Introduction
Intervertebral disc degeneration (IDD) is a major
risk factor for lower back pain that is a worldwide problem. It
badly affects human life and causes a serious socioeconomic burden
(1–4). An imbalance between anabolic and
catabolic genes expressed by chondrocytes that belong to the
nucleus pulposus (NP) result in IDD. The etiology of IDD is very
complicated. Numerous cellular events, including cell apoptosis,
inflammation, autophagy and increased matrix catabolism, have been
confirmed to be involved in the progress of IDD (5,6). It has
previously been reported that NP cells, which share the common
features of chondrocytes, play critical roles in maintaining
integrity of intervertebral discs (IVDs) (7,8). One of
the major features of IDD is the loss of proteoglycan (PG) content
from IVDs. Lipopolysaccharide (LPS), a strong promoter of
inflammation, can reduce PG content and cause the occurrence of IDD
(9,10). Furthermore, pro-inflammatory factors
such as interleukin (IL)-1β, IL-6 and tumor necrosis factor (TNF)-α
also serve critical functions in IDD. However, the underlying
molecular and cellular mechanisms of IDD remain unclear.
MicroRNAs (miRs) are small (20–22 nucleotides in
length), non-coding, single-stranded RNAs that serve important
functions in post-transcriptionally regulating gene expression
(11,12). Previous studies have demonstrated
that miRs are critical in cancer progress, as well as in
inflammatory, neurodegenerative and most degenerative disorders
(7,8). Previous reports have demonstrated that
miRs serve important functions in degenerative disc diseases, such
as IDD (13–16). It has also been indicated that
miR-148a is downregulated in IDD compared with spinal cord injury
(17), but the underlying mechanisms
are still unknown.
Thus, the aim of the present study was to verify the
expression and investigate the role of miR-148a in IDD and explore
the underlying mechanisms.
Materials and methods
Materials
LPS was purchased from Sigma-Aldrich (Merck KGaA,
Darmstadt, Germany). ELISA kits for TNF-α (cat no. E-CL-M0047c),
IL-1β (cat no. E-EL-M0037c) and IL-6 (cat no. E-EL-M0044c) were
obtained from Elabscience Biotechnology Co., Ltd. (Wuhan, China).
Antibodies against p38 (cat no. 9212)/p-p38 (cat no. 4631) and
GAPDH (cat no. 97166) and secondary antibodies (anti-rabbit IgG,
horseradish peroxidase (HRP)-linked antibody; cat no. 7074;
anti-mouse IgG HRP-linked antibody; cat no. 7076) were supplied by
Cell Signaling Technology, Inc. (Danvers, MA, USA). miR-148a mimic
(cat no. miR10000243-1-5)/inhibitor (cat no. miR20000243-1-5) were
purchased from Guangzhou Ribobio Co., Ltd. (Guangzhou, China); all
other chemicals and reagents were purchased from Sinopharm Chemical
Reagent Co., Ltd. (Shanghai, China).
Patients
The study was approved by the Human Ethics
Committees Review Board at the Affiliated Hospital of Xuzhou
Medical College (Xuzhou, China) and written informed consent was
obtained from each patient prior to enrollment. A total of 30
patients with IDD and 30 healthy volunteers were enrolled from the
Department of Spinal Surgery at the Affiliated Hospital of Xuzhou
Medical College (Xuzhou, China) from January 2014 to January 2015.
Peripheral blood mononuclear cells (PBMCs) were obtained from the
peripheral blood of patients and healthy volunteers as described
previously (18). Peripheral blood
was centrifuged at 650 × g for 25 min at room temperature. Middle
phases containing Human PBMCs were further centrifuged (650 × g for
10 min at room temperature) by density using Lymphoprep (Stemcell
Technologies, Inc., Beijing, China) (19).
Cell culture
Human NP cells were purchased from ScienCell
Research Laboratories, Inc. (San Diego, CA, USA; cat. no. 4800) and
cultured in Dulbecco's Modified Eagle's medium (DMEM; Gibco; Thermo
Fisher Scientific, Inc., Waltham, MA, USA) containing 10% fetal
bovine serum (Gibco), 1% L-glutamine (Gibco), 100 mg/ml
streptomycin and 100 U/ml penicillin and then incubated in a 5%
CO2 incubator at 37°C. The culture medium was changed
every 2 days and cells were passaged until they reached 90%
confluence.
