expression of anammox and
ammonia-oxidizing archaea and bacteria in soybean and rice paddy
soils of Northeast China. Appl Microbiol Biotechnol. 98:2675–2686.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Nosrat IV, Smith CA, Mullally P, Olson L
and Nosrat CA: Dental pulp cells provide neurotrophic support for
dopaminergic neurons and differentiate into neurons in vitro;
implications for tissue engineering and repair in the nervous
system. Eur J Neurosci. 19:2388–2398. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Király M, Porcsalmy B, Pataki A, Kádár K,
Jelitai M, Molnár B, Hermann P, Gera I, Grimm WD, Ganss B, et al:
Simultaneous PKC and cAMP activation induces differentiation of
human dental pulp stem cells into functionally active neurons.
Neurochem Int. 55:323–332. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang W, Walboomers XF, Shi S, Fan M and
Jansen JA: Multilineage differentiation potential of stem cells
derived from human dental pulp after cryopreservation. Tissue Eng.
12:2813–2823. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Petrovic V and Stefanovic V: Dental
tissue-new source for stem cells. ScientificWorldJournal.
9:1167–1177. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sun SS, Zhang L, Yang J and Zhou X: Role
of runt-related transcription factor 2 in signal network of tumors
as an inter-mediator. Cancer Lett. 361:1–7. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wojtowicz AM, Templeman KL, Hutmacher DW,
Guldberg RE and Garcia AJ: Runx2 overexpression in bone marrow
stromal cells accelerates bone formation in critical-sized femoral
defects. Tissue Eng Part A. 16:2795–2808. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ducy P, Zhang R, Geoffroy V, Ridall AL and
Karsenty G: Osf2/Cbfa1: A transcriptional activator of osteoblast
differentiation. Cell. 89:747–754. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Xu N, Guan L, He Y, Li D, Han B and Yang
F: Dynamic expression of Runx2 during the differnetiation of
odontoblsts and osteoblasts. Chin J Conser Dent. 23:209–13.
2013.(In Chinese).
|
|
20
|
Nasu K, Tsuno A, Yuge A and Narahara H:
Mevalonate-Ras homology (rho)/rho-associated coiled-coil-forming
protein kinase (ROCK)-mediated signaling pathway as a therapeutic
target for the treatment of endometriosis-associated fibrosis. Curr
Signal Transduct Ther. 5:141–148. 2010. View Article : Google Scholar
|
|
21
|
Niu LN, Sun JQ, Li QH, Jiao K, Shen LJ, Wu
D, Tay F and Chen JH: Intrafibrillar-silicified collagen scaffolds
enhance the osteogenic capacity of human dental pulp stem cells. J
Dent. 42:839–849. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Gronthos S, Arthur A, Bartold PM and Shi
S: A method to isolate and culture expand human dental pulp stem
cells. Methods Mol Biol. 698:107–121. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bruderer M, Richards RG, Alini M and
Stoddart MJ: Role and regulation of RUNX2 in osteogenesis. Eur Cell
Mater. 28:269–286. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gronthos S, Mankani M, Brahim J, Robey PG
and Shi S: Postnatal human dental pulp stem cells (DPSCs) in vitro
and in vivo. Proc Natl Acad Sci USA. 97:13625–13630. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
He WX, Niu ZY, Zhao SL, Jin WL, Gao J and
Smith AJ: TGF-beta activated Smad signalling leads to a
Smad3-mediated down-regulation of DSPP in an odontoblast cell line.
