Introduction
Chronic rhinosinusitis (CRS) is a multifactorial,
heterogeneous inflammatory disease of the nose and paranasal
sinuses characterized by 12 weeks of persistent symptoms, such as
congestion, pain or facial pressure, stuffiness, cough, impairment
or loss of the sense of smell (anosmia), nasal discharge and
fatigue (1). CRS affects ~12% of
the worldwide population and severely impairs quality of life
(2). Based on the absence or
presence of nasal polyps (NP), CRS can be categorized into two main
forms, CRS without NP (CRSsNP) and CRS with NP (CRSwNP) (1). CRSwNP is the most commonly studied
among both types. Polyps are outgrowths of edematous inflammatory
tissues that have grown into the middle meatus (3). CRSwNP causes anosmia more readily than
CRSsNP and is less likely to respond to antibiotics (1,4).
CRSwNP can further be divided into two subtypes based on the
histological features: Eosinophilic CRSwNP (ECRSwNP) and
non-eosinophilic CRSwNP (non-ECRSwNP). In Western countries, 67-88%
of patients with CRSwNP exhibit mucosal eosinophilic infiltration,
which is characterized by type II inflammation (5-7).
However, in East Asian countries, such as China, Korea and Japan,
only 21.7-59.6% of patients exhibit mucosal eosinophilia, which is
characterized by T helper cell (Th)1/Th17-dominant inflammation
(7-11).
There was a significant increase in the number of patients with
ECRSwNP compared with non-ECRSwNP from 1999 to 2011 in Thailand
(12). ECRSwNP is higher in CT
images and polyp scores compared with non-ECRSwNP and has a poor
response to medical and surgical therapy (9,13). The
pathogenesis and pathophysiological mechanisms of ECRSwNP and
non-ECRSwNP remain poorly understood.
Immune cell infiltration is the characteristic
manifestation of chronic inflammation. A histological study on a
large sample of CRS tissues demonstrated that CRSwNP presented a
higher amount of immune cell infiltration compared with CRSsNP, and
there were higher levels of cell infiltration of cytotoxic
CD8+ T cells, B cells and macrophages in CRSwNP, as
determined by immunohistochemistry (14). In addition, ECRSwNP has been
reported to be associated with fewer M1 macrophages and more M2
macrophages compared with non-ECRSwNP, and the number of
IL-10+CD68+ macrophages was decreased in
ECRSwNP (15). Specific chemokines
and cytokines, such as MIP-1α, IL-5 and RANTES, are crucial to
eosinophil recruitment, survival and differentiation (16-18).
Based on gene expression profiles, Th2-related molecules (IL-4 and
colony-stimulating factor 2), cell cycle regulators (cyclin
dependent kinase inhibitor 1A and cyclin D1), and tissue
fibrosis-related molecules (TGF-β) are upregulated in ECRSwNP; on
the other hand, interferons (IFNs) and acute inflammatory cytokines
are upregulated in non-ECRSwNP (19). The long non-coding RNA XLOC_010280
was demonstrated to be specifically expressed in ECRSwNP and found
to regulate chemokine (C-C motif) ligand 18 (CCL18), which may
explain the markedly higher expression of CCL18 in ECRSwNP compared
with that in non-ECRSwNP (20).
To reveal the immune cell infiltration and key genes
of CRSwNP, 22 immune cell types and the pathophysiology of CRSwNP
were analyzed by CIBERSORT (21)
and Weighted Gene Correlation Network Analysis (WGCNA) (22) methods according to eosinophilic
pathology characteristics. The combined analysis of immune cell
infiltration and key genes of CRSwNP provided a deeper
understanding of the immune and inflammatory response exhibited in
CRS.
Materials and methods
Acquisition and processing of transcriptome and gene
expression profile data. The transcriptome gene expression profile
data used in the present study were obtained from a public domain.
The expression matrix and sample information (GSE72713) (20) were downloaded from the Gene
Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/). Genes with similar
names were merged and homogenized using the limma package (version
3.11; https://www.bioconductor.org/packages/release/bioc/html/limma.html).
Genes that were not expressed in all samples were omitted; a total
of 21,014 expressed genes were further analyzed in all samples.
Evaluation of immune cell infiltration
in CRSwNP
The nine samples obtained from GSE72713 was used to
investigate the 22 immune cell types in CRSwNP by CIBERSORT
(20,21). CIBERSORT is an analytical tool in
which 547 gene expression signatures were used to quantify the 22
immune cells. The expression data were submitted to the CIBERSORT
official website (https://cibersort.stanford.edu/). The algorithm used a
signature matrix at 2,500 permutations to improve the accuracy of
the deconvolution. Kruskal-Wallis test was used to analyze the
differences among the three groups (Normal control, ECRSwNP and
non-ECRSwNP) using R software (version 3.6.3; https://www.r-project.org/). The correlation of
infiltrated immune cells was analyzed by a Pearson's test using
corrplot R package (Version 0.84; https://github.com/taiyun/corrplot) (22).
Construction of the WGCNA phenotype
co-expression network
The nine samples obtained from GSE72713 were divided
into four groups: Normal control (CTRL; n=3; GSM1868859;
GSM1868860; GSM1868861), ECRSwNP (n=3; GSM1868853; GSM1868854;
GSM1868855), non-ECRSwNP (n=3; GSM1868856; GSM1868857; GSM1868858),
and NP, which included ECRSwNP and non-ECRSwNP samples. The tissue
eosinophil number per high power field (HPF) of ECRSwNP was >20,
counted in the lamina propria of the polyps in five random
microscopic HPFs at x400 magnification. Subjects who did not
fulfill the criteria were categorized as non-ECRSwNP. Overall,
21,014 genes from nine samples were analyzed by WGCNA (23). The samples were clustered and the
pickSoftThreshold was calculated (24). Power value 16 was used to construct
the block-wise modules (24). The
color row beneath the dendrogram illustrates the 28-module
assignment obtained using the dynamicTreeCut package (version 1.63)
(25). The correlation between
phenotype and block-wise modules was then analyzed using Pearson's
test.
