Introduction
Eugenol is the major bioactive component of clove,
which has been previously documented to exhibit anti-oxidant,
anti-mutagenic, anti-microbial, anti-inflammatory and anti-tumor
properties (1). In particular, the
anti-oxidant activity of eugenol is garnering significant attention
(2). Since the discovery of
anti-oxidative effects of eugenol suppressed copper-mediated lipid
peroxidation on erythrocyte membranes (3), analogous anti-oxidative effects of
eugenol have also been documented in a number of different cell
types, including hepatocytes (4),
macrophages (5) and cancer cells
(6). Numerous in vivo
studies have also reported the ability of eugenol in eliminating
reactive oxygen species (ROS), where the anti-inflammatory and
anti-cancer effects of eugenol may be due to its capability in
scavenging ROS (2,5). Other studies have also previously
suggested that the mechanism underlying the anti-oxidant activity
of eugenol may involve cyclooxygenase-2 (COX-2) inhibition or
direct trapping of ROS molecules (7,8).
Aspirin eugenol ester (AEE), synthesized by combining aspirin with
eugenol, has been reported to attenuate oxidative injury of
vascular endothelial cells by regulating nitric oxide synthase and
Nrf2 signaling (9). However, the
molecular mechanism by which eugenol suppresses the intracellular
concentration of ROS and free radicals remains to be fully
elucidated.
Nuclear transcription factor erythroid 2p45-related
factor 2 (Nrf2) belongs to a family of basic leucine zipper protein
transcription factors, which contributes to cellular defense
against oxidative stress induced by external stimuli by regulating
the transcription of a number of anti-oxidative factors (10). Accumulating evidence has suggested
that Nrf2 serves a pivotal role in the initiation and maintenance
of the protective response against oxidative stress in normal and
neoplastic cells by reducing intracellular ROS levels (11). Therefore, the identification of
natural Nrf2 activator is currently the topic of extensive
investigation, with the aim of developing novel therapeutic
interventions for diseases associated with oxidative stress
(12). Curcumin, a natural product
that is enriched in Curcuma longa and Oleanolic acid, an
active component widely distributed in plants, have both been
reported to exert anti-oxidative activity by inducing Nrf2
activation (13,14). In addition, methyleugenol, a
derivative of eugenol, has been documented to serve a protective
role against tert-Butyl hydroperoxide (t-BHP)-induced cytotoxicity
by activating the 5'AMP-activated protein kinase (AMPK)/Glycogen
synthase kinase 3β (GSK3β) and ERK-Nrf2 signaling pathways
(15).
Therefore, the aim of the present study was to
investigate the mechanism underlying the ROS-eliminating activity
of eugenol and any potential effects on Nrf2 signaling.
Materials and methods
Cell lines and reagents
293 cells and NIH-3T3 cells were obtained from the
American Type Culture Collection. Cells were cultured in DMEM
supplemented with 10% FBS, 2 mM glutamine and 100 U/ml
penicillin-streptomycin (Life Technologies; Thermo Fisher
Scientific, Inc.) at 37˚C in 5% CO2 and were
sub-cultured every 3-4 days.
Eugenol, methyleugenol, acetyleugenol,
tert-butylhydroquinone (tBHQ) and diamide were purchased from
Sigma-Aldrich (Merck KGaA). Nrf2 antibody was purchased from Abcam
(dilution 1:1,000, cat. no. ab62352). GAPDH (dilution 1:1,000, cat.
no. 2118), human influenza hemagglutinin (HA)-Tag (dilution
1:1,000, cat. no. 3724), lamin A/C (dilution 1:1000, cat. no. 2032)
and cleaved caspase-3 (dilution 1:1,000, cat. no. 9664) antibodies
were purchased from Cell Signaling Technology, Inc.
Plasmids
The NC16 pcDNA3.1 FLAG NRF2 plasmid was a gift from
Professor Randall Moon (cat. no. 36971; Addgene, Inc.). A total of
1 µg plasmid was used per transfection reaction in a 60-mm dish.
The pGV113-shNRF2 and pGV113-shControl were purchased from Shanghai
GeneChem Co., Ltd. A total of 1 µg plasmid was used per
transfection reaction in a 60-mm dish. Short hairpin sequence
targeting the Nrf2 coding region was 5'-AGCAAACAAGAGATGGCAA-3',
which was the sequence incorporated into the pGV113-shNRF2 plasmid.