Cell treatment
Human NP cells (5×104 cells per well)
were added to a 6-well plate and transiently transfected with
miR-148a mimic (50 nM)/inhibitor (100 nM) using Lipofectamine with
Plus reagent (Thermo Fisher Scientific, Inc.) and control
transfections were performed lacking miR-148a. The nucleotide
sequences used were as follows: miR-148a mimic forward,
5′-UCAGUGCAUGACAGAACUUGG-3′ and reverse,
5′-AAGTTCUGUCAUGCACUGAUU-3′; NC forward,
5′-UUCUCCGAACGUGUCACGUTT-3′ and reverse,
5′-ACGUGACACGUUCGGAGAATT-3′; and miR-148a inhibitor,
5′-ACAAAGUUCUGUAGUGCACUGA-3′. At 24 h following the transfection,
5×104 cells were stimulated with LPS (10 µM) in
serum-free medium (DMEM) for 24 h at 37°C under 5% CO2.
The suspensions were then centrifuged at 1,000 × g for 10 min at
4°C and collected 24 h after the initiation of each treatment.
Cells were then harvested for subsequent experimentation. The
groups were divided into the following: A control group (NP cells
without any treatment); an NC group (NP cells transfected with NC
oligonucleotides and stimulated with LPS); a mimic group (NP cells
transfected with miR148a mimic and then stimulated with LPS) and an
inhibitor group (NP cells transfected with miR148a inhibitor and
then stimulated with LPS).
RNA isolation and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from human PBMCs and human
NP cells using TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocol. Each
sample (1 µg RNA) was then reverse-transcribed to cDNA using a
reverse-transcription (RT) reagent kit (Takara Biotechnology Co.,
Ltd., Dalian, China). A 7500 real-time PCR instrument (Applied
Biosystems; Thermo Fisher Scientific, Inc.) was used to quantify
miR-148a, TNF-α, IL-1β and IL-6 mRNA levels. Amplification
conditions were as follows: 16°C for 30 min, 42°C for 30 min, 85°C
for 5 min and hold at 4°C. qPCR was performed using the QuantiTect
SYBR-Green PCR kit (Takara Biotechnology Co., Ltd.). All reactions
were performed in triplicate with the following conditions: 95°C
for 10 min, followed by 37 cycles at 95°C for 15 sec and 72°C for
30 sec. All quantitative miRNA data were normalized to U6
expression and quantitative mRNA data were normalized to GAPDH. The
comparative Cq method was used for the relative amounts of each
transcript (20). The primers for
qPCR were as presented in Table
I.
 | Table I.Primer sequences for polymerase chain
reaction analysis. |
Table I.
Primer sequences for polymerase chain
reaction analysis.
| Gene | Forward primer
(5′-3′) | Reverse primer
(5′-3′) |
|---|
| TNF-α |
CCTGTCTCTTCCTACCCAACC |
GCAGGAGTGTCCGTGTCTTC |
| IL-1β |
CTGTGACTCGTGGGATGATG |
AGGGATTTTGTCGTTGCTTG |
| IL-6 |
GTGCTCCTGGTATTGCTGGT |
GGCTCCTCGTTTTCCTTCTT |
| miR-148a |
ATGCTCAGTGCACTACAGAA | GTGCAGGGTCCGAGGT |
| U6 |
CTCGCTTCGGCAGCACA |
AACGCTTCACGAATTTGCGT |
| GAPDH |
CTTTGGTATCGTGGAAGGACTC |
GTAGAGGCAGGGATGATGTTCT |
Western blot analysis
In brief, total cellular protein from NP cells was
extracted using RIPA buffer (Cell Signaling Technology, Inc.) and
separated by SDS-PAGE analysis. Sample protein concentrations were
determined using the BCA protein assay (Thermo Fisher Scientific,
Inc.). Each lane was loaded with 25 µg protein and resolved using a
10% SDS-PAGE gel and transferred to a polyvinylidene fluoride
membrane. Membranes were then blocked using Tris-buffered saline
with 0.1% Tween-20 (TBST) containing 5% non-fat milk for 1 h at
room temperature. Membranes were subsequently incubated overnight
at 4°C with primary antibodies against p-p38 and p38 (1:1,000) or
GAPDH (1:2,000). Membranes were incubated with the correspondent
secondary antibody (1:5,000) at room temperature for 2 h. Protein
bands were observed with enhanced chemiluminescence and imaged.
