Introduction
Subclinical hypothyroidism (SCH) is closely
associated with disturbances in lipid metabolism, and is
characterized by serum thyroid-stimulating hormone (TSH) levels
that are above the reference range, while the serum total or free
thyroid hormone levels remain within the reference range (1). The prevalence of SCH ranges between 4
and 20% of the population in different regions of the world
(2). In recent years, a growing body
of evidence has indicated that SCH is an independent risk factor
for lipid metabolic disorders, such as cardiovascular diseases and
nonalcoholic fatty liver disease (NAFLD) (3). Although there have been studies on the
SCH molecular mechanism, mainly focusing on ligand-receptor
interactions and the biological effects at the cellular or
molecular levels (4), the underlying
mechanism of this condition currently remains unclear.
MicroRNAs (miRNAs/miRs) are small non-coding RNAs
with a length of ~18–23 nucleotides, which interact with mRNAs upon
specific base-pairing in the 3′-untranslated region to repress the
mRNA expression via translational inhibition or mRNA degradation.
miRNAs repress multiple target genes in linear whole pathways or
network nodes, thereby simultaneously exerting a larger cumulative
effect (5). These small molecules
are the focus of basic research on regulating biological processes,
as well as of applied research for their potential application as
biomarkers and therapeutic agents (6). Recently, accumulating evidence has
supported the importance of hepatic miRNAs in the physiological
process of hepatic lipid metabolism and a wide spectrum of
diseases, including viral hepatitis, cirrhosis, hepatoma and NAFLD
(7). However, little is known on the
role of miRNAs in hepatic lipid metabolic disorders associated with
SCH.
Proteomics analysis has been widely used to identify
and quantify proteins associated with biological functions that are
regulated by miRNAs (8). miRNA
target regulatory modules have previously been identified and
studied in liver fibrosis (9).
However, to the best of our knowledge, no previous study has
investigated the regulatory modules in SCH. In the present study,
miRNAs alterations and proteome profiles in SCH were compared and
integrated. in SCH mice. The integration was achieved by targeting
predictions at the sequence level.
Materials and methods
Research animals
Male C57BL/6 mice (age, 7 weeks, weight, 20–21 g)
were purchased from Vital River Corporation (Beijing, China) and
housed in designated specific-pathogen-free cages on a 12 h
light–dark cycle at 23°C and 60% humidity. Mice were allowed free
access to an irradiated chow and sterilized water. After
acclimatization for 1 week, the mice were randomly divided into two
groups, including the SCH model (n=9) and control (CON, n=7)
groups. The SCH and CON groups were given methimazole (MMI; 0.08
mg/kg body weight per day) or an equal volume of vehicle,
respectively, in their drinking water for 16 weeks. Finally, the
mice were euthanized using pentobarbital sodium (concentration=20
mg/ml; dose=120 mg/kg body weight) through the intraperitoneal
route. Following cervical dislocation to ensure death, 600 µl blood
samples and the liver tissues of mice were collected. All animal
experiments were performed according to the relevant guidelines and
institutional policies (10). The
animal protocol was approved by the Shandong Provincial Hospital
Animal Care and Use Committee (no. 2015-003; Jinan, China).
Serum TSH, free thyroxine (FT4), lipid
profile and liver function assay
Serum TSH level was determined using a mouse ELISA
kit (MyBioSource), following the product manual. Serum FT4
concentrations were determined using specific radioimmunoassay kits
(Jiuding Diagnostic). Serum triglyceride (TG), total cholesterol
(TC), high-density lipoprotein cholesterol (HDL-C), low-density
lipoprotein cholesterol (LDL-C), alanine transaminase (ALT) and
aspartate aminotransferase (AST) levels were analyzed using an
Olympus AU5400 automatic biochemical analyzer (Olympus Corporation)
at Shandong Provincial Hospital.
Hepatic TG and TC assays
The hepatic TG and TC content was analyzed using a
TG assay kit (E1013; Applygen Technologies, Inc.) and a TC assay
kit (E1015; Applygen Technologies, Inc.), respectively. Briefly,
approximately 50 mg of liver samples were cut, homogenized using a
manual homogenizer and lysed in TG kit or TC kit lysis buffer for
10 min at room temperature. After centrifugation at 2,000 × g for 5
min at room temperature, the supernatants were divided into two
parts, one for TG or TC measurements and the other for protein
concentration assay. A bicinchoninic acid (BCA) protein assay was
performed using the BCA protein assay kit (Pierce; Thermo Fisher
Scientific, Inc.) to determine protein concentration. The final TG
or TC levels were standardized by protein concentration and were
expressed as µmol per gram of protein.
Oil red O staining
To determine the levels of hepatic lipids, frozen
liver sections (8 µm) were mounted on slides and air dried for 10
min at room temperature, then sections were fixed with 10% buffered
formalin for 30 min at room temperature. After washing with PBS,
sections were stained with Oil red O for 10 min at room
temperature, and then counterstained with hematoxylin for 20 sec.
Photomicrographs were captured using an imaging system under a
microscope (Axiovert 100M; Zeiss AG).
miRNA extraction, reverse
transcription (RT) into cDNA and quantitative polymerase chain
reaction (qPCR) for miRNA
Liver miRNAs were extracted from the tissues using
the MiRcute miRNA isolation kit (Tiangen Biotech Co., Ltd.),
according to the manufacturer's protocol. Next, the concentration
of RNA was quantified using a NanoDrop 1000 system (NanoDrop
Technologies; Thermo Fisher Scientific, Inc.). For RT into cDNA,
the RNA was poly(A)-tailed and reverse transcribed into cDNA using
random primers and the miRcute miRNA First-Strand cDNA Synthesis
Kit (Tiangen Biotech Co., Ltd.), following which qPCR was performed
with the miRcute miRNA qPCR Detection kit (Tiangen Biotech Co.,
Ltd.) following the manufacturer's protocol with the forward
primers presented in Table I. The
universal reverse primers were synthesized and provided by Tiangen
Biotech Co., Ltd. The PCR thermocycling conditions were as
following: Denaturation at 95°C for 15 min, followed by 45 cycles
of 94°C for 20 sec and 60°C for 34 sec. The cycle quantification
(Cq) value was calculated using the second derivative maximum
method (11). U6 small nuclear RNA
was used as the internal control. The relative expression of each
miRNA following normalization was determined as follows: Cq (U6)-Cq
(miRNA).
 | Table I.Forward primer sequences used in
reverse transcription-quantitative polymerase chain reaction. |
Table I.