The gene expression levels of related modules were
verified by quantitative PCR (qPCR). GAPDH was as the reference
gene. The mRNA expression for the selected 12 genes was measured
using SYBR® Premix Ex Taq™ (Takara Bio, Inc.) under the
following thermocycling conditions: Initial denaturation at 95˚C
for 5 min, followed by 40 cycles at 95˚C for 10 sec and 55˚C for 30
sec. The expression levels in the samples were determined by the
relative quantity curve method 2-ΔΔCq (26). Primer sequences are listed in
Table SI. The new collected qPCR
verified samples (non-eosinophil polyps, eosinophil and nasal
mucosa tissues), which are distinct from the nine samples
aforementioned, were collected by the Department of Otolaryngology,
Head and Neck Surgery, Tongde Hospital of Zhejiang Province
(Zhejiang, China) between April 2018 and December 2019. The Ethics
Committee of Tongde Hospital of Zhejiang Province (approval no.
XMSB2018013) approved the present study and written informed
consent was obtained from nine patients (mean age: 34±5 years; age
range: 25-40 years; nine males and three females). The RNA was
extracted from the nine tissue samples of patients using RNeasy
Mini Kit (Qiagen GmbH) according to manufacturer's protocol and
cDNA was synthesized using First Strand cDNA Synthesis Kit (cat.
no. K1612; Thermo Fisher Scientific, Inc.). The 12 related genes
were amplified by qPCR using Roche LightCycler® 480II
(Roche diagnostics).
Module gene function analysis
The gene functions of modules that were found to be
significantly related to the phenotypes (CTRL, ECRSwNP, non-ECRSwNP
and NP) were enriched in Gene Ontology (GO; http://geneontology.org/) and Kyoto Encyclopedia of
Genes and Genomes (KEGG) (https://www.kegg.jp/). P<0.05 was set as the
cut-off value. Connections between the significant GO and KEGG
terms were visualized using a clusterProfiler package with a
network diagram (27). The heatmap
was drawn using the pheatmap package (version 1.0.12; https://cran.r-project.org/web/packages/pheatmap/index.html).
Results
Composition of immune cells in
CRSwNP
Initially, the fractions of infiltrated immune cells
among the CTRL, ECRSwNP and non-ECRSwNP groups were investigated
based on the gene expression profile data by CIBERSORT. The
percentage of infiltrated cells was shown and 17 types of immune
cells were found to infiltrate the nasal mucosa and NP tissues
(Table SII). A total of five types
of immune cells (memory B cells, naive CD4+ T cells, follicular
helper T cells, regulatory T cells and resting NK cells) were not
found in the CTRL, ECRSwNP and non-ECRSwNP groups (Fig. 1 and Table SII). A notable difference was
observed in eosinophil infiltration among the CTRL, ECRSwNP and
non-ECRSwNP groups (Fig. 1). In
addition, the heatmap depicted plasma and CD4+ memory
resting cells as the major cell groups using the pheatmap package
(Fig. 1B). The percentage of
eosinophils in the infiltrated cells was 12.26±5.08% in the ECRSwNP
group, but no eosinophils infiltrated the CTRL and non-ECRSwNP
groups (Fig. 1 and Table SII). The activated natural killer
(NK) cells were also only present in the ECRSwNP group (Fig. 1B). Although the number of
infiltrated resting CD4+ T cells in CTRL tissues
(45.86±1.67%) was higher compared with that in ECRSwNP
(25.83±2.32%) and non-ECRSwNP (29.62±3.56%), the infiltrated
percentage of activated CD4+ T cells in the non-ECRSwNP
group (6.46±1.84%) was higher compared with that in the ECRSwNP
(2.68±1.11%) and CTRL (2.74±2.18%) groups (Fig. 1C and Table SII). Among infiltrated macrophages
(M0, M1 and M2), the number of infiltrated M2 macrophages in the
ECRSwNP group (12.56±0.77%) was higher compared with that in the
CTRL (4.12±1.98%) and non-ECRSwNP (3.24±1.21%) groups.
Additionally, the number of infiltrated M1 macrophages in the
non-ECRSwNP group (7.06±0.9%) was higher compared with that in the
CTRL (2.58±1.43%) and ECRSwNP (0.31±0.26%) groups. M0 macrophage
infiltration was found in lesser amounts in the CTRL group, but not
in the other two CRSwNP groups. Furthermore, the activated
dendritic cells (2.06±0.72%) and resting mast cells (12.86±5.92%)
in the ECRSwNP group were higher compared with those in the CTRL
(activated dendritic cells 0.56±0.78%; resting mast cells
6.29±2.81%) and non-ECRSwNP groups (activated dendritic cells
6.79±2.86%; resting mast cells 0.43±0.31%; Fig. 1 and Table SII).
Correlation among infiltrated immune
cells in CRSwNP
A total of five immune cell types, including
eosinophils, activated NK cells, activated dendritic cells, resting
mast cells and M2 macrophages, were found in higher fractions in
the ECRSwNP group compared with those in the CTRL and non-ECRSwNP
groups (Fig. 1C and Table SII). The correlation between these
infiltrated cells was further analyzed. Eosinophils were found to
be significantly positively correlated with activated mast cells,
activated NK cells and M2 macrophages (r>0.75; Fig. 2). The M2 macrophages were
significantly positively correlated with eosinophils, activated NK
cells and activated dendritic cells (r>0.75), but significantly
negatively correlated with γδT cells and M1 macrophages
(r<-0.75; Fig. 2). The M1
macrophages were significantly positively correlated with γδT cells
and activated CD4+ T cells (r>0.75; Fig. 2). These three cell types mostly
infiltrated the non-ECRSwNP group (Fig.