ELISA assay
In order to determine levels of TNF-α, IL-1β and
IL-6 in the NP cellular supernatant, ELISA was performed with ELISA
kits, following the manufacturer's protocol.
Statistical analysis
Data are presented as the mean ± SD. All statistical
analyses were performed using SPSS 17.0 statistical software (SPSS,
Inc., Chicago, IL, USA). A t-test or one-way analysis of variance
followed by Tukey's test was used to evaluate differences between
groups. P<0.05 was considered to indicate a statistically
significant difference.
Results
Downregulation of miR-148a in IDD
To investigate the potential role of miR-148a in
IDD, the relative expression of miR-148a in PBMCs separated from
IDD patients and control subjects was compared. miR-148a expression
level was also detected in NP cells with or without LPS
stimulation. As shown in Fig. 1A,
the expression level of miR-148a was significantly decreased in
PBMCs of patients with IDD compared with control subjects.
Furthermore, miR-148a expression level in LPS-stimulated NP cells
was significantly lower compared with the untreated NP cells
(Fig. 1B). These data suggested that
miR-148a is downregulated in IDD.
miR-148a affects pro-inflammatory
cytokines levels in LPS-stimulated NP cells
In order to evaluate the effects of miR-148a in IDD,
miR-148a expression was induced in LPS-stimulated NP cells using
miR-148a mimic and inhibited using miR-148a inhibitor, while
negative control (NC) oligonucleotides were used as the control. As
shown in Fig. 1B, compared with the
NP cells transfected with NC oligonucleotides, miR-148a expression
was significantly upregulated by miR-148a mimic transfection and
significantly inhibited by miR-148a inhibitor transfection. Then,
the mRNA expression level and quantity of pro-inflammatory
cytokines TNF-α, IL-1β and IL-6 was measured by RT-qPCR and ELISA,
respectively. The results indicated that, compared with the NC
group, the mRNA expression level of TNF-α, IL-1β and IL-6
significantly decreased in NP cells transfected with miR-148a mimic
(Fig. 2A) and significantly
increased in NP cells transfected with miR-148a inhibitor (Fig. 2B). In addition, the ELISA results
indicated that, compared with the NC group, cellular supernatant
TNF-α, IL-1β and IL-6 levels significantly decreased in NP cells
transfected with miR-148a mimic and significantly increased in NP
cells transfected with miR-148a inhibitor (Fig. 3). These results indicated that
miR-148a inhibits pro-inflammatory cytokine levels in
LPS-stimulated NP cells. This suggests that miR-148a may inhibit
pro-inflammatory factors released by IVD cells.