Forward primer sequences used in
reverse transcription-quantitative polymerase chain reaction.
| Name | Sequence
(5′-3′) | GenBank no. | Tm (°C) |
|---|
| U6 |
CTCGCTTCGGCAGCACA | NR002082.1 | 58 |
| mmu-miR-10b-5p |
TACCCTGTAGAACCGAATTTGG | LM378853.1 | 60 |
| mmu-miR-24-3p |
TGGCTCAGTTCAGCAGGAACAG | LM378864.1 | 60 |
| mmu-miR-29a-3p |
TAGCACCATCTGAAATCGGTTA | LM379104.1 | 60 |
| mmu-miR-30b-5p |
TGTAAACATCCTACACTCAGCT | LM378817.1 | 60 |
| mmu-miR-30c-5p |
TGTAAACATCCTACACTCTCAGC | LM379083.1 | 60 |
| mmu-miR-34a-5p |
TGGCAGTGTCTTAGCTGGTTGT | LM379111.1 | 60 |
|
mmu-miR-125b-5p |
TCCCTGAGACCCTAACTTGTGA | LM378823.1 | 60 |
|
mmu-miR-130a-3p |
CAGTGCAATGTTAAAAGGGCAT | LM378828.1 | 60 |
|
mmu-miR-148a-3p |
TCAGTGCACTACAGAACTTTGT | LM379085.1 | 60 |
| mmu-miR-155 |
TTAATGCTAATTGTGATAGGGGT | LM608259.1 | 60 |
|
mmu-miR-199a-5p |
CCCAGTGTTCAGACTACCTGTTC | LM378873.1 | 60 |
| mmu-miR-206 |
TGGAATGTAAGGAAGTGTGTGG | LM379180.1 | 60 |
| mmu-miR-210-3p |
CTGTGCGTGTGACAGCGGCTGA | LM379180.1 | 60 |
|
mmu-miR-291b-3p |
AAAGTGCATCCATTTTGTTTGT | LM379852.1 | 60 |
| mmu-miR-370-3p |
GCCTGCTGGGGTGGAACCTGGT | LM379368.1 | 60 |
|
mmu-miR-467b-5p |
GTAAGTGCCTGCATGTATATG | LM380560.1 | 60 |
|
mmu-miR-486a-5p |
TCCTGTACTGAGCTGCCCCGAG | LM379823.1 | 60 |
Protein preparation and liquid
chromatography-tandem mass spectrometry (LC-MS/MS) analysis
Hepatic protein was extracted as previously
described (12). Next, protein
concentration was determined using the BCA Assay kit (Thermo Fisher
Scientific, Inc.). The protein samples from three mice were pooled
with the ratio 1:1:1 as a biological sample to avoid erroneous
conclusions due to individual variations. Each pool was analyzed in
duplicate. A total of 200 µg protein from each pool was reduced,
alkylated and digested with trypsin. Subsequently, the dried
peptides were labeled following the manufacturer's recommendations
of the isobaric tags for relative and absolute quantitation (iTRAQ)
4-plex kits (SCIEX, Framingham, MA, USA) with iTRAQ tags, as
follows: iTRAQ 114 for CON, iTRAQ 115 for SCH, iTRAQ 116 for CON
replicate and iTRAQ 117 for SCH replicate. The eluted peptides were
analyzed by LC-MS/MS, according to a previously described protocol
(13).
Functional enrichment analysis
For functional analysis of the altered proteins, the
proteins were imported into the Database for Annotation,
Visualization and Integrated Discovery (DAVID version 6.7;
http://david.abcc.ncifcrf.gov/) which
involves an integrated biological knowledge base and analytical
tools designed to systematically extract biological meaning from
large gene/protein lists in order to perform a Gene Ontology (GO)
functional enrichment analysis and a Kyoto Encyclopedia of Genes
and Genomes (KEGG) pathway enrichment analysis. Path mining tools
such as gene function classification, functional annotation table
or clustering were used for analysis. Furthermore, TargetScan
(Release 7.2; http://www.targetscan.org/vert_72/) and miRanda
(August 2010 release; http://www.microrna.org/microrna/microrna/home.do)
database analyses were employed to identify putative targets of
miRNAs among the differentially expressed proteins (14).
Statistical analysis
The data were analyzed using SPSS software (version
23.0; IBM Corp.) and are expressed as the mean ± standard
deviation. Differences between two groups were compared using an
unpaired Student's t-test. Cluster version 3.0 and Java TreeView
version 1.60 (Stanford University) were used to perform
agglomerative hierarchical cluster analysis. All of the calculated
P-values were two-sided, and P≤0.05 was considered to indicate a
statistically significant difference.
Results
Generation of an SCH mouse model and
determination of lipid parameters
An SCH mouse model was established by MMI
administration for 16 weeks. The SCH mice exhibited normal serum
FT4 levels (Fig. 1A) and higher TSH
levels (Fig. 1B) as compared with
those in CON mice. The serum ALT (Fig.
1C) and AST (Fig. 1D) levels in
SCH mice were similar to those exhibited by the CON group. In
addition, serum TG (Fig. 1E), TC
(Fig. 1F) and LDL-C (Fig. 1G) levels in SCH mice were all
significantly higher when compared with those in CON mice, whereas
there was no marked difference in serum HDL-C levels (Fig. 1H). Oil red O staining of liver
tissues indicated greater lipid droplet accumulation in the livers
of SCH mice in comparison with the CON mice (Fig. 1I). Furthermore, the hepatic TG
(Fig. 1J) and TC (Fig. 1K) content were markedly increased in
the SCH mice.