1 and Table SII). Thus, these
results suggested that ECRSwNP and non-ECRSwNP may differ
significantly in immune cell infiltration and core gene expression
profiles.
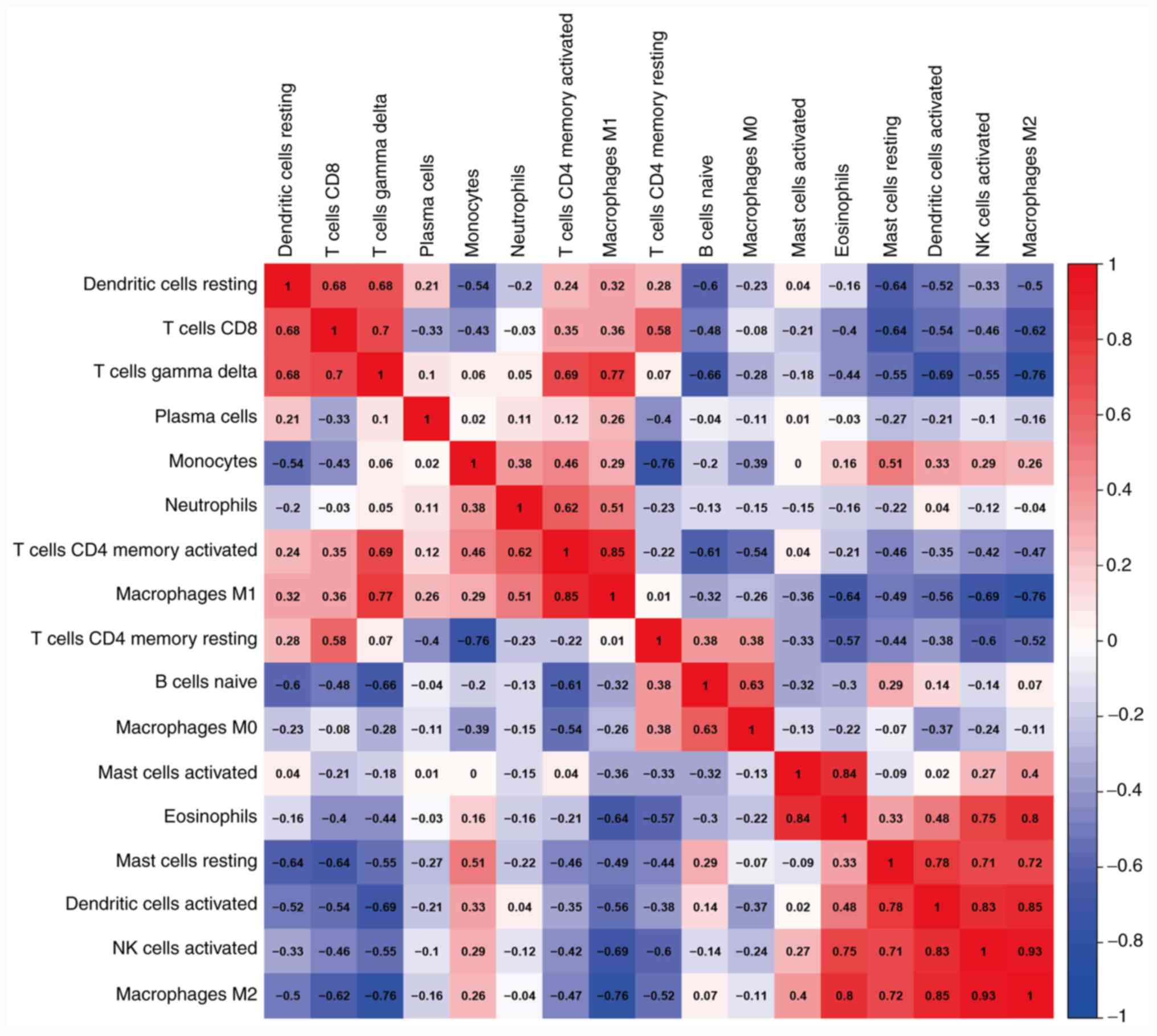 | Figure 2Correlation matrix of the proportion
of 22 immune cells in the CTRL, ECRSwNP and non-ECRSwNP groups.
Variables are ordered in the heatmap matrix. Eosinophils were
significantly positively correlated with activated mast cells,
activated NK cells and M2 macrophages. The M2 macrophages were
significantly positively correlated with eosinophils, activated NK
cells and activated dendritic cells, and significantly negatively
correlated with γδT cells and M1 macrophages. The M1 macrophages
were significantly positively correlated with γδT cells and
activated CD4+ T cells. In the heatmap, the blue color
represents low adjacency (negative correlation), while the red
represents high adjacency (positive correlation). ECRSwNP,
eosinophilic chronic rhinosinusitis with nasal polyps; non-ECRSwNP,
non-eosinophilic chronic rhinosinusitis with nasal polyps; CTRL,
control; NK, natural killer. |
Core gene expression modules in
CRSwNP
To detect different core genes in ECRSwNP and
non-ECRSwNP, gene expression profiles of the three group samples
(CTRL, ECRSwNP and non-ECRSwNP) involving ~21,014 genes were
clustered by hierarchical cluster analysis. Good expression
clustering was observed in all three groups (Fig. 3A). In addition, power value 16 was
used to construct block-wise modules using WGCNA. The 21,014
expressed genes were divided into 28 modules according to
expression (Fig. 3B and C). The grey module was the unknown module.