miR-148a affects activation of the
p38/mitogen-activated protein kinase (MAPK) pathway in
LPS-stimulated NP cells
To investigate the underlying mechanism by which
miR-148a affects the inflammatory response, the involvement of the
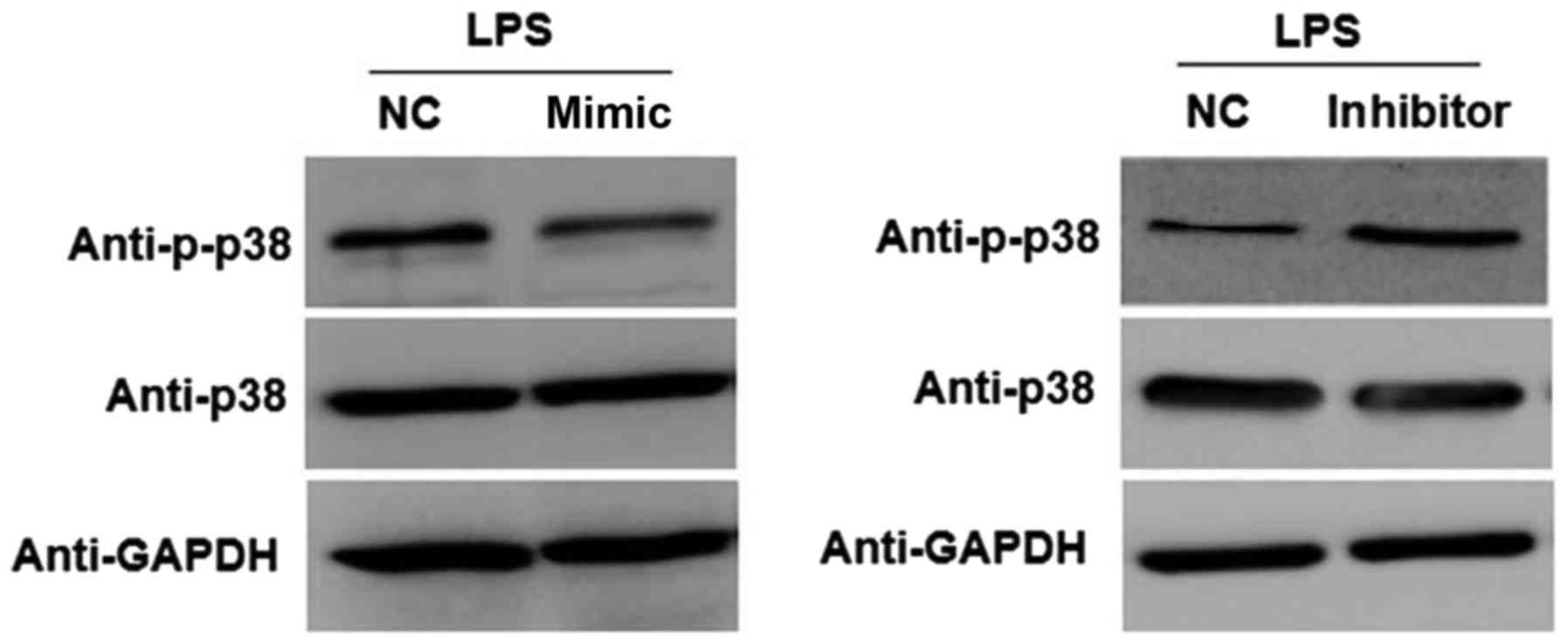
p38/MAPK pathway was evaluated. As shown in Fig. 4, compared with the NC group,
overexpression of miR-148a markedly reduced the protein expression
level of p-p38 in LPS-stimulated NP cells transfected with miR-148a
mimic. Furthermore, transfection with miR-148a inhibitor resulted
in a marked increase in the expression of p-p38 in LPS-stimulated
NP cells compared with the NC group. These results indicated that
miR-148a overexpression inhibits activation of the p38/MAPK pathway
in LPS-stimulated NP cells.
Discussion
Numerous studies have indicated that miRs serve key
functions in the development of IDD (13,14,16,21).
However the expression and role of miR-148a in IDD remains unknown.
In the present study, the role of miR-148a in IDD was investigated
and the pathological links between miR-148a, IDD and inflammatory
pathways were investigated.
In order to evaluate the role of miR-148a in IDD,
the expression level of miR-148a was measured in PBMCs isolated
from IDD patients and control subjects. miR-148a expression level
was also detected in NP cells with or without LPS stimulation. The
findings suggested that miR-148a was significantly downregulated in
IDD patients compared with the healthy controls, as well as in the
LPS-stimulated NP cells compared with non-stimulated cells. Similar
to these results, previous studies on miRNA expression in
degenerative discs have reported that numerous miRNAs are
abnormally expressed in IDD (22–25).