Construction of the plasmid-antioxidant responsive element
(pARE)-TI-luciferase reporter (ARE-luciferase) plasmid was
performed according to protocols previously described (16,17).
The original backbone plasmid is pGL3 Luciferase Reporter Vectors
(Promega Corporation). A total of 200 ng plasmid was used per
transfection reaction in 24-well plates. HA-ubiquitin was a gift
from Edward Yeh (car. no. 18712; Addgene, Inc.). A total of 1 µg
HA-ubiquitin was used per transfection reaction in a 60-mm
dish.
ARE-derived luciferase activity
assay
The construction of pARE-TI-luciferase reporter
(ARE-luciferase) was completed as previously described (16,17).
293 cells or NIH-3T3 cells (2x105 cells per well in
24-well plates) were transfected with the Renilla, which was
used for normalizing all luciferase activity (Promega Corporation)
and ARE-luciferase plasmids using Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.). The cells were lysed
24 h after transfection and assayed for luciferase activity using
Dual-Luciferase® Assay system (Promega Corporation)
according to the manufacturer's protocols. Relative light units
were measured using a SpectraMax M5 microplate reader (Molecular
Devices, LLC).
Reverse transcription-quantitative PCR
(RT-qPCR)
Total cellular RNA was isolated using RNeasy Mini
Kit (Qiagen China Co., Ltd.), following which cDNA was synthesized
using SuperScript IV Reverse Transcriptase (Invitrogen; Thermo
Fisher Scientific, Inc.) according to the manufacturer's protocol.
qPCR was performed in a ABI 7500 Real-Time PCR system (Applied
Biosystems; Thermo Fisher Scientific, Inc.) using FastStart
SYBR® Green Master (Sigma-Aldrich; Merck KGaA) according
to the manufacturer's protocols at the following thermocycling
conditions: Initial denaturation at 95˚C for 15 sec, annealing at
60˚C for 40 cycles and extension at 72˚C for 30 sec. Primer
sequences used were as follows: NRF2-forward,
5'-AACCAGTGGATCTGCCAACTACTC-3' and reverse,
5'-CTGCGCCAAAAGCTGCAT-3'; glutamate-cysteine ligase modifier
regulatory subunit (GCL-M) forward, 5'-GCTGTATCAGTGGGCACAG-3' and
reverse, 5'-CGCTTGAATGTCAGGAATGC-3'; glutathione S-transferase A1
(GSTA1) forward, 5'-CCTGCCTTTGAA AAAGTCTTAAAG-3' and reverse,
5'-AAGTTCCACCA GGTGAATGTCA-3' and GAPDH forward, 5'-GGGAAG
GTGAAGGTCGGAGT-3' and reverse, 5'-TGTAGTTGAGGT CAATGAAGGGG-3'. All
reactions were performed in triplicate, and repeated at least
twice.
Western blotting
Following treatments, 293 cells or NIH-3T3 cells
were washed with ice-cold PBS and harvested using RIPA lysis buffer
(Thermo Fisher Scientific, Inc.) containing protease inhibitors
(Roche Diagnostics). Cell lysates were vigorously vortexed,
homogenized in an ultrasonicator at 15-25 kHz for 10 sec and left
on ice for 30 min. The homogenates were centrifuged at 12,000 x g
for 15 min at 4˚C. The supernatant was subsequently collected and
equal amounts (30 µg) of total protein per sample, as determined by
Bicinchoninic Acid protein assay (Pierce; Thermo Fisher Scientific,
Inc.), was mixed with 4X loading buffer and heated at 95˚C for 5
min. The samples were then separated by 7.5% SDS-PAGE at 120 V and
transferred onto polyvinylidene difluoride membranes (Immobilon-P;
EMD Millipore) for 1.5 h. The membranes were blocked with 5% BSA
(Sigma-Aldrich; Merck KGaA) dissolved in 1X TBS-0.1% Tween 20
buffer for 1 h at room temperature and incubated with indicated
primary antibodies overnight at 4˚C. Following primary antibody
incubation, the membranes were washed three times with TBST before
being incubated with the following secondary antibodies: Goat
anti-mouse immunoglobulin G (IgG)-horseradish peroxidase (HRP)
(1:3,000; cat. no. sc-2005; Santa Cruz Biotechnology, Inc.) and
goat anti-rabbit IgG-HRP (1:3,000; cat. no. sc-2004; Santa Cruz
Biotechnology, Inc.) for 1 h at room temperature and then washed
with TBST 3 times. Protein bands were visualized using SuperSignal™
West PICO (Pierce; Thermo Fisher Scientific, Inc.) followed by
exposure to X-ray films, where exposure scans varied from 5 sec to
60 min. Quantitative data normalized against the reference gene
(GAPDH for most samples and lamin A/C for nuclear samples) were
obtained by densitometric analysis using the Bio-Rad Quantity One
software (version 4.6.2; Bio-Rad Laboratories, Inc.).