The expression levels of each module gene were clustered and
correlated. All gene modules were clustered into three big clades
(Fig. 3B and C). The gene expression modules and clades
may be closely related to the phenotype of CRSwNP.
Correlation of gene expression modules
with the phenotype of CRSwNP
To clarify the correlation between the gene module
and phenotype, the obtained samples were divided into four groups
(CTRL, ECRSwNP, non-ECRSwNP and NP groups). Two gene modules (pink
and turquoise) were found to be significantly negatively correlated
with the nasal polyp phenotype (P<0.05; r<-0.80; Fig. 4). The gene function clusters of the
two modules were analyzed by clusterProfiler for GO and KEGG. The
pink module showed no significant enrichment of GO functions and
KEGG pathways. The turquoise module was significantly enriched
(P<0.05) in 64 GO terms and seven KEGG pathways (Fig. 5 and Table SIII). The ‘olfactory receptor
activity’ (GO:0004984) and ‘olfactory transduction’ (hsa04740) were
major functional enrichments (Fig.
5A and C). The G
protein-coupled receptor activity and channel proteins activity in
turquoise module have extensive networks of shared genes (Fig. 5B). The expression of the olfactory
related genes, including OR10A3 and OR1A1, was higher in CTRL
groups compared with that in the NP group, especially in ECRSwNP
(Figs. 5D and S1). These results revealed the molecular
mechanism by which CRSwNP affects olfaction.
Correlation of gene expression modules
with ECRSwNP and non-ECRSwNP
The gene module and phenotype correlation analysis
showed that the brown module was significantly positively
correlated with ECRSwNP (P=0.003; r=0.86; Fig. 4). In the brown module, 20 GO terms
and 16 KEGG pathways were significantly enriched (P<0.05;
Fig. 6 and Table SIV). The ‘CCR chemokine receptor
binding’ (GO:0048020), ‘cytokine activity’ (GO:0005125), ‘cytokine
receptor binding’ (GO:0005126), ‘receptor ligand activity’
(GO:0048018), ‘growth factor receptor binding’ (GO:0070851) and
‘epidermal growth factor receptor binding’ (GO:0005154) function
(Fig. 6A) formed a functional
network that was significantly positively associated (Figs. 4 and 6B). In the brown module, many genes,
including CDH5, SHC1 and ITGA5, were found to be associated with
‘cytokine receptor binding’ and ‘growth factor receptor binding’
double GO function (Fig. 6C).
Multiple inflammatory-related signaling pathways, including,
PI3K-Akt signaling, JAK-STAT signaling, TNF and IL-17 signaling,
were significantly enriched in the brown module (Fig. 6D). The gene expression of the brown
module in the ECRSwNP group was higher compared with that in the
CTRL and non-ECRSwNP groups (Fig.
6E and F). The GO function and
enriched pathways illustrated the molecular mechanism of ECRSwNP
formation.
The correlation analysis did not demonstrate the
significant positive correlation of a gene module with non-ECRSwNP,
although it justified the significant negative correlation of the
blue module with non-ECRSwNP (P<0.05; r<-0.80; Fig. 4). The GO analysis revealed that
genes correlated with non-ECRSwNP mainly involved a variety of
transferases (GO:0034212, GO:0004674, GO:0018024, GO:0019787 and
GO:0004842), GTPases (GO:0031267, GO:0030695, GO:0017016 and
GO:0005096) and receptor binding functions (GO:0046965, GO:0046966,
and GO:0035257; Fig. 7A and
Table SV). These functional genes
formed four major functional networks (Fig. 7B). Numerous cell
proliferation-related and cancer signaling pathways, such as the
‘MAPK signaling pathway’, ‘Wnt signaling pathway’, ‘mTOR signaling
pathway’, ‘hepatocellular carcinoma’ and ‘breast cancer’ in the
blue module and some suppressor genes (such as P53 and DUSP8)
involved in these pathways were downregulated in non-ECRSwNP group
compared with those in the CTRL and ECRSwNP groups (Fig. 7C and E). There were extensive networks of
interactions between these gene signaling pathways (Fig. 7D). These results suggested that the
formation of non-ECRSwNP was similar to the growth of tumors
(Fig. 7E). However, the ‘autophagy’
(hsa04136, hsa04140 and hsa04150) was found to be negatively
associated with non-ECRSwNP (Figs.
4 and 7C), where some
autophagy-related genes (such as CAMKK2, ATG2A and MAPK9) were
downregulated in non-ECRSwNP compared with those in the CTRL and
ECRSwNP groups (Fig. 7F). The
dysfunction of autophagy in nasal mucosa may be the primary cause
of non-ECRSwNP.
Discussion
CRS is defined as inflammation of the nasal mucosa;
CRSwNP represents 20-25% of all cases worldwide (3). The two main histological types of
CRSwNP (ECRSwNP and non-ECRSwNP) have been identified based on
eosinophil infiltration, geographical location and ethnicity
(5-11).
The immune cell infiltration of CRSwNP is higher compared with that
of CRSsNP (14); however, the rate
of infiltration of different types of immune cells, other than
eosinophils and macrophages, is still unclear in ECRSwNP and
non-ECRSwNP (15). In the present
study, the proportion of 22 immune cell types in immune
microenvironment of ECRSwNP and non-ECRSwNP was analyzed by
CIBERSORT based on the gene expression profile.
