Introduction
Atherosclerosis is a chronic progressive
inflammatory vascular disease, which poses a major threat to human
health and has been attracting increasing attention (1). Thrombotic events associated with acute
rupture or erosion of an unstable plaque, rather than gradual
narrowing of the lumen, have been shown to be responsible for the
majority of the clinical consequences of atherosclerosis (2). It is well established that vascular
smooth muscle cells (VSMCs) serve an important role in the
pathogenesis of atherosclerosis (2,3).
VSMCs, as one of the key components of the plaques, are derived
from the medial layer of the vessel wall, which acts as a regulator
of the atherosclerotic plaque (4-6).
Furthermore, excess proliferation or dysfunction of VSMCs
contributes to atherogenesis as a response to vascular injury,
inflammation and lipoprotein accumulation during disease
progression (1,7). Oxidized low-density lipoprotein
(ox-LDL) contributes to the atherosclerotic lesion through several
mechanisms, such as the dysfunction of endothelial cells, and the
excess migration and proliferation of VSMCs, as well as
contributing to plaque instability (8). Therefore, in the present study VSMCs
were treated with ox-LDL and used as the cell model for
atherosclerosis in order to investigate the specific mechanisms
underlying the pathogenesis of atherosclerosis.
Previous studies have revealed that long non-coding
RNAs (lncRNAs), a class of non-coding RNAs >200 nucleotides in
length, serve an important role in the onset and development of
multiple human diseases, including cancer, diabetes, inflammatory
diseases and cardiovascular diseases (9-11).
The expression levels of the lncRNA zinc finger antisense 1 (ZFAS1)
were revealed to be upregulated in the plaques of patients with
atherosclerosis compared with in controls, as well as in
atherosclerosis rat models (12,13),
indicating that ZFAS1 is closely associated with the progression of
atherosclerosis. A recent study reported that ZFAS1 upregulation
was observed in the cytoplasm and sarcoplasmic reticulum of mouse
cardiomyocytes challenged with hypoxic stimulation, and that it
impaired cardiac function in a mouse model of acute myocardial
infarction, and these effects were readily reversed by
ZFAS1-knockdown (14). Furthermore,
lncRNA ZFAS1 promotes the proliferation, invasion and migration of
various cancer cells, including nasopharyngeal carcinoma (15), cervical carcinoma (16) and colorectal cancer (17). Additionally, ZFAS1 may promote the
proliferation and migration of chondrocytes, and suppress apoptosis
and matrix synthesis in osteoarthritis (18). Overall, the aforementioned findings
indicate that ZFAS1 serves a key role in promoting cell
proliferation and invasion. Thus, it was hypothesized that ZFAS1
may serve as a potential biomarker in atherosclerosis induced by
ox-LDL, and may promote the proliferation and invasion of VSMCs
under pathological conditions.
In the present study, VSMCs were treated with
multiple doses of ox-LDL to induce a cell model of atherosclerosis,
in order to investigate whether ZFAS1 expression was upregulated by
ox-LDL treatment in a dose-dependent manner and to determine
whether ZFAS1 may be of value as a novel biomarker for dysfunction
of VSMCs in the pathological condition of atherosclerosis.
Materials and methods
Cell culture and transfection
The VSMC cell line was obtained from the China
Infrastructure of Cell Line Resources, Institute of Basic Medical
Sciences (Chinese Academy of Sciences). The cells were cultured in
DMEM (HyClone; Cytiva) with 10% fetal bovine serum (FBS; Gibco;
Thermo Fisher Scientific, Inc.), 1% penicillin (100 U/ml) and 1%
streptomycin (100 mg/ml) (Beyotime Institute of Biotechnology) at
37˚C in a humidified incubator with 5% CO2. VSMCs were
cultured in 6-well plates for 12 h, and then transfected with
indicated plasmids. ZFAS1-1 small interfering (si)RNA
(5'-CTGGCTGAACCAGTTCCACAAGGTT-3'), ZFAS1-2 siRNA
(5'-TACTTCTCCTAGTTGCAGTCAGG-3') and the scramble negative control
siRNA (si-NC; 5'-ACGTGACACGTTCGGAGAATT-3') were obtained from
Shanghai GenePharma Co., Ltd., and 100 nM of each siRNA was
transfected into VSMCs for ZFAS1-knockdown using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) following the manufacturer's instructions. In
addition, ZFAS1 transcript cDNA
(5'-UGCGUGCCAAGCGCGACAUGGCGCGGAAGCCGAGAAGCCCCGGAGGCCC-3') was
inserted into the pCDNA3.1 vector [Jiman Biotechnology (Shanghai)
Co., Ltd.] and constructed by Biotech Integrated Solutions, and
then 2 mg/l ZGAS1 pcDNA3.1 or empty pcDNA3.1 was transfected into
VSMCs to achieve ZFAS1 overexpression (oe-ZFAS1) using
Lipofectamine® 2000. After transfection for 8 h at 37˚C,
the VSMCs were exposed to ox-LDL (25, 50 and 100 mg/l; Beijing
Solarbio Science & Technology Co., Ltd.) for 12, 24 and 48 h at
37˚C.