Nrf2 half-life experiments
293 cells or NIH-3T3 cells (seeded into 6-well
plates at 3x105 cells/well) were transfected with the
NC16 pCDNA3.1 FLAG NRF2 plasmid using Lipofectamine®
2000 (Invitrogen; Thermo Fisher Scientific, Inc.). At 24 h
following transfection, the cells were incubated with either
vehicle or three eugenol derivatives for 6 h before being treated
with 30 µg/ml cycloheximide (Sigma-Aldrich; Merck-KGaA), a protein
synthesis inhibitor. Cells were harvested for analysis at 0, 5, 15
and 20 min.
Ubiquitination assay
293 cells (seeded into 6-well plates at
3x105 cells/well) were co-transfected with pcDNA3.1
expression vectors encoding HA-ubiquitin and Nrf2 using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.). At 24 h following transfection, the cells were
treated with eugenol at 50 µM and MG132 at a final concentration of
2 µM (Sigma-Aldrich; Merck KGaA) for 12 h, 37˚C to inhibit
proteasome activity. Whole-cell extracts were subsequently prepared
by lysis with RIPA buffer and subjected to purification procedures.
A total of 20 µl agarose beads coupled with anti-FLAG antibody
(1:1,000; cat. no. F3165; Sigma-Aldrich; Merck KGaA) was added to
the cell lysates and incubated with gentle agitation for 1-3 h at
4˚C. The lysates were microcentrifuged at 7,000 x g for 30 seconds
at 4˚C. Subsequently, the pellet was washed 5 times on ice with 500
µl 1X RIPA buffer. Precipitates were visualized by western blot
analysis using the aforementioned anti-HA antibodies.
Cytoplasmic and nuclear protein
extraction
293 cells with pcDNA3.1-Nrf2 or empty pcDNA3.1
plasmid were treated with eugenol (50 µg/ml), tBHQ (100 µg/ml) or
diamide (20 µg/ml) at 37˚C for 6 h. Cytoplasmic and nuclear protein
were extracted using NE-PER™ Nuclear and Cytoplasmic Extraction
Reagents (Pierce; Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocol.
ROS detection
HEK-293 cells or NIH-3T3 cells were seeded into
6-well plates at a density of 3x105 cells/well in fully
supplemented DMEM for 24 h before they were treated with 50 µg/ml
eugenol for 6 h at 37˚C. Untreated and eugenol-treated cells were
subsequently harvested in fully supplemented DMEM, centrifuged for
5 min at 200 x g and 4˚C and suspended in Dulbecco's PBS (DPBS)
containing the 10 µM CM-H2DCFDA dye (Invitrogen; Thermo Fisher
Scientific, Inc.) with or without 50 µM H2O2,
at 37˚C in the dark for 30 min. The cells were then pelleted and
resuspended in ice-cold DPBS prior to the addition of propidium
iodide (1 µg/ml) to the cells for 5 min at room temperature, for
analysis by flow cytometry (MoFlo® flow cytometer;
Beckman Coulter, Inc.) by measuring fluorescence emission at 525 nm
(PI) following excitation at 488 nm (DCFDA/FITC). Since live cells
are impermeant to propidium iodide, only cells negative for
propidium iodide staining were assessed for ROS production. Data
was analyzed by Kaluza C Analysis software 2.1 (Beckman Coulter,
Inc.).