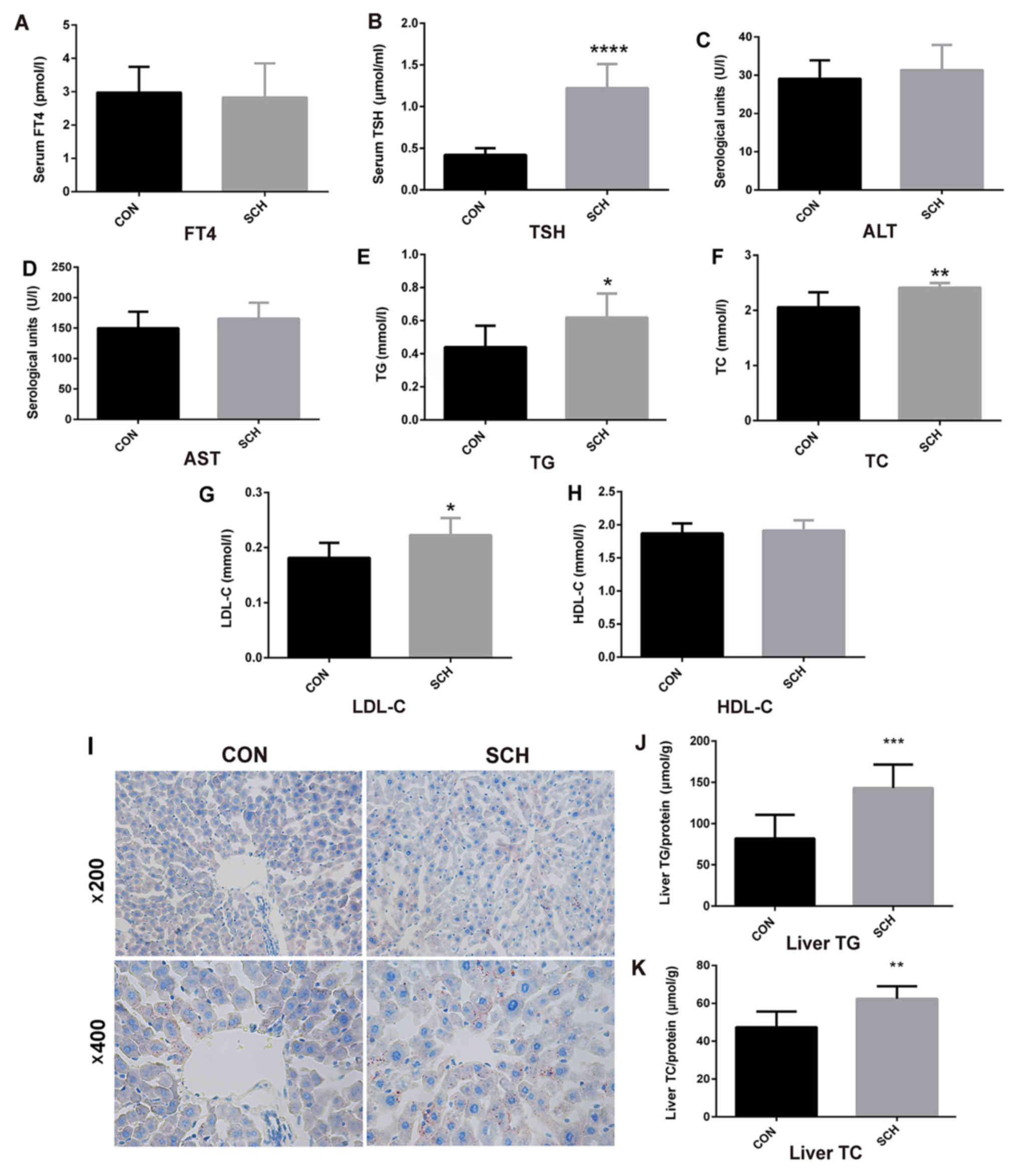 | Figure 1.Generation of SCH mouse model and
determination of lipid parameters. C57BL/6 mice were administered
methimazole (0.08 mg/kg BW per day) in the SCH group (n=9) or an
equal volume of vehicle in the CON group (n=7) for 16 weeks. (A)
FT4, (B) TSH, (C) ALT, (D) AST, (E) TG, (F) TC, (G) LDL-C and (H)
HDL-C levels in the serum of mice were detected. (I) Oil red O
staining of liver tissues (magnification, ×200 and ×400). (J) TG
and (K) TC content in liver tissues of mice. The data are presented
as the mean ± standard deviation. The TG and TC contents were
standardized to the corresponding total protein content in the
liver. *P<0.05, **P<0.01, ***P<0.001 and ****P<0.0001
vs. CON group. SCH, subclinical hypothyroidism; CON, control; FT4,
free thyroxine; TSH, thyroid-stimulating hormone; ALT, alanine
transaminase; AST, aspartate aminotransferase; TG, triglyceride;
TC, total cholesterol; LDL-C, low density lipoprotein cholesterol;
HDL-C, high density lipoprotein cholesterol. |
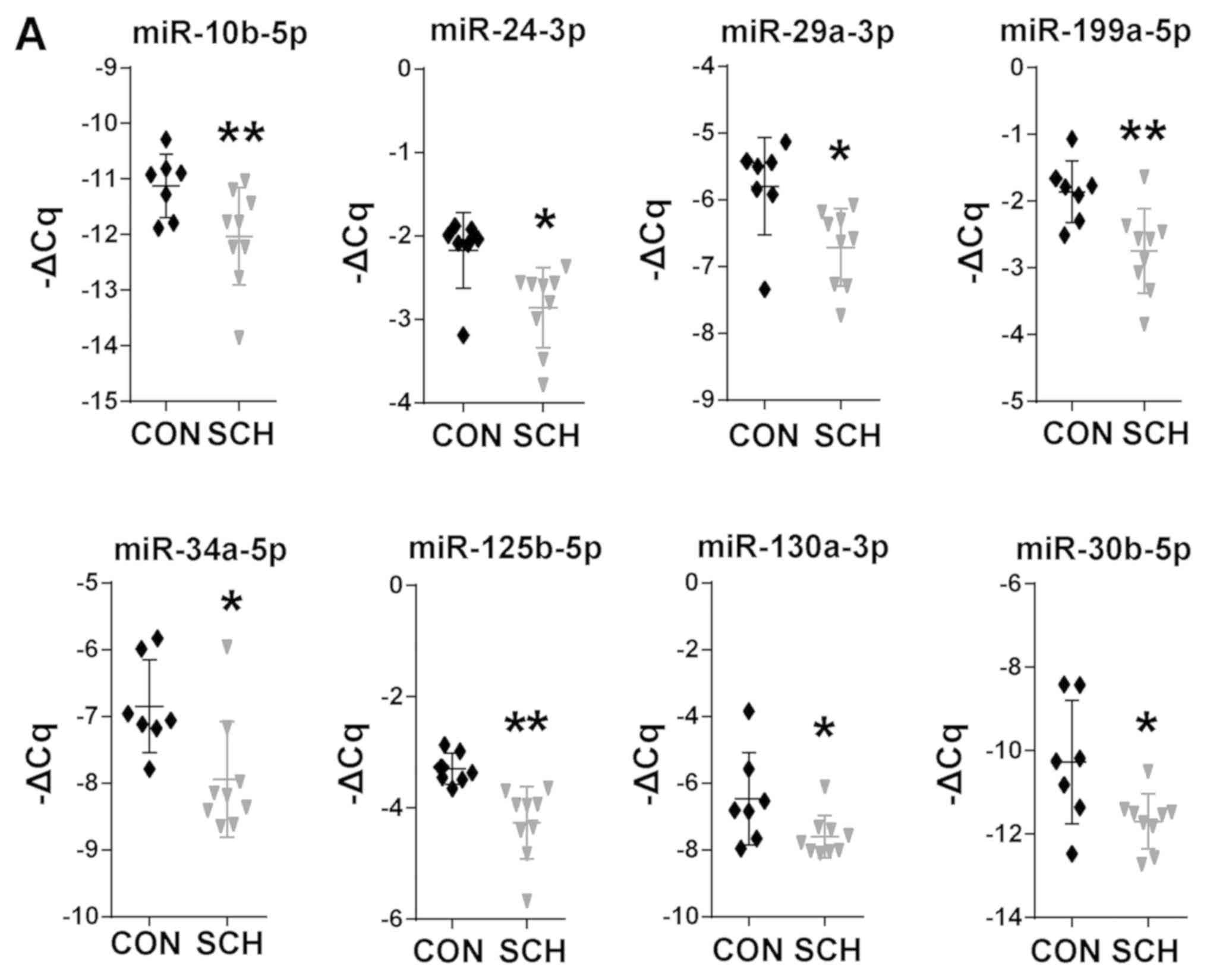
Differentially expressed miRNAs in the
livers of SCH mice
17 candidate miRNAs involved in hepatic lipid
metabolic were selected based on previous studies (15–32) and
non-coding RNA sequencing (data not shown). The present study then
screened these miRNAs by RT-qPCR. Among these 17 miRNAs, 8 miRNAs
were found to be significantly dysregulated in SCH mice, including