To investigate the underlying mechanism of the role
of miR-148a in IDD, a stable
miR-148a-overexpression/underexpression human NP cell line was
established by transfection with miR-148a mimic/inhibitor and an
IDD cell model was established by LPS stimulation. A previous study
confirmed that inflammation drives the degeneration of IVD
(26). The increased expression
level of pro-inflammatory cytokines secreted by IVD cells is the
major characteristic of IDD and these cytokines promote
extracellular matrix degradation, chemokine production and changes
in IVD cell phenotype (27). Thus,
in the present study, the mRNA expression level of pro-inflammatory
cytokines (TNF-α, IL-1β and IL-6) was evaluated in NP cells. The
results revealed that miR-148a mimic reduced the mRNA expression
level of pro-inflammatory cytokines in LPS-stimulated NP cells and
miR-148a inhibitor presented the opposite effect. Furthermore, the
cellular supernatant levels of TNF-α, IL-1β and IL-6 were detected
by ELISA and the results indicated the same trends as mRNA
expression levels.
Finally, in order to investigate whether the
p38/MAPK pathway is involved in the regulation of inflammatory
responses by miR-148a in IDD, activation of the p38/MAPK pathway
was evaluated in the present study. It was identified that
upregulation of miR-148a in LPS-stimulated NP cells decreased the
phosphorylation of p38, while miR-148a inhibitor increased the
phosphorylation of p38. These results indicated that activation of
the p38/MAPK pathway is associated with the function of miR-148a in
regulating inflammatory response in LPS-stimulated NP cells.
In conclusion, the present findings suggest that
miR-148a serves a function in IDD progress and miR-148a
overexpression suppresses pro-inflammatory factors released by IVD
cells though regulating the p38/MAPK pathway. Thus, miR-148a may
have potential therapeutic applications in the treatment of
IDD.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Millecamps M, Tajerian M, Naso L, Sage EH
and Stone LS: Lumbar intervertebral disc degeneration associated
with axial and radiating low back pain in ageing SPARC-null mice.
Pain. 153:1167–1179. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Vos T, Flaxman AD, Naghavi M, Lozano R,
Michaud C, Ezzati M, Shibuya K, Salomon JA, Abdalla S, Aboyans V,
et al: Years lived with disability (YLDs) for 1160 sequelae of 289
diseases and injuries 1990–2010: A systematic analysis for the
global burden of disease study 2010. Lancet. 380:2163–2196. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gore M, Sadosky A, Stacey BR, Tai KS and
Leslie D: The burden of chronic low back pain: Clinical
comorbidities, treatment patterns and health care costs in usual
care settings. Spine (Phila Pa 1976). 37:E668–E677. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Takatalo J, Karppinen J, Taimela S,
Niinimäki J, Laitinen J, Sequeiros RB, Samartzis D, Korpelainen R,
Näyhä S, Remes J and Tervonen O: Association of abdominal obesity
with lumbar disc degeneration-a magnetic resonance imaging study.
PLoS One. 8:e562442013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kepler CK, Ponnappan RK, Tannoury CA,
Risbud MV and Anderson DG: The molecular basis of intervertebral
disc degeneration. Spine J. 13:318–330. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Risbud MV and Shapiro IM: Role of
cytokines in intervertebral disc degeneration: Pain and disc
content. Nat Rev Rheumatol. 10:44–56. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Teague EM, Print CG and Hull ML: The role
of microRNAs in endometriosis and associated reproductive
conditions. Hum Reprod Update. 16:142–165. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Croce CM: Causes and consequences of
microRNA dysregulation in cancer. Nat Rev Genet. 10:704–714. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ellman MB, Kim JS, An HS, Chen D, KC R, An
J, Dittakavi T, van Wijnen AJ, Cs-Szabo G, Li X, et al: Toll-like
receptor adaptor signaling molecule MyD88 on intervertebral disk
homeostasis: In vitro, ex vivo studies. Gene. 505:283–290. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Iwata M, Ochi H, Asou Y, Haro H, Aikawa T,
Harada Y, Nezu Y, Yogo T, Tagawa M and Hara Y: Variations in gene
and protein expression in canine chondrodystrophic nucleus pulposus
cells following long-term three-dimensional culture. PLoS One.