Cell viability
293 cells or NIH-3T3 cells were transfected with
shRNA in 60-mm dishes. At 12 h after transfection, cells were
seeded into 96-well plates (1x103 cells per well in a
96-well plate. 293 cells were treated at 37˚C with eugenol (0, 25,
50 or 100 µg/ml) and H2O2 (0, 50, 100 or 250
µM) for 24 h, while NIH-3T3 cells were treated with 100 µg/ml
eugenol. Before absorbance measurement, cells were washed with PBS
followed by the addition of MTT (1 mg/ml/well) and incubation for 4
h at 37˚C. The medium was subsequently discarded from each well
where the formazan crystals were dissolved in DMSO. Absorbance was
measured at 570 nm using a SpectraMax M5 Plate reader (Molecular
Devices, LLC). All experiments were performed in triplicate. The
IC50 of each group was measured using an IC50 Calculator
(https://www.aatbio.com/tools/ic50-calculator/).
Statistical analysis
Each experiment was reproduced at least three times
with consistent results. Pairwise comparisons were made using
Student's t-test or a non-parametric U Mann-Whitney test using the
SPSS version 18.0 (SPSS, Inc.) and Microsoft Excel Office 2011
(Microsoft Corporation) softwares. Multiple group analyses were
performed using one-way ANOVA followed by Tukey's post hoc test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Eugenol increases the concentration of
Nrf2 proteins and its transcriptional activity
Nrf2 serves an important role in protective
responses against oxidative stress (16,17).
Therefore, for the present study, the effect of eugenol and its
derivatives, methyleugenol and acetyleugenol (Fig. 1), on the expression of Nrf2 and its
transcriptional activity on target gene transcription were
investigated in HEK-293 cells and the mouse fibroblast cell line
NIH-3T3. Luciferase reporter assay based on plasmids containing
ARE, a putative Nrf2-responsive cis-acting element, revealed that
transcriptional activity of Nrf2 was significantly increased in
both HEK-293 and NIH-3T3 cells treated with eugenol compared with
control vehicle cells treated with DMSO (Fig. 2A); with dose-dependency observed for
HEK-293 cells. However, no significant differences in Nrf2
transcriptional activity were observed between
acetyleugenol-treated and control groups (Fig. 2A and B). For methyleugenol treatment, it appears
to be weaker than eugenol in HEK-293 cells but similar to eugenol
at high doses; but had no effect on 3T3 cells.
To verify the effect of these three eugenol
derivatives on Nrf2-mediated transcription further, RT-qPCR was
performed on HEK-293 cells to determine the expression levels of
target genes downstream of Nrf2 activation (GCL-M, GSTA1 and
NFE2L2). Of the three eugenol derivatives tested, only
eugenol treatment significantly increased mRNA expression of all
three genes tested, whereas treatment with the other two
derivatives only elevated one or two of the Nrf2 target genes
tested (Fig. 3A-C).
 | Figure 3Eugenol enhanced the expression levels
of Nrf2 target genes in 293 cells. (A) After 24 h treatment with 50
µg/ml eugenol and its two derivatives, mRNA expression of GCL-M and
(B) GSTA1, target genes for Nrf2, was quantified by reverse
transcription-quantitative PCR. (C) mRNA expression of NFE2L2, as
measured by reverse transcription-quantitative PCR, after treatment
with 50 µg/ml eugenol and its two derivatives. In all instances, 50
µg/ml tBHQ was applied as positive control. Data are presented as
mean ± SD from three independent experiments. *P<0.05
vs. Mock. Nrf2, nuclear factor erythroid 2-related factor 2; GCL-M,
glutamate-cysteine ligase modifier regulatory subunit; GSTA1,
glutathione S-transferase A1; tBHQ, tert-butylhydroquinone. |
Western blotting revealed that the expression levels
of Nrf2 protein were increased by treatment with eugenol, but not
with the other two derivatives in HEK-293 cells (Fig. 4A). Similar observations were
obtained in NIH-3T3 cells (Fig.
4B). These data suggest that eugenol can promote the expression
and transcriptional activity of Nrf2 in HEK-293 and NIH-3T3
cells.