miR-10b-5p, miR-24-3p, miR-29a-3p, miR-30b-5p, miR-34a-5p,
miR-125b-5p, miR-130a-5p and miR-199a-5p (Fig. 2A; P<0.05). The expression of the
remaining 9 miRNAs was not markedly different between the SCH and
CON mice (Fig. 2B). These results
suggested that the 8 dysregulated miRNAs may be involved in the
development of hepatic lipid metabolic disorders in SCH.
Functional analysis of proteins
identified by proteomics
The present study performed iTRAQ-based proteomic
analysis in the livers of SCH mice. A fold-change >1.8 or
<0.56 was deemed significantly upregulated or downregulated,
respectively. A total of 1,969 proteins were quantified, of which
36 were found to be differentially expressed in the livers of SCH
mice, including 22 upregulated and 14 downregulated proteins, as
compared with their expression in control tissues. Next, all the
differentially expressed proteins were further categorized by DAVID
analysis.
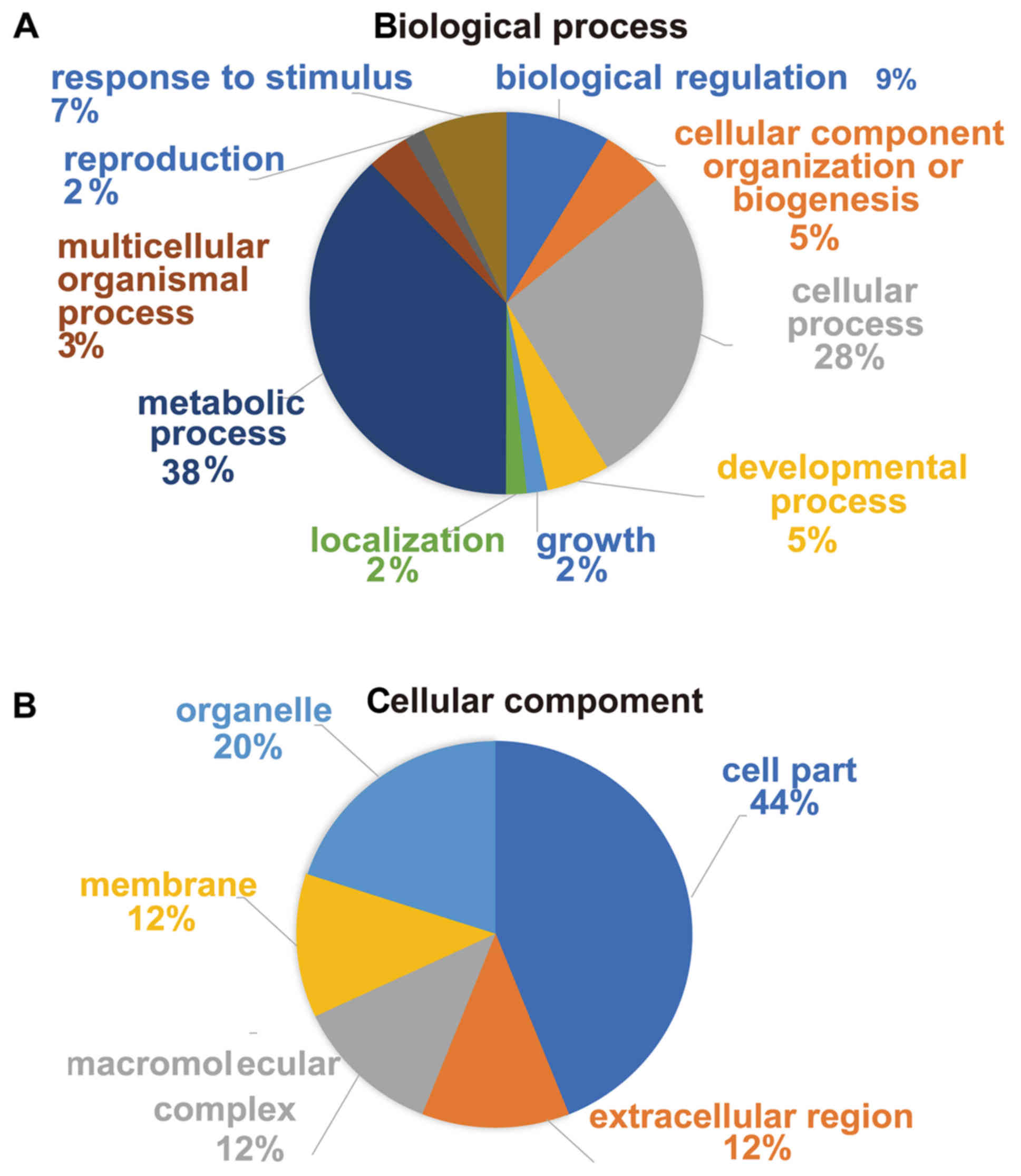
By GO functional classification, the identified
proteins were clustered into three groups, including proteins
involved in biological processes, cell components and molecular
functions. In the biological process cluster, the majority of
proteins were assigned to ‘metabolic process’ (38%) and ‘cellular
process’ (28%; Fig. 3A). The top two
enriched terms in the cellular component cluster included ‘cell
part’ (44%) and ‘organelle’ (20%; Fig.