The resting CD4+ memory T cells were
found in the highest proportion in the CTRL group, whereas the
activated CD4+ T cells were mainly infiltrated in the
non-ECRSwNP group. Memory T cells are essential components of
immunological memory, and memory CD4 cells could help protect
against infections (28). The
memory CD4+ T cells also outnumber memory
CD8+ T cells in the lung, skin and mucosal surfaces, and
function to direct protective responses in addition to coordinating
the recruitment of immune cells to tissue sites (29-31).
The resident CD4+ memory T cells remodel epithelial
responses to accelerate neutrophil recruitment during pneumonia
(32). These findings indicated
that the occurrence of non-ECRSwNP may be closely related to the
activation of CD4+ cells. The activated CD4+
T cells were positively correlated with M1 macrophages and γδT
cells. Non-ECRSwNP has been reported to be characterized by
Th1/Th17-dominant inflammation, and infiltration by a greater
number of neutrophils and M1 macrophages (1,14,15).
IFN-γ, which promotes the polarization of M1 macrophages, is highly
expressed in non-ECRSwNP (33). The
functionality of γδT cells is induced upon the recognition of
stress antigens, which promotes cytokine production, and regulates
pathogen clearance, inflammation and tissue homeostasis (34).
In the ECRSwNP group, the eosinophil infiltration
was significantly correlated with the infiltration of M2
macrophages, activated mast cells and activated NK cells. Enhanced
accumulation of M2 macrophages has previously been demonstrated in
ECRSwNP compared with in non-ECRSwNP (15,35).
The crucial role of mast cells was evident in a mouse model of
eosinophilic CRS, where none of the mast cell-deficient mice
subjected to chronic allergen challenge developed cystic changes or
polypoid changes in the nose or sinuses (36,37).
Furthermore, in a previous study, patients with CRS presented a
significant decrease in eosinophil apoptosis mediated by NK cells
compared with healthy controls; eosinophilic inflammation and NK
cell dysfunction were responsible for this decrease (38).
Gene expression was correlated with phenotype to
obtain gene expression modules for NP, ECRSwNP and non-ECRSwNP
groups in the present study. One previous study revealed that
olfactory dysfunction is very frequent (~90%) in CRSwNP and does
not depend on nasal obstruction, as assessed by both polyp size and
nasal airflow limitation (39). A
previous study established that superior turbinate eosinophilia is
correlated with an olfactory deficit in patients with CRS (40). The enriched results of olfactory and
channel activity genes suggested that NP may affect the expression
of olfactory receptors and channel activity genes to impair the
olfactory signaling and neuroactive ligand-receptor pathways. To
the best of our knowledge, this is the first time that NP have been
linked to the expression of olfactory genes.
The present study showed that in ECRSwNP, functions
including the ‘binding of cell adhesion molecules’ (GO:0050839),
‘DNA-binding transcription activator activity’ (GO:0001228),
‘cytokine receptor binding’ (GO:0005126), ‘cytokine activity’
(GO:0005125), ‘growth factor receptor binding’ (GO:0005154) and
‘glucocorticoid receptor binding’ (GO:0035259), and ‘inflammatory
signaling pathways’ (hsa04151, hsa04630 and hsa04668) were enriched
and expressed a positive correlation. Cytokines are involved in the
pathogenesis of CRS (41). IL-5
serves an important role in regulating eosinophil development, and
is essential for its maturation and release into circulation
(42-44).
IL-13 can increase the levels of β-catenin, which contributes to
cell-cell adhesion in CRS (45).
The C-C chemokines, RANTES/CCL5, monocyte chemotactic protein-4
(MCP-4/CCL13), and eotaxin/CCL26 have also been found to be
increased in atopic dermatitis skin lesions, and likely contribute
to the chemotaxis of eosinophils and Th2 lymphocytes into the skin
(46). Oncostatin M, a member of
the IL-6 family, and IL-6, have been reported to serve an important
role in the pathophysiology of eosinophilic inflammation in ECRSwNP
(20). Nevertheless, controversy
remains concerning IL-6 function because IL-6 has been predicted as
an upstream regulator in non-ECRSwNP (19). The aforementioned cytokines were
expressed in higher amounts in ECRSwNP compared with those in the
CTRL and non-ECRSwNP groups. Topical corticosteroids have been
demonstrated to exert a beneficial effect on ECRS and nasal
polyposis, and the mechanism of action of nasal steroids appears to
be multi-factorial, being initiated by their binding to the
glucocorticoid receptor (47,48).
Glucocorticoid resistance to glucocorticoids may be the major cause
of therapy failure in ECRSwNP (49).
Previous studies have demonstrated that in
non-ECRSwNP, serum amyloid A (SAA) levels were significantly
upregulated compared with those in ECRSwNP (19,20);
SAA triggers the production of cytokines associated with
neutrophilic inflammation and improves neutrophil chemotaxis
(50,51). In the present study, the blue gene
module was negatively correlated with non-ECRSwNP and established
that genes correlated with non-ECRSwNP were mainly associated with
a variety of transferases (GO:0034212, GO:0004674, GO:0018024,
GO:0019787 and GO:0004842), GTPases (GO:0031267, GO:0030695,
GO:0017016 and GO:0005096) and receptor binding functions
(GO:0046965, GO:0046966, and GO:0035257) and cancer cell pathways
(hsa05224, hsa05213, hsa05217 and hsa05215). These results
suggested that the formation of non-ECRSwNP may be closely related
to epithelial cell injury. Besides, it was identified that the
autophagy pathway was negatively associated with non-ECRSwNP, and
some autophagy genes were downregulated. It has recently been
established that deficient autophagy in myeloid cells exacerbates
CRS and that IFN-γ-induced insufficient autophagy adds to
p62-dependent apoptosis of epithelial cells in CRSwNP (52,53).