Eugenol stabilizes Nrf2 and increases
its nuclear accumulation
To explore the molecular mechanism underlying the
effect of eugenol on Nrf2, changes in the stability of Nrf2
following exposure to eugenol were tested. Nrf2 proteins persisted
for markedly longer in the presence of eugenol in both 293
(Fig. 5A) and NIH-3T3 cells
(Fig. 5B). Ubiquitination assay
also showed that Nrf2 ubiquitination was markedly reduced following
treatment with eugenol (Fig. 6).
Samples without UB and NRF2 plasmid transfection acted as a
negative control.
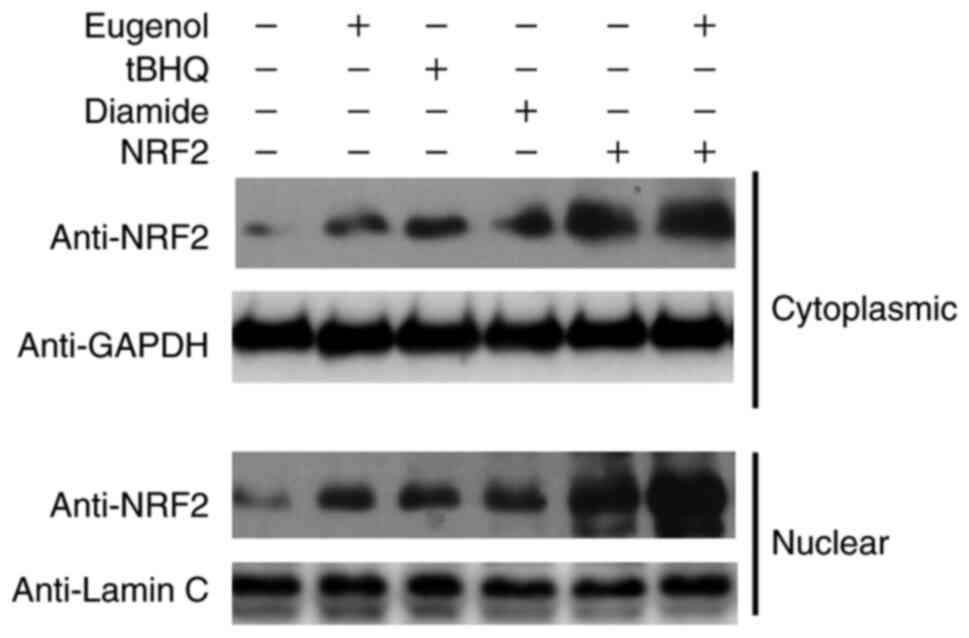
Subsequently, further western blotting was performed
to assess Nrf2 expression in the cytosol and nucleus of 293 cells.
Diamide and tBHQ was reported to increase NRF2 protein expression
(15). The nuclear and cytoplasmic
content of Nrf2 was found to be markedly increased following
treatment with eugenol (Fig. 7).
Taken together, these observations suggest that eugenol was able to
induce the stabilization and nuclear accumulation of Nrf2.
Eugenol protects cells against
H2O2-induced oxidative damage
To investigate the effects of eugenol on oxidative
stress, DCF assays were conducted on 293 cells, which were loaded
with the ROS-sensitive dye CM-H2DCFDA following eugenol treatment.
After exposure to H2O2, cells treated with
eugenol exhibited significantly lower intracellular ROS levels
compared with mock cells (Fig.
8).
MTT assay was used to assess the protective effects
of eugenol on H2O2-treated 293 and 3T3 cells.
Eugenol was found to significantly increase cell viability in the
presence of H2O2 (Fig. 9). These results suggest that eugenol
protects cells from H2O2-induced
cytotoxicity.
Nrf2 induction is required for the
protective effects of eugenol on cells against oxidative
stress
To study the role of increased Nrf2 expression and
transcriptional activity in the enhanced cell viability induced by
eugenol, Nrf2 expression was knocked down using plasmids expressing
short hairpin RNA (Fig. 11A). Nrf2
knockdown significantly abrogated the protective effects of eugenol
on HEK-293 and 3T3 cells following exposure to
H2O2 (Fig.