RNA isolation and reverse
transcription-quantitative (RT-q)PCR
Total RNA from VSMCs was isolated using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) following the manufacturer's instructions. RNA (1 µg) was
reverse transcribed into first-strand cDNA using the Reverse
Transcriptase kit according to the manufacturer's protocol
(TransGen Biotech Co., Ltd.). qPCR was performed using SYBR Green
Mixture (Takara Bio, Inc.) in the ABI Prism 7300 Sequence Detection
System (Applied Biosystems; Thermo Fisher Scientific, Inc.). The
thermocycling conditions were: Initial denaturation for 10 min at
94˚C, followed by 40 of cycles of denaturation for 30 sec at 94˚C,
annealing for 30 sec at 55˚C and extension for 30 sec at 72˚C The
2-ΔΔCq method (19) was
applied to determine the relative target gene expression. The
sequences for the qPCR primers were as follows: ZFAS1 forward,
5'-AGCGTTTGCTTTGTTCCC-3' and reverse, 5'-CTCCCTCGATGCCCTTCT-3';
GAPDH forward, 5'-GGTCTCCTCTGACTTCAACA-3' and reverse,
5'-AGCCAAATTCGTTGTCATAC-3'. GAPDH was employed as an internal
control.
Cell Counting Kit-8 (CCK-8) assay
The cell viability of VSMCs was determined using the
CCK-8 assay (Dojindo Molecular Technologies, Inc.). Briefly, VSMCs
transfected with or without oe-ZFAS1 or si-ZFAS1 were seeded at the
density of 2x103 cells/well into 96-well plates and
treated with ox-LDL. After transfection for 12, 24 and 48 h, CCK-8
reagent (10 µl) was added into each well and incubated with VSMCs
for another 2 h. The absorbance at 450 nm was measured using an
ELISA plate reader (Bio-Rad Laboratories, Inc.).
Western blotting
After treatment, proteins were isolated from VSMCs
transfected with or without oe-ZFAS1 or si-ZFAS1 using RIPA lysis
buffer (Beyotime Institute of Biotechnology) and quantified using a
BCA assay kit (Thermo Fisher Scientific, Inc.). Protein samples (25
µg/lane) were loaded and separated via 10% SDS-PAGE and transferred
onto PVDF membranes (EMD Millipore). After blocking with 5% skimmed
milk for 1 h at room temperature, the membrane was incubated with
primary antibodies against Ki67 (cat. no. ab21700; 1:1,000; Abcam),
proliferating cell nuclear antigen [PCNA; cat. no. 13110; 1:1,000;
Cell Signaling Technology, Inc. (CST)], matrix metallopeptidase
(MMP)2 (cat. no. 4022; 1:1,000; CST), MMP9 (cat. no. 3852; 1:1,000;
CST) and GAPDH (cat. no. 8884; 1:2,000; CST) overnight at 4˚C.
After washing with PBS, the membrane was incubated with horseradish
peroxidase-conjugated secondary antibody (cat. no. ab97080;
1:10,000; Abcam) for 2 h at room temperature. ECL Reagent was used
to develop color of protein bands (SuperSignal West Atto; Thermo
Fisher Scientific, Inc). The gray values of the protein bands were
determined using ImageJ software (version 1.48; National Institutes
of Health). Wound healing assay. Wound healing assay was performed
to assess the migration of VSMCs. Cells were plated in 6-well
plates to generate a confluent monolayer. A scratch was created
using a 200-µl sterile pipette tip, followed by washing with PBS
three times. Subsequently, VSMCs were cultured with fresh
serum-free DMEM for 24 h at 37˚C. Finally, the width of the scratch
wound was observed, and images were captured at x100 magnification
using a fluorescence microscope (Olympus IX53; Olympus
Corporation). The recovered wound area (%) at the indicated time
point (24 h) was calculated according to the following formula:
[(wound width at 0 h) - (wound width at 24 h)] / wound width at 0
h.