Thus, these results suggested the significance of autophagy in
non-ECRSwNP.
The composition and characteristics of infiltrated
immune cells in the immune microenvironment of ECRSwNP and
non-ECRSwNP were systematically analyzed in the present study
(Fig. 8). A total of four types of
immune cells (resting mast cells, activated dendritic cells, M2
macrophages and activated NK cells) were found to have a direct and
indirect correlation with eosinophils in ECRSwNP. Notably, M1
macrophages and activated CD4+ memory T cells were
correlated in non-ECRSwNP. The findings suggested that NP may
affect the expression of olfactory receptors and channel activity
genes to impair olfactory signaling pathways and neuroactive
ligand-receptor pathways. In addition, the cell adhesion molecules,
cytokines and glucocorticoid receptors may serve a vital role in
ECRSwNP. On the other hand, epithelial cell injury and autophagy
may serve an essential role in non-ECRSwNP. These results provided
a good basis for the elucidation of the underlying mechanism and
treatment of CRSwNP. However, the findings of this study were
limited, as only a few high-quality sequencing samples were
available in the public database. Thus, further analysis involving
a large clinical sample is required.
Supplementary Material
Expression levels of 12 related genes
of non-eosinophil polyps, eosinophil polyps and nasal mucosa
tissues. Blue represents non-ECRSwNP, red represents non-nasal
mucosa tissues (CTRL) and green represents ECRSwNP. ECRSwNP,
eosinophilic chronic rhinosinusitis with nasal polyps; non-ECRSwNP,
non-eosinophilic chronic rhinosinusitis with nasal polyps; CTRL,
control; OR10A3, olfactory receptor 10A3; OR1A1, olfactory receptor
1A1; PPARD, peroxisome proliferator-activated receptor δ; ABCC8,
ATP-binding cassette sub-family C member 8; PPARG, peroxisome
proliferator-activated receptor γ; CCL.26, C-C motif chemokine 26;
AQP10, aquaporin-10.
The qPCR primers of 12 related genes
of non-eosinophil polyps, eosinophil polyps and nasal mucosa
tissues
The results of 22 immune cell
infiltration of non-eosinophil polyps, eosinophil polyps and nasal
mucosa tissues.
Go term and KEGG pathway of brown
module
Go term and KEGG pathway of brown
module
Go term and KEGG pathway of blue
module
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from the
National Natural Science Foundation of China (grant no. 81673809),
the Scientific and Technological Program of Zhejiang province
(grant no. 2017F10024), the Traditional Chinese Medicine Scientific
Program of Zhejiang province (grant no. 2016ZQ002), the National
Natural Science Foundation of Zhejiang province (grant no.
LY19H280005) and the Medical and health Scientific and
Technological Program of Zhejiang province (grant no.
2019KY347).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
GX, XX, QW, YZ, YG, WL and ML designed the study and
analyzed the data. ML, GX and XX wrote the manuscript. All authors
read and approved the final manuscript.
Ethics approval and consent to
participate
This study was approved by the Ethics Committee of
Tongde Hospital of Zhejiang Province (approval no. XMSB2018013) and
written informed consent was obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Fokkens WJ, Lund VJ, Mullol J, Bachert C,
Alobid I, Baroody F, Cohen N, Cervin A, Douglas R, Gevaert P, et
al: European position paper on rhinosinusitis and nasal polyps
2012. Rhinol Suppl. 23:1–298. 2012.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Kwah JH and Peters AT: Nasal polyps and
rhinosinusitis. Allergy Asthma Proc. 40:380–384. 2019.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Schleimer RP: Immunopathogenesis of
chronic rhinosinusitis and nasal polyposis. Annu Rev Pathol.
12:331–357. 2017.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Meltzer EO, Hamilos DL, Hadley JA, Lanza
DC, Marple BF, Nicklas RA, Bachert C, Baraniuk J, Baroody FM,
Benninger MS, et al: Rhinosinusitis: Establishing definitions for
clinical research and patient care. J Allergy Clin Immunol.
114:155–212. 2004.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Soler ZM, Sauer D, Mace J and Smith TL:
Impact of mucosal eosinophilia and nasal polyposis on quality-of
life outcomes after sinus surgery. Otolaryngol Head Neck Surg.
142:64–71. 2010.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Vlaminck S, Vauterin T, Hellings PW,
Jorissen M, Acke F, Van Cauwenberge P, Bachert C and Gevaert P: The
importance of local eosinophilia in the surgical outcome of chronic
rhinosinusitis: A 3-year prospective observational study. Am J
Rhinol Allergy. 28:260–264. 2014.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Wang X, Zhang Bo M, Holtappels G, Zheng M,
Lou H, Wang H, Zhang L and Bachert C: Diversity of TH cytokine
profiles in patients with chronic rhinosinusitis: A multicenter
study in Europe, Asia, and Oceania. J Allergy Clin Immunol.
138:1344–1353. 2016.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Lou H, Meng Y, Piao Y, Wang C, Zhang L and
Bachert C: Predictive significance of tissue eosinophilia for nasal
polyp recurrence in the Chinese population. Am J Rhinol Allergy.
29:350–356. 2015.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Nakayama T, Yoshikawa M, Asaka D, Okushi
T, Matsuwaki Y, Otori N, Hama T and Moriyama H: Mucosal
eosinophilia and recurrence of nasal polyps-new classification of
chronic rhinosinusitis. Rhinology. 49:392–396. 2011.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Cho SW, Kim DW, Kim JW, Lee CH and Rhee
CS: Classification of chronic rhinosinusitis according to a nasal
polyp and tissue eosinophilia: Limitation of current classification
system for Asian population. Asia Pac Allergy. 7:121–130.