10). In addition, western blot analysis of cleaved caspase-3, a
key regulator of apoptosis, suggested that eugenol treatment
abolished the pro-apoptotic activity of H2O2
on HEK-293 and 3T3 cells, as evidenced by reduced levels of cleaved
caspase-3 in cells transfected with shControl after
H2O2 treatment. However, the expression
levels of cleaved Caspase-3 after exposure to
H2O2 was not affected by eugenol treatment
following Nrf2 knockdown (Fig.
11B). These results suggest that increased Nrf2 expression is
required for the protective effects of eugenol on
H2O2-induced oxidative stress.
Discussion
The present study revealed that the transcriptional
activity and expression of Nrf2 is increased by the eugenol
treatment, resulting in increased expression of target genes.
Although the two other derivatives of eugenol tested also exerted
similar effects on Nrf2 function, their potency appeared to be
weaker compared with that of eugenol. This result may be explained
by the importance of the hydroxyl group attached to aromatic ring,
which may mediate the anti-oxidant properties of eugenol. In the
other two derivatives, the hydroxyl group was replaced by methoxyl
and acetyl-O groups, respectively. This finding suggests that this
hydroxyl group should be preserved in subsequent optimizations of
eugenol for the enhancement of bioactivity.
Mechanistically, the present study demonstrated that
eugenol increases the expression level and activity of Nrf2 by
stabilizing the Nrf2 protein through suppressing ubiquitination.
The attachment of ubiquitin to Nrf2 by Kelch-like ECH associated
protein 1, an E3 ligase that specifically recognizes Nrf2, lead to
its degradation in a proteasome-dependent manner (18,19).
This event is believed to be the major regulatory mechanism
upstream of Nrf2(18). Although the
effect of eugenol on Nrf2 levels has been confirmed by the present
study, available data remain insufficient to conclude if Keap1 or
Nrf2 itself is a target of eugenol, which warrant further
study.
A number of molecular mechanisms have been proposed
to be associated with the anti-oxidant properties of eugenol. For
instance, COX-2 mRNA expression was reduced by eugenol treatment in
LPS-stimulated macrophages in a previous study (7), whilst eugenol was reported to
sequester hydroxyl radicals in another study in an in vitro
system (8). Subsequent experiments
in the present study revealed that eugenol treatment can rescue
cells from oxidative stress induced by H2O2
exposure in a Nrf2 dependent manner. The near complete reversal of
this protective effect by eugenol on cells by Nrf2 knockdown
suggest that the activation of Nrf2 signaling is a novel mechanism
by which eugenol counters against oxidative stress. These data
confirmed the pivotal role of Nrf2 signaling in the regulation of
cellular Redox systems and further support Nrf2 as a promising
target for the development of novel anti-oxidative drugs.
Huang et al (9) previously found that AEE protects
vascular endothelial cells from oxidative injury by regulating NOS
and Nrf2 signaling pathways, whilst Zhou et al (15) reported that methyleugenol may
exhibit a protective role against t-BHP-induced cytotoxicity
through the activation of the AMPK/GSK3β- and ERK-Nrf2 signaling
pathways. Comparing the present study with these two previous
studies aforementioned, the present study revealed that eugenol,
but not AEE or methyleugenol, exhibited anti-oxidant
properties.
In conclusion, the present study provided evidence
that eugenol activates Nrf2 signaling resulting in the protection
of cells from damage from oxidative stress. These data elucidated
the molecular mechanism underlying the anti-oxidative activity of
Nrf2 signaling, supporting the notion that eugenol may be a
promising lead compound for the development of novel potent
anti-oxidant drugs.
Acknowledgements
Not applicable.
Funding
This work was supported by the National Natural
Science Foundation of China (grant nos. 81272264, 81172013,
81101505 and 81672926) and Zhejiang Provincial Natural Science
Foundation of China (grant no. LQ16C090001).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
LM and YG performed the cell experiments. LM, JL and
QL performed the in vitro and in vivo assays. LM and WY designed
the study, interpreted the results and wrote and edited the
manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Kamatou GP, Vermaak I and Viljoen AM:
Eugenol--from the remote Maluku Islands to the international market
place: A review of a remarkable and versatile molecule. Molecules.
17:6953–6981. 2012.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Srinivasan K: Antioxidant potential of
spices and their active constituents. Crit Rev Food Sci Nutr.