Transwell chamber assay
The Transwell chamber assay was used to determine
the invasive ability of VSMCs. Briefly, after transfection with or
without oe-ZFAS1 or si-ZFAS1, the VSMCs were resuspended in 200 µl
serum-free DMEM, and 4x104 cells were loaded into the
upper chambers of Transwell plates precoated with Matrigel Mix for
5 h at 37˚C (BD Biosciences), and the lower chambers were filled
with 500 µl DMEM with 10% FBS as a chemoattractant. After
incubation for 24 h at 37˚C, the membrane was fixed with 4%
paraformaldehyde for 25 min at room temperature, and the cells on
the lower surface of the membrane were stained with 0.1% crystal
violet solution for 30 min at room temperature, and finally
examined under a fluorescence microscope at x100 magnification
(Olympus IX53; Olympus Corporation). The invasion rate was
calculated according to the following formula: Number of cells in
tested group / number of cells in control group.
Statistical analysis
Statistical analyses were performed using SPSS 19.0
(IBM Corp.), and data are presented as the mean ± SEM from at least
three repeated experiments. Differences between multiple groups
were analyzed by one-way ANOVA followed by Tukey's post hoc test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
ZFAS1 expression is upregulated by
ox-LDL treatment in VSMCs
To verify the role of ZFAS1 in atherosclerosis,
VSMCs were incubated with ox-LDL to simulate the high blood lipid
environment, and ZFAS1 mRNA expression was detected by RT-qPCR. The
results revealed that ZFAS1 mRNA expression was significantly
higher with increasing doses of ox-LDL compared with the control
group, and the increase was dose-dependent (Fig. 1A). Hence, 100 mg/l ox-LDL was
selected for further experimentation. Moreover, VSMCs were
stimulated with ox-LDL (100 mg/l) for 12, 24 and 48 h. The results
of RT-qPCR revealed that ZFAS1 mRNA expression was significantly
increased by ox-LDL stimulation at 24 and 48 h in a time-dependent
manner (Fig. 1B). Thus, VSMCs
treated with 100 mg/l ox-LDL for 48 h were selected for subsequent
experiments.
ZFAS1-knockdown inhibits the
ox-LDL-induced excessive proliferation of VSMCs
To explore the effect of ZFAS1 expression on
cellular behaviors, ZFAS1 expression was knocked down, and the
proliferation of VSMCs was analyzed. First, si-ZFAS1 was used to
achieve ZFAS1-knockdown. The RT-qPCR results demonstrated that
ZFAS1 mRNA expression in the si-ZFAS1-1 and si-ZFAS1-2 groups was
significantly lower compared with that in the control group,
particularly in the si-ZFAS1-2 group (Fig. 2A). Hence, si-ZFAS1-2 was selected
for further experiments. The CCK-8 assay results indicated that
ox-LDL treatment significantly increased the viability of VSMCs at
24 and 48 h compared with the control group, while ZFAS1-knockdown
significantly decreased VSMC viability when compared with the
ox-LDL group (Fig. 2B). Finally,
the expression levels of proteins associated with cell
proliferation was quantified by western blotting. PCNA and Ki67
were identified as proliferation markers. As shown in Fig. 2C, ox-LDL stimulation caused
significant upregulation of Ki67 and PCNA expression. Notably,
ZFAS1-knockdown in ox-LDL-treated cells led to a significant
decrease in Ki67 and PCNA expression (Fig. 2C). Hence, these results indicated
that ZFAS1-knockdown inhibited the ox-LDL-induced excessive
proliferation of VSMCs.
ZFAS1 overexpression promotes the
proliferation of VSMCs stimulated by ox-LDL
To explore the effect of ZFAS1 expression on cell
proliferation, VSMC proliferation was determined following ZFAS1
overexpression. The RT-qPCR results revealed that ZFAS1 mRNA
expression in VSMCs was significantly increased by oe-ZFAS1
transfection (Fig. 3A). The CCK-8
assay results revealed that ZFAS1 overexpression significantly
increased the viability of VSMCs induced by ox-LDL (Fig. 3B). Finally, the western blotting
results demonstrated that ZFAS1 overexpression significantly
upregulated Ki67 and PCNA expression in ox-LDL-induced VSMCs
(Fig. 3C). Thus, ZFAS1
overexpression promoted the proliferation of VSMCs stimulated by
ox-LDL.