2017.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Zhang N, Van Zele T, Perez-Novo C, Van
Bruaene N, Holtappels G, DeRuyck N, Van Cauwenberge P and Bachert
C: Different types of T-effector cells orchestrate mucosal
inflammation in chronic sinus disease. J Allergy Clin Immunol.
122:961–968. 2008.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Katotomichelakis M, Tantilipikorn P,
Holtappels G, De Ruyck N, Feng L, Van Zel T, Muangsomboon S,
Jareonchasri P, Bunnag C, Danielides V, et al: Inflammatory
patterns in upper airway disease in the same geographical area may
change over time. Am J Rhinol Allergy. 27:354–360. 2013.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Sakuma Y, Ishitoya J, Komatsu M, Shiono O,
Hirama M, Yamashita Y, Kaneko T, Morita S and Tsukuda M: New
clinical diagnostic criteria for eosinophilic chronic
rhinosinusitis. Auris Nasus Larynx. 38:583–588. 2011.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Cao PP, Li HB, Wang BF, Wang SB, You XJ,
Cui YH, Wang DY, Desrosiers M and Liu Z: Distinct immunopathologic
characteristics of various types of chronic rhinosinusitis in adult
Chinese. J Allergy Clin Immunol. 124:478–484. 2009.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Wang ZC, Yao Y, Wang N, Liu JX, Ma J, Chen
CL, Deng YK, Wang MC, Liu Y, Zhang XH and Liu Z: Deficiency in
interleukin-10 production by M2 macrophages in eosinophilic chronic
rhinosinusitis with nasal polyps. Int Forum Allergy Rhinol.
8:1323–1333. 2018.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Perić A, Baletić N, Sotirović J and
Špadijer-Mirković C: Macrophage inflammatory protein-1 production
and eosinophil infiltration in chronic rhinosinusitis with nasal
polyps. Ann Otol Rhinol Laryngol. 124:266–272. 2015.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Bachert C, Wagenmann M, Hauser U and
Rudack C: IL-5 synthesis is upregulated in human nasal polyp
tissue. J Allergy Clin Immunol. 99:837–842. 1997.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Meyer JE, Bartels J, Görögh T, Sticherling
M, Rudack C, Ross DA and Maune S: The role of RANTES in nasal
polyposis. Am J Rhinol. 19:15–20. 2005.PubMed/NCBI
|
|
19
|
Okada N, Nakayama T, Asaka D, Inoue N,
Tsurumoto T, Takaishi S, Otori N, Kojima H, Matsuda A, Oboki K, et
al: Distinct gene expression profiles and regulation networks of
nasal polyps in eosinophilic and non-eosinophilic chronic
rhinosinusitis. Int Forum Allergy Rhinol. 8:592–604.
2018.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Wang W, Gao Z, Wang H, Li T, He W, Lv W
and Zhang J: Transcriptome analysis reveals distinct gene
expression profiles in eosinophilic and noneosinophilic chronic
rhinosinusitis with nasal polyps. Sci Rep. 6(26604)2016.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Newman AM, Liu CL, Green MR, Gentles AJ,
Feng W, Xu Y, Hoang CD, Diehn M and Alizadeh AA: Robust enumeration
of cell subsets from tissue expression profiles. Nat Methods.
12:453–457. 2015.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Wei TY and Simko V: R package ‘corrplot’:
Visualization of a Correlation Matrix (version 0.84). urihttps://github.com/taiyun/corrplotsimplehttps://github.com/taiyun/corrplot.
|
|
23
|
Langfelder P and Horvath S: WGCNA: An R
package for weighted correlation network analysis. BMC
Bioinformatics. 9(559)2008.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Zhang B and Horvath S: A general framework
for weighted gene co-expression network analysis. Stat Appl Genet
Mol Biol. 4(17)2005.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Langfelder P, Zhang B and Horvath S:
Defining clusters from a hierarchical cluster tree: The dynamic
tree cut package for R. Bioinformatics. 24:719–720. 2008.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Yu G, Wang LG, Han Y and He QY:
ClusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012.PubMed/NCBI View Article : Google Scholar
|
|
28
|
MacLeod MK, Clambey ET, Kappler JW and
Marrack P: CD4 memory T cells: What are they and what can they do?
Semin Immunol. 21:53–61. 2009.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Turner DL, Bickham KL, Thome JJ, Kim CY,
D'Ovidio F, Wherry EJ and Farber DL: Lung niches for the generation
and maintenance of tissue-resident memory T cells. Mucosal Immunol.
7:501–510. 2014.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Teijaro JR, Turner D, Pham Q, Wherry EJ,
Lefrançois L and Farber DL: Cutting edge: Tissue-Retentive lung
memory CD4 T cells mediate optimal protection to respiratory virus
infection. J Immunol. 187:5510–5514. 2011.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Nakanishi Y, Lu B, Gerard C and Iwasaki A:
CD8(+) T lymphocyte mobilization to virus-infected tissue requires
CD4(+) T-cell help. Nature. 462:510–513. 2009.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Shenoy AT, Wasserman GA, Arafa EI, Wooten
AK, Smith NM, Martin IM, Jones MR, Quinton LJ and Mizgerd JP: Lung
CD4+ resident memory T cells remodel epithelial responses to
accelerate neutrophil recruitment during pneumonia. Mucosal
Immunol. 13:334–343. 2019.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Wu D, Wang J and Zhang M: Altered
Th17/Treg ratio in nasal polyps with distinct cytokine profile
association with patterns of inflammation and mucosal remodeling.