Arch Oral Biol. 49:911–918. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Rocha ÉD, de Brito NJ, Dantas MM, Silva
Ade A, Almeida Md and Brandão-Neto J: Effect of zinc
supplementation on GH, IGF1, IGFBP3, OCN, and ALP in
non-zinc-deficient children. J Am Coll Nutr. 34:290–299. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Vimalraj S, Arumugam B, Miranda PJ and
Selvamurugan N: Runx2: Structure, function, and phosphorylation in
osteoblast differentiation. Int J Biol Macromol. 78:202–208. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Feng Y, LoGrasso PV, Defert O and Li R:
Rho kinase (ROCK) inhibitors and their therapeutic potential. J Med
Chem. 59:2269–2300. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Julian L and Olson MF: Rho-associated
coiled-coil containing kinases (ROCK): Structure, regulation, and
functions. Small GTPases. 5:e298462014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Tripathi BK, Grant T, Mertins P, Qian X,
Wang D, Papageorge AG, Carr SA and Lowy DR: AKT positively
regulates Rho-GTP by attenuating the GAP activity of the DLC1 tumor
suppressor: A mechanistic study with translational implications.
Cancer Res. 76 14 Suppl:S43772016. View Article : Google Scholar
|
|
31
|
Menna L, Pablo and Cardama: PROTEINAS DE
LIGACAO AO GTP [Descritor de assunto]. Front Psychol. 6:623–626.
2015.PubMed/NCBI
|
|
32
|
Nour-Eldine W, Ghantous CM, Zibara K, et
al: Adiponectin Attenuates Angiotensin II-Induced Vascular Smooth
Muscle Cell Remodeling through Nitric Oxide and the RhoA/ROCK
Pathway[J]. Frontiers in Pharmacology. 7:862016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Prowse PD, Elliott CG, Hutter J and
Hamilton DW: Inhibition of Rac and ROCK signalling influence
osteoblast adhesion, differentiation and mineralization on titanium
topographies. PLoS One. 8:e588982013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Tang Z, Shi D, Jia B, Chen J, Zong C, Shen
D, Zheng Q, Wang J and Tong X: Exchange protein activated by cyclic
adenosine monophosphate regulates the switch between adipogenesis
and osteogenesis of human mesenchymal stem cells through increasing
the activation of phosphatidylinositol 3-kinase. Int J Biochem Cell
Biol. 44:1106–1120. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Santos A, Bakker AD, de Blieck-Hogervorst
JM and Klein-Nulend J: WNT5A induces osteogenic differentiation of
human adipose stem cells via rho-associated kinase ROCK.
Cytotherapy. 12:924–932. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kusuyama J, Bandow K, Shamoto M, Kakimoto
K, Ohnishi T and Matsuguchi T: Low intensity pulsed ultrasound
(LIPUS) influences the multilineage differentiation of mesenchymal
stem and progenitor cell lines through ROCK-Cot/Tpl2-MEK-ERK
signaling pathway. J Biol Chem. 289:10330–10344. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Li G, Liu L, Shan C, Cheng Q, Budhraja A,
Zhou T, Cui H and Gao N: RhoA/ROCK/PTEN signaling is involved in
AT-101-mediated apoptosis in human leukemia cells in vitro and in
vivo. Cell Death Dis. 5:e9982014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Chen S, Gluhak-Heinrich J, Wang YH, Wu YM,
Chuang HH, Chen L, Yuan GH, Dong J, Gay I and MacDougall M: Runx2,
osx, and dspp in tooth development. J Dent Res. 88:904–909. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Li S, Kong H, Yao N, Yu Q, Wang P, Lin Y,
Wang J, Kuang R, Zhao X and Xu J: The role of runt-related
transcription factor 2 (Runx2) in the late stage of odontoblast
differentiation and dentin formation. Biochem Biophys Res Commun.
410:698–704. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Gao RT, Zhan LP, Meng C, Zhang N, Chang
SM, Yao R and Li C: Homeobox B7 promotes the osteogenic
differentiation potential of mesenchymal stem cells by activating
RUNX2 and transcript of BSP. Int J Clin Exp Med. 8:10459–10470.
2015.PubMed/NCBI
|
|
41
|
Zhang X, Ma Y, Fu X, Liu Q, Shao Z, Dai L,
Pi Y, Hu X, Zhang J, Duan X, et al: Runx2-modified adipose-derived
stem cells promote tendon graft integration in anterior cruciate
ligament reconstruction. Sci Rep. 6:190732016. View Article : Google Scholar : PubMed/NCBI
|