3B). In the molecular function cluster, a great number of
proteins were assigned to ‘catalytic activity’ (58%) and ‘binding’
(24%; Fig. 3C). Furthermore,
according to KEGG pathway analysis, the majority of the identified
proteins were involved in KEGG pathways such as ‘Alzheimer's
disease’, ‘oxidative phosphorylation’, ‘Parkinson's disease’ and
‘NAFLD’ (Fig. 3D). Taken together,
the results suggested that these differentially expressed proteins
may be important effectors associated with hepatic lipid metabolic
disorders in SCH.
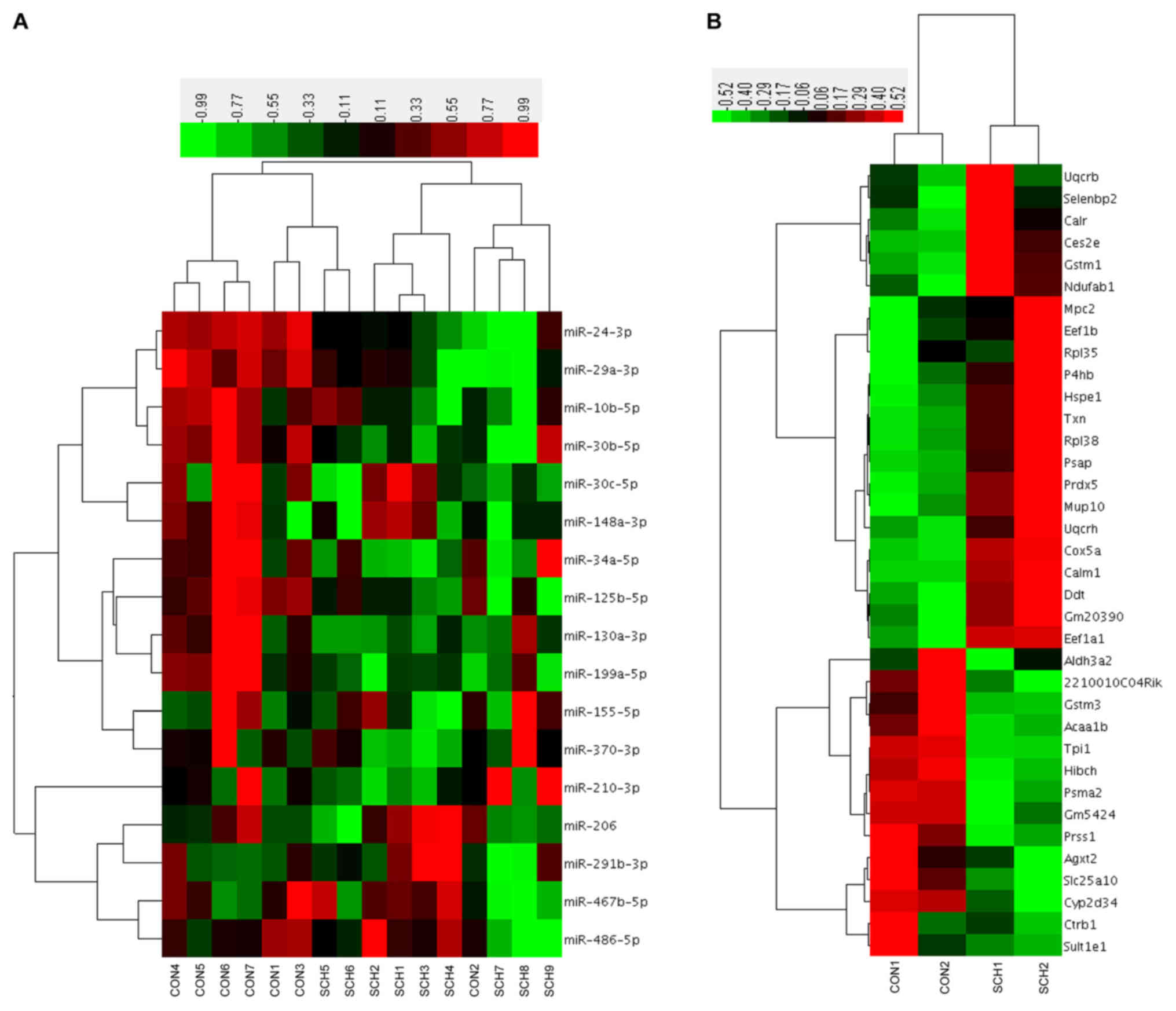
Hierarchical clustering based on the
17 candidate miRNAs and 36 proteins
Hierarchical clustering was performed based on the
17 candidate hepatic miRNAs (Fig.
4A). Of the 7 CON mice, 6 were clustered together with 2 SCH
mice. Of the 9 SCH mice, 7 were clustered together with only 1 CON
mouse. Next, a hierarchical cluster was constructed using the
expression values of the 36 identified proteins (Fig. 4B). The two groups exhibited marked
separation, indicating that the liver proteins expressed in SCH
mice were distinct from those in CON mice.