54:352–372. 2014.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Nagashima K: Inhibitory effect of eugenol
on Cu2+-catalyzed lipid peroxidation in human erythrocyte
membranes. Int J Biochem. 21:745–749. 1989.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Nagababu E and Lakshmaiah N: Inhibition of
microsomal lipid peroxidation and monooxygenase activities by
eugenol. Free Radic Res. 20:253–266. 1994.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Joe B and Lokesh BR: Role of capsaicin,
curcumin and dietary n-3 fatty acids in lowering the generation of
reactive oxygen species in rat peritoneal macrophages. Biochim
Biophys Acta. 1224:255–263. 1994.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Jaganathan SK and Supriyanto E:
Antiproliferative and molecular mechanism of eugenol-induced
apoptosis in cancer cells. Molecules. 17:6290–6304. 2012.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Kim SS, Oh OJ, Min HY, Park EJ, Kim Y,
Park HJ, Nam Han Y and Lee SK: Eugenol suppresses cyclooxygenase-2
expression in lipopolysaccharide-stimulated mouse macrophage
RAW264.7 cells. Life Sci. 73:337–348. 2003.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Ogata M, Kaneya D, Shin-Ya K, Li L, Abe Y,
Katoh H, Seki S, Seki Y, Gonda R, Urano S, et al: Trapping effect
of eugenol on hydroxyl radicals induced by L-DOPA in vitro. Chem
Pharm Bull (Tokyo). 53:1167–1170. 2005.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Huang MZ, Yang YJ, Liu XW, Qin Z and Li
JY: Aspirin eugenol ester attenuates oxidative injury of vascular
endothelial cells by regulating NOS and Nrf2 signalling pathways.
Br J Pharmacol. 176:906–918. 2019.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Gold R, Kappos L, Arnold DL, Bar-Or A,
Giovannoni G, Selmaj K, Tornatore C, Sweetser MT, Yang M, Sheikh
SI, et al: DEFINE Study Investigators: Placebo-controlled phase 3
study of oral BG-12 for relapsing multiple sclerosis. N Engl J Med.
367:1098–1107. 2012.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Niture SK, Khatri R and Jaiswal AK:
Regulation of Nrf2-an update. Free Radic Biol Med. 66:36–44.
2014.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Kumar H, Kim IS, More SV, Kim BW and Choi
DK: Natural product-derived pharmacological modulators of Nrf2/ARE
pathway for chronic diseases. Nat Prod Rep. 31:109–139.
2014.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Xie Y, Zhao QY, Li HY, Zhou X, Liu Y and
Zhang H: Curcumin ameliorates cognitive deficits heavy ion
irradiation-induced learning and memory deficits through enhancing
of Nrf2 antioxidant signaling pathways. Pharmacol Biochem Behav.
126:181–186. 2014.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Lu YF, Liu J, Wu KC and Klaassen CD:
Protection against phalloidin-induced liver injury by oleanolic
acid involves Nrf2 activation and suppression of Oatp1b2. Toxicol
Lett. 232:326–332. 2015.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Zhou J, Ma X, Cui Y, Song Y, Yao L, Liu Y
and Li S: Methyleugenol protects against t-BHP-triggered oxidative
injury by induction of Nrf2 dependent on AMPK/GSK3β and ERK
activation. J Pharmacol Sci. 135:55–63. 2017.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Miltenberger RJ, Cortner J and Farnham PJ:
An inhibitory Raf-1 mutant suppresses expression of a subset of
v-raf-activated genes. J Biol Chem. 268:15674–15680.
1993.PubMed/NCBI
|
|
17
|
Wasserman WW and Fahl WE: Functional
antioxidant responsive elements. Proc Natl Acad Sci USA.
94:5361–5366. 1997.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Jaramillo MC and Zhang DD: The emerging
role of the Nrf2-Keap1 signaling pathway in cancer. Genes Dev.
27:2179–2191. 2013.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Liu J, Shaik S, Dai X, Wu Q, Zhou X, Wang
Z and Wei W: Targeting the ubiquitin pathway for cancer treatment.
Biochim Biophys Acta. 1855:50–60. 2015.PubMed/NCBI View Article : Google Scholar
|