8:e631202013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lee RC, Feinbaum RL and Ambros V: The
C. elegans heterochronic gene lin-4 encodes small RNAs with
antisense complementarity to lin-14. Cell. 75:843–854. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yu X, Li Z, Shen J, Wu WK, Liang J, Weng X
and Qiu G: MicroRNA-10b promotes nucleus pulposus cell
proliferation through RhoC-Akt pathway by targeting HOXD10 in
intervetebral disc degeneration. PLoS One. 8:e830802013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ohrt-Nissen S, Døssing KB, Rossing M,
Lajer C, Vikeså J, Nielsen FC, Friis-Hansen L and Dahl B:
Characterization of miRNA expression in human degenerative lumbar
disks. Connect Tissue Res. 54:197–203. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu G, Cao P, Chen H, Yuan W, Wang J and
Tang X: MiR-27a regulates apoptosis in nucleus pulposus cells by
targeting PI3K. PLoS One. 8:e752512013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liu H, Huang X, Liu X, Xiao S, Zhang Y,
Xiang T, Shen X, Wang G and Sheng B: miR-21 promotes human nucleus
pulposus cell proliferation through PTEN/AKT signaling. Int J Mol
Sci. 15:4007–4018. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhao B, Yu Q, Li H, Guo X and He X:
Characterization of microRNA expression profiles in patients with
intervertebral disc degeneration. Int J Mol Med. 33:43–50. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Diaz-Morales N, Iannantuoni F,
Escribano-Lopez I, Bañuls C, Rovira-Llopis S, Sola E, Rocha M,
Hernandez-Mijares A and Victor VM: Does metformin modulate
endoplasmic reticulum stress and autophagy in type 2 diabetic
peripheral blood mononuclear cells? Antioxid Redox Signal.
2017.(Epub ahead of print).
|
|
19
|
Nakanishi R, Kitao H, Kiniwa M, Morodomi
Y, Iimori M, Kurashige J, Sugiyama M, Nakashima Y, Saeki H, Oki E
and Maehara Y: Monitoring trifluridine incorporation in the
peripheral blood mononuclear cells of colorectal cancer patients
under trifluridine/tipiracil medication. Sci Rep. 7:169692017.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang HQ, Yu XD, Liu ZH, Cheng X, Samartzis
D, Jia LT, Wu SX, Huang J, Chen J and Luo ZJ: Deregulated miR-155
promotes Fas-mediated apoptosis in human intervertebral disc
degeneration by targeting FADD and caspase-3. J Pathol.
225:232–242. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Gruber HE, Hoelscher GL, Ingram JA and
Hanley EN Jr: Genome-wide analysis of pain-, nerve- and
neurotrophin-related gene expression in the degenerating human
annulus. Mol Pain. 8:632012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xu YQ, Zhang ZH, Zheng YF and Feng SQ:
Dysregulated miR-133a mediates loss of typeII collagen by directly
targeting matrix metalloproteinase 9 (MMP9) in human intervertebral
disc degeneration. Spine (Phila Pa 1976). 41:E714–E724. 2016.
View Article : Google Scholar
|
|
24
|
Wang B, Wang D, Yan T and Yuan H:
MiR-138-5p promotes TNF-α-induced apoptosis in human intervertebral
disc degeneration by targeting SIRT1 through PTEN/PI3K/Akt
signaling. Exp Cell Res. 345:199–205. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ma JF, Zang LN, Xi YM, Yang WJ and Zou D:
MiR-125a Rs12976445 Polymorphism is associated with the apoptosis
status of nucleus pulposus cells and the risk of intervertebral dis
degeneration. Cell Physiol Biochem. 38:295–305. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Teixeira GQ, Pereira CL, Ferreira JR, Maia
AF, Gomez-Lazaro M, Barbosa MA, Neidlinger-Wilke C and Goncalves
RM: Immunomodulation of human mesenchymal stem/stromal cells in
intervertebral disc degeneration: insights from a
proinflammatory/degenerative ex vivo model. Spine (Phila Pa 1976).
2017.(Epub ahead of print).
|
|
27
|
Maidhof R, Jacobsen T, Papatheodorou A and
Chahine NO: Inflammation induces irreversible biophysical changes
in isolated nucleus pulposus cells. PLoS One. 9:e996212014.
View Article : Google Scholar : PubMed/NCBI
|