To investigate the correlation between altered
miRNAs and proteins in the livers of SCH mice, the present study
established a module between miRNAs and proteins using TargetScan
and miRanda for prediction (Table
II). The regulatory module contained 3 miRNAs, 4 proteins and 4
miRNA-protein connections. It was observed that miR-34a-5p targets
thioredoxin (Txn), miR-130a-3p targets elongation factor 1-beta
(Eef1b) and prosaposin (Psap), while miR-24-5p targets
selenium-binding protein 2 (Selenbp2).
 | Table II.Targets of differentially expressed
miRNAs in subclinical hypothyroidism mice. |
Table II.
Targets of differentially expressed
miRNAs in subclinical hypothyroidism mice.
| miRNA | Putative target
gene | Definition |
|---|
| mmu-miR-24-3p | Selenbp2 | Selenium-binding
protein 2 |
| mmu-miR-34a-5p | Txn | Thioredoxin |
|
mmu-miR-130a-3p | Eef1b | Elongation factor
1-β |
|
| Psap | Prosaposin |
Discussion
In previous clinical studies, a positive association
has been reported between TSH and serum TG levels (33). TG synthesis could be induced by TSH
through GPAT3 in adipocytes (34),
while TSH has been demonstrated to promote hepatic TG accumulation
by increasing SREBP-1c activity (12). In order to further investigate the
effect of TSH on hepatic lipid metabolism, a noninvasive method of
MMI administration in the drinking water to successfully establish
SCH mouse model (4). miRNAs have
been studied in a variety of liver diseases, including viral
hepatitis, cirrhosis, hepatoma and NAFLD (7,35).
However, their application is challenging, as miRNAs have different
and intersecting target genes (5).
Previous research has revealed that modules containing genes and
targeting regulators could be used as diagnostic and therapeutic
tools (36). Therefore, it was
hypothesized that miRNA and proteome profiles could be integrated
to form an miRNA-protein regulatory module, which may be associated
with hepatic lipid metabolism disorders in SCH and thereby be used
to explore potential therapeutic targets.
In the present study, a total of 17 hepatic miRNAs
that have been confirmed as crucial gene regulators of hepatic
lipid metabolism were selected to explore their profiles in SCH,
among them, miR-10b regulates hepatocyte steatosis by targeting
peroxisome proliferator-activated receptor-α (15). In addition, miR-24 and miR-125b
regulate hepatic lipid accumulation by targeting insulin-induced
gene 1 and fatty acid synthase (FAS), respectively (16,20).
miR-29 and miR-486 are reported to regulate cholesterol metabolism
by targeting hydroxy-3-methyl-glutaryl-CoA reductase and histone
acetyltransferase-1, respectively (17,28).
Furthermore, miR-30b-5p and miR-30c-5p belong to the same family of
miRNAs regulating fatty acid synthesis genes, targeting elongation
of very long chain fatty acids protein 5 (ELOVL5) and fatty acid
synthase (FAS), respectively (18,19).
miR-34a has been reported to participate in proinflammatory NAFLD
(24), while miR-130a-3p directly
targets transforming growth factor-β receptors 1 and 2, which may
contribute to hepatic fibrosis (21). Additionally, miR-148a regulates
cholesterol and TG homeostasis by controlling multiple metabolic
regulatory circuits (30). miR-155
and miR-467b modulate hepatic steatosis by targeting liver X
receptor α and hepatic lipoprotein lipase (22,23). It
has also been demonstrated that miR-199a-5p and miR-370 were
implicated in fatty acid β-oxidation in mitochondria (25,27). In
addition, by simultaneously regulating insulin signaling and
adipogenesis, miR-206 reduced lipid and glucose production in the
liver of obese mice (29). In liver,
miR-291b-3p promotes lipogenesis by suppressing AMPKα1 expression
and activity (31). Hepatic miR-210
is elevated in cholestatic mice and PBC patients, promoting bile
acids-induced liver injury by targeting mixed-lineage leukemia-4
(MLL4) (32). In the present study,
it was observed that 8 (miR-10b-5p, miR-24-3p, miR-29a-3p,
miR-30b-5p, miR-34a-5p, miR-125b-5p, miR-130a-5p and miR-199a-5p)
out of these 17 miRNAs associated with hepatic lipid metabolic
disorders were downregulated in the livers of SCH mice, suggesting
that these miRNAs may be involved in hepatic lipid metabolic
disorders in SCH.
The present study subsequently conducted iTRAQ
labeling analysis and identified 36 proteins with altered
expression levels. Targeting predictions of miRNAs by TargetScan
and miRanda were used to identify the potential targets (37). Of the significantly altered proteins,
four were found to be potential targets of 3 differentially
expressed miRNAs, namely the Txn, Eef1b, Psap and Selenbp2
proteins. It has been reported that the the Txn protein, has
notable properties as a crucial defense against oxidative stress
(38). Txn forms a system with
thioredoxin reductase and nicotinamide adenine dinucleotide
phosphate, and further eliminates reactive oxygen species, the
excessive production of which leads to oxidative stress
contributing to cardiac dysfunction and insulin resistance in NAFLD
(39). TSH is known to directly
produce oxidative stress, and oxidative damage to lipid
peroxidation has been reported in SCH patients (40). The present study demonstrated marked
alterations in miR-34a-5p and Txn levels in SCH mice, which may be
involved in the development of hepatic lipid metabolism disorders
in these mice via oxidative stress.
Eef1b, an enzyme that may be localized in the
endoplasmic reticulum, has been reported to promote protein
synthesis, and protect Leishmania major from chemical and
oxidative stress (41). Furthermore,
it is known that oxidative damage leads to lipid peroxidation,
which is a key component of SCH and NAFLD (42). Thus, Eef1b, as the target of
miR-130a-3p, may be involved in hepatic lipid metabolism disorders
in SCH mice through lipid peroxidation.
Several minerals and trace elements are essential
for normal thyroid hormone metabolism, such as iodine, iron and
selenium, and hypothyroidism can easily arise in regions of severe
iodine and selenium deficiency (43). However, to the best of our knowledge,
the roles of Selenbp2 and Psap in hepatic lipid metabolism in SCH
have not yet been studied; thus, the underlying mechanism requires
further exploration.