Medicine (Baltimore). 95(e2998)2016.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Bonneville M, O'Brien RL and Born WK:
Gammadelta T cell effector functions: A blend of innate programming
and acquired plasticity. Nat Rev Immunol. 10:467–478.
2010.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Shapouri-Moghaddam A, Mohammadian S,
Vazini H, Taghadosi M, Esmaeili SA, Mardani F, Seifi B, Mohammadi
A, Afshari JT and Sahebkar A: Macrophage plasticity, polarization,
and function in health and disease. J Cell Physiol. 233:6425–6440.
2018.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Hua X, Naselsky WC, Jania CM, Chason KD,
Huang JJ, Doerschuk CM, Graham SM, Senior BA and Tilley SL: Mast
cell deficiency limits the development of chronic rhinosinusitis in
mice. Ann Otol Rhinol Laryngol. 125:290–296. 2016.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Gröger M, Bernt A, Wolf M, Mack B,
Pfrogner E, Becker S and Kramer MF: Eosinophils and mast cells: A
comparison of nasal mucosa histology and cytology to markers in
nasal discharge in patients with chronic sino-nasal diseases. Eur
Arch Otorhinolaryngol. 270:2667–2676. 2013.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Kim JH, Choi GE, Lee BJ, Kwon SW, Lee SH,
Kim HS and Jang YJ: Natural killer cells regulate eosinophilic
inflammation in chronic rhinosinusitis. Sci Rep.
6(27615)2016.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Gelardi M, Piccininni K, Quaranta N,
Quaranta V, Silvestri M and Ciprandi G: Olfactory dysfunction in
patients with chronic rhinosinusitis with nasal polyps is
associated with clinical-cytological grading severity. Acta
Otorhinolaryngol Ital. 39:329–335. 2019.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Lavin J, Min JY, Lidder AK, Huang JH, Kato
A, Lam K, Meen E, Chmiel JS, Norton J, Suh L, et al: Superior
turbinate eosinophilia correlates with olfactory deficit in chronic
rhinosinusitis patients. Laryngoscope. 127:2210–2218.
2017.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Shah SA, Ishinaga H and Takeuchi K:
Pathogenesis of eosinophilic chronic rhinosinusitis. J Inflamm
(Lond). 13(11)2016.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Clutterbuck E, Shields JG, Gordon J, Smith
SH, Boyd A, Callard RE, Campbell HD, Young IG and Sanderson CJ:
Recombinant human interleukin 5 is an eosinophil differentiation
factor but has no activity in standard human B cell growth factor
assays. Eur J Immunol. 17:1743–1750. 1987.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Clutterbuck EJ, Hirst EM and Sanderson CJ:
Human interleukin-5 (IL-5) regulates the production of eosinophils
in human bone marrow cultures: Comparison and interaction with
IL-1, IL-3, IL-6, and GMCSF. Blood. 73:1504–1512. 1989.PubMed/NCBI
|
|
44
|
Clutterbuck EJ and Sanderson CJ:
Regulation of human eosinophil precursor production by cytokines: A
comparison of recombinant human interleukin-1 (rhIL-1), rhIL-3,
rhIL-5, rhIL-6, and rh granulocyte-macrophage colony-stimulating
factor. Blood. 75:1774–1779. 1990.PubMed/NCBI
|
|
45
|
Sauter A, Stern-Straeter J, Chang RC,
Hörmann K and Naim R: Influence of interleukin-13 on beta-catenin
levels in eosinophilic chronic rhinosinusitis cell culture. Int J
Mol Med. 21:447–452. 2008.PubMed/NCBI
|
|
46
|
Novak N and Donald YM: Role of barrier
dysfunction and immune response in atopic dermatitis. In: Pediatric
Allergy: Principles and Practice. 3rd edition. Elsevier, pp438-447,
2016.
|
|
47
|
Alobid I and Mullol J: Role of medical
therapy in the management of nasal polyps. Curr Allergy Asthma Rep.
12:144–153. 2012.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Takeda K, Takeno S, Hirakawa K and Ishino
T: Expression and distribution of glucocorticoid receptor isoforms
in eosinophilic chronic rhinosinusitis. Auris Nasus Larynx.
37:700–707. 2010.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Pujols L, Mullol J, Benítez P, Torrego A,
Xaubet A, de Haro J and Picado C: Expression of the glucocorticoid
receptor alpha and beta isoforms in human nasal mucosa and polyp
epithelial cells. Respir Med. 97:90–96. 2003.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Wang H, Bai J, Ding M, Liu W, Xu R, Zhang
J, Shi J and Li H: Interleukin-17A contributes to the expression of
serum amyloid A in chronic rhinosinusitis with nasal polyps. Eur
Arch Otorhinolaryngol. 270:1867–1872. 2013.PubMed/NCBI View Article : Google Scholar
|
|
51
|
He R, Sang H and Ye RD: Serum amyloid A
induces IL-8 secretion through a G protein-coupled receptor,
FPRL1/LXA4R. Blood. 101:1572–1581. 2003.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Choi GE, Yoon SY, Kim JY, Kang DY, Jang YJ
and Kim HS: Autophagy deficiency in myeloid cells exacerbates
eosinophilic inflammation in chronic rhinosinusitis. J Allergy Clin
Immunol. 141:938–950. 2018.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Wang BF, Cao PP, Wang ZC, Li ZY, Wang ZZ,
Ma J, Liao B, Deng YK, Long XB, Xu K, et al: Interferon-γ-induced
insufficient autophagy contributes to p62-dependent apoptosis of
epithelial cells in chronic rhinosinusitis with nasal polyps.
Allergy. 72:1384–1397. 2017.PubMed/NCBI View Article : Google Scholar
|






