In conclusion, the present study identified a
miRNA-protein regulatory module, which included 3 miRNAs and 4
proteins, that may be associated with hepatic lipid metabolism by
integrating miRNA and proteome profiles in SCH mice. To the best of
our knowledge, no previous studies have investigated miRNAs that
are involved in hepatic lipid metabolism in SCH, and the present
study may thus provide potential therapeutic targets and
significant evidence for researchers to better understand the
underlying pathogenesis of hepatic lipid metabolism in SCH.
However, further investigation will be necessary in the future to
verify these findings.
Acknowledgements
The abstract of this study was presented at the
Endocrine Society Annual Meeting ENDO 2019, 22-23rd March 2019, New
Orleans, LA, USA and was published as abstract number SAT-047.
Funding
This study was supported by grants from the National
Key Research and Development Program of China (no. 2017YFC1309800),
the National Basic Research Program (no. 2012CB524900), the
National Natural Science Foundation (nos. 81230018, 81471006,
81430020 and 81500595) and the Taishan Scholar's Special Expert
Plan.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
LZ performed the animal experiments and wrote the
draft of the manuscript. KW, TB and LZ performed the molecular
biology experiments. LG helped to develop the manuscript and
interpreted the results. XZ and WC designed the study and performed
data analysis. All authors approved the final manuscript for
publication.
Ethics approval and consent to
participate
All animal experiment procedures were approved by
the Shandong Provincial Hospital Animal Care and Use Committee
(approval no. 2015-003) and were in compliance with the Guide for
the Care and Use of Laboratory Animals.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Surks MI, Ortiz E, Daniels GH, Sawin CT,
Col NF, Cobin RH, Franklyn JA, Hershman JM, Burman KD, Denke MA, et
al: Subclinical thyroid disease: Scientific review and guidelines
for diagnosis and management. JAMA. 291:228–238. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Quinn TJ, Gussekloo J, Kearney P, Rodondi
N and Stott DJ: Subclinical thyroid disorders. Lancet. 380:335–337.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Xu L, Ma H, Miao M and Li Y: Impact of
subclinical hypothyroidism on the development of non-alcoholic
fatty liver disease: A prospective case-control study. J Hepatol.
57:1153–1154. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhou L, Ding S, Li Y, Wang L, Chen W, Bo
T, Wu K, Li C, Liu X, Zhao J, et al: Endoplasmic reticulum stress
may play a pivotal role in lipid metabolic disorders in a novel
mouse model of subclinical hypothyroidism. Sci Rep. 6:313812016.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Rottiers V and Näär AM: MicroRNAs in
metabolism and metabolic disorders. Nat Rev Mol Cell Biol.
13:239–250. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Filipowicz W, Bhattacharyya SN and
Sonenberg N: Mechanisms of post-transcriptional regulation by
microRNAs: Are the answers in sight? Nat Rev Genet. 9:102–114.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Baffy G: MicroRNAs in nonalcoholic fatty
liver disease. J Clin Med. 4:1977–1988. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lu Y, Wang J, Guo X, Yan S and Dai J:
Perfluorooctanoic acid affects endocytosis involving clathrin light
chain A and microRNA-133b-3p in mouse testes. Toxicol Appl
Pharmacol. 318:41–48. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chen W, Zhao W, Yang A, Xu A, Wang H, Cong
M, Liu T, Wang P and You H: Integrated analysis of microRNA and
gene expression profiles reveals a functional regulatory module
associated with liver fibrosis. Gene. 636:87–95. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li Y, Wang L, Zhou L, Song Y, Ma S, Yu C,
Zhao J, Xu C and Gao L: Thyroid stimulating hormone increases
hepatic gluconeogenesis via CRTC2. Mol Cell Endocrinol. 446:70–80.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yan F, Wang Q, Lu M, Chen W, Song Y, Jing
F, Guan Y, Wang L, Lin Y, Bo T, et al: Thyrotropin increases
hepatic triglyceride content through upregulation of SREBP-1c
activity. J Hepatol. 61:1358–1364. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Calderon-Gonzalez KG, Valero Rustarazo ML,
Labra-Barrios ML, Bazán-Méndez CI, Tavera-Tapia A, Herrera-Aguirre
ME, Sánchez del Pino MM, Gallegos-Pérez JL, González-Márquez H,
Hernández-Hernández JM, et al: Determination of the protein
expression profiles of breast cancer cell lines by quantitative
proteomics using iTRAQ labelling and tandem mass spectrometry. J
Proteomics. 124:50–78. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zheng L, Lv GC, Sheng J and Yang YD:
Effect of miRNA-10b in regulating cellular steatosis level by
targeting PPAR-alpha expression, a novel mechanism for the
pathogenesis of NAFLD. J Gastroenterol Hepatol. 25:156–163. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ng R, Wu H, Xiao H, Chen X, Willenbring H,
Steer CJ and Song G: Inhibition of microRNA-24 expression in liver
prevents hepatic lipid accumulation and hyperlipidemia. Hepatology.
60:554–564. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu MX, Gao M, Li CZ, Yu CZ, Yan H, Peng
C, Li Y, Li CG, Ma ZL, Zhao Y, et al: Dicer1/miR-29/HMGCR axis
contributes to hepatic free cholesterol accumulation in mouse
non-alcoholic steatohepatitis. Acta Pharmacol Sin. 38:660–671.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang M, Li CC, Li F, Li H, Liu XJ, Loor
JJ, Kang XT and Sun GR: Estrogen promotes hepatic synthesis of
long-chain polyunsaturated fatty acids by regulating ELOVL5 at
post-transcriptional level in laying hens. Int J Mol Sci.
18:E14052017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Fan J, Li H, Nie X, Yin Z, Zhao Y, Chen C
and Wen Wang D: MiR-30c-5p ameliorates hepatic steatosis in leptin
receptor-deficient (db/db) mice via down-regulating FASN.
Oncotarget. 8:13450–13463. 2017.PubMed/NCBI
|
|
20
|
Zhang ZC, Liu Y, Xiao LL, Li SF, Jiang JH,
Zhao Y, Qian SW, Tang QQ and Li X: Upregulation of miR-125b by
estrogen protects against non-alcoholic fatty liver in female mice.
J Hepatol. 63:1466–1475. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang Y, Du J, Niu X, Fu N, Wang R, Zhang
Y, Zhao S, Sun D and Nan Y: MiR-130a-3p attenuates activation and
induces apoptosis of hepatic stellate cells in nonalcoholic
fibrosing steatohepatitis by directly targeting TGFBR1 and TGFBR2.
Cell death Dis. 8:e27922017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ahn J, Lee H, Chung CH and Ha T: High fat
diet induced downregulation of microRNA-467b increased lipoprotein
lipase in hepatic steatosis. Biochem Biophys Res Commun.
414:664–669. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Miller AM, Gilchrist DS, Nijjar J, Araldi
E, Ramirez CM, Lavery CA, Fernández-Hernando C, McInnes IB and
Kurowska-Stolarska M: MiR-155 has a protective role in the
development of non-alcoholic hepatosteatosis in mice. PLoS One.
8:e723242013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Castro RE, Ferreira DM, Afonso MB,
Borralho PM, Machado MV, Cortez-Pinto H and Rodrigues CM:
miR-34a/SIRT1/p53 is suppressed by ursodeoxycholic acid in the rat
liver and activated by disease severity in human non-alcoholic
fatty liver disease. J Hepatol. 58:119–125. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li B, Zhang Z, Zhang H, Quan K, Lu Y, Cai
D and Ning G: Aberrant miR199a-5p/caveolin1/PPARα axis in hepatic
steatosis. J Mol Endocrinol. 53:393–403. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhang B, Wang R, Du J, Niu J, Zhang R, Xu
S, Niu X, Zhang Q and Nan Y: Upregulated microRNA-199a-5p inhibits
nuclear receptor corepressor 1 translation in mice with
nonalcoholic steatohepatitis. Mol Med Rep. 10:3080–3086. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Iliopoulos D, Drosatos K, Hiyama Y,
Goldberg IJ and Zannis VI: MicroRNA-370 controls the expression of
microRNA-122 and Cpt1alpha and affects lipid metabolism. J Lipid
Res. 51:1513–1523. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Liu D, Zhang M, Xie W, Lan G, Cheng HP,
Gong D, Huang C, Lv YC, Yao F, Tan YL, et al: MiR-486 regulates
cholesterol efflux by targeting HAT1. Biochem Biophys Res Commun.
472:418–424. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wu H, Zhang T, Pan F, Steer CJ, Li Z, Chen
X and Song G: MicroRNA-206 prevents hepatosteatosis and
hyperglycemia by facilitating insulin signaling and impairing
lipogenesis. J Hepatol. 66:816–824. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wagschal A, Najafi-Shoushtari SH, Wang L,
Goedeke L, Sinha S, deLemos AS, Black JC, Ramírez CM, Li Y, Tewhey
R, et al: Genome-wide identification of microRNAs regulating
cholesterol and triglyceride homeostasis. Nat Med. 21:1290–1297.
2015. View
Article : Google Scholar : PubMed/NCBI
|
|
31
|
Meng X, Guo J, Fang W, Dou L, Li M, Huang
X, Zhou S, Man Y, Tang W, Yu L and Li J: Liver MicroRNA-291b-3p
promotes hepatic lipogenesis through negative regulation of
adenosine 5′-monophosphate (AMP)-activated protein kinase α1. J
Biol Chem. 291:10625–10634. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kim YC, Jung H, Seok S, Zhang Y, Ma J, Li
T, Kemper B and Kemper JK: MicroRNA-210 promotes bile acid-induced
cholestatic liver injury by targeting mixed-lineage leukemia-4
methyltransferase in mice. Hepatology. Sep 24–2019.(Epub ahead of
print). View Article : Google Scholar
|
|
33
|
Wanjia X, Chenggang W, Aihong W, Xiaomei
Y, Jiajun Z, Chunxiao Y, Jin X, Yinglong H and Ling G: A high
normal TSH level is associated with an atherogenic lipid profile in
euthyroid non-smokers with newly diagnosed asymptomatic coronary
heart disease. Lipids Health Dis. 11:442012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ma S, Jing F, Xu C, Zhou L, Song Y, Yu C,
Jiang D, Gao L, Li Y, Guan Q and Zhao J: Thyrotropin and obesity:
Increased adipose triglyceride content through glycerol-3-phosphate
acyltransferase 3. Sci Rep. 5:76332015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Louten J, Beach M, Palermino K, Weeks M
and Holenstein G: MicroRNAs expressed during viral infection:
Biomarker potential and therapeutic considerations. Biomarker
Insights. 10 (Suppl 4):S25–S52. 2015.
|
|
36
|
Barabasi AL, Gulbahce N and Loscalzo J:
Network medicine: A network-based approach to human disease. Nat
Rev Genet. 12:56–68. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Peterson SM, Thompson JA, Ufkin ML,
Sathyanarayana P, Liaw L and Congdon CB: Common features of
microRNA target prediction tools. Front Genet. 5:232014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lu J and Holmgren A: The thioredoxin
antioxidant system. Free Radic Biol Med. 66:75–87. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Yang J, Hamid S, Cai J, Liu Q, Xu S and
Zhang Z: Selenium deficiency-induced thioredoxin suppression and
thioredoxin knock down disbalanced insulin responsiveness in
chicken cardiomyocytes through PI3K/Akt pathway inhibition. Cell
Signal. 38:192–200. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Haribabu A, Reddy VS, Pallavi CH, Bitla
AR, Sachan A, Pullaiah P, Suresh V, Rao PV and Suchitra MM:
Evaluation of protein oxidation and its association with lipid
peroxidation and thyrotropin levels in overt and subclinical
hypothyroidism. Endocrine. 44:152–157. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Sasikumar AN, Perez WB and Kinzy TG: The
many roles of the eukaryotic elongation factor 1 complex. Wiley
Interdiscip Rev RNA. 3:543–555. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Seki S, Kitada T, Yamada T, Sakaguchi H,
Nakatani K and Wakasa K: In situ detection of lipid peroxidation
and oxidative DNA damage in non-alcoholic fatty liver diseases. J
Hepatol. 37:56–62. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zimmermann MB and Kohrle J: The impact of
iron and selenium deficiencies on iodine and thyroid metabolism:
Biochemistry and relevance to public health. Thyroid. 12:867–878.
2002. View Article : Google Scholar : PubMed/NCBI
|