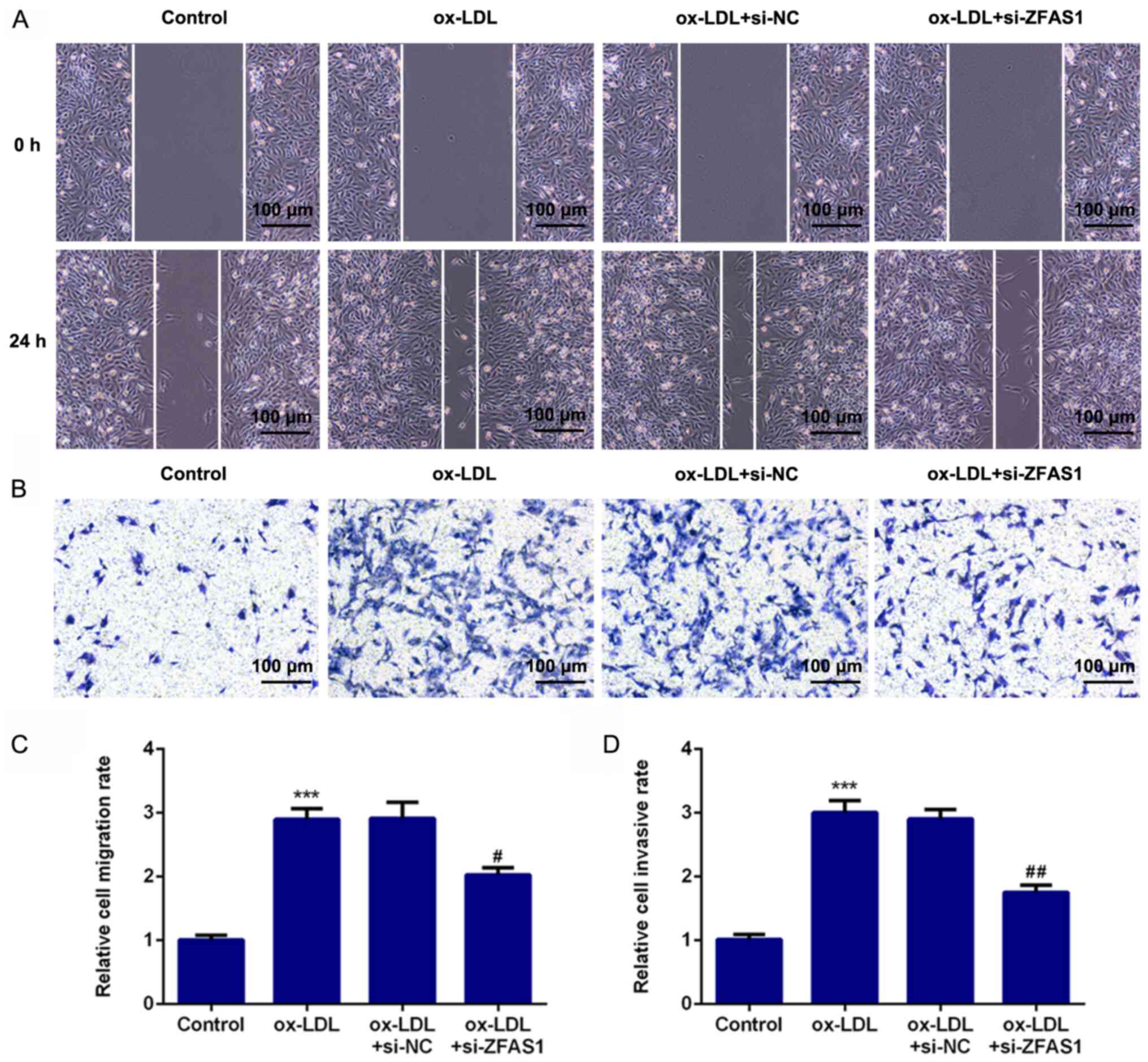
ZFAS1-knockdown suppresses the
excessive migration and invasion of VSMCs induced by ox-LDL
To investigate the effect of ZFAS1 expression on
cellular behavior, ZFAS1 expression was inhibited, and the
migration and invasion of VSMCs were analyzed. The results of the
wound healing assay revealed that ox-LDL treatment significantly
promoted the migration of VSMCs, while ZFAS1-knockdown
significantly suppressed the migration of ox-LDL-induced VSMCs
(Fig. 4A and C). Furthermore, the results of the
Transwell chamber assay demonstrated that ox-LDL stimulation
significantly promoted the invasion of VSMCs, whereas
ZFAS1-knockdown significantly inhibited the invasion of
ox-LDL-treated VSMCs (Fig. 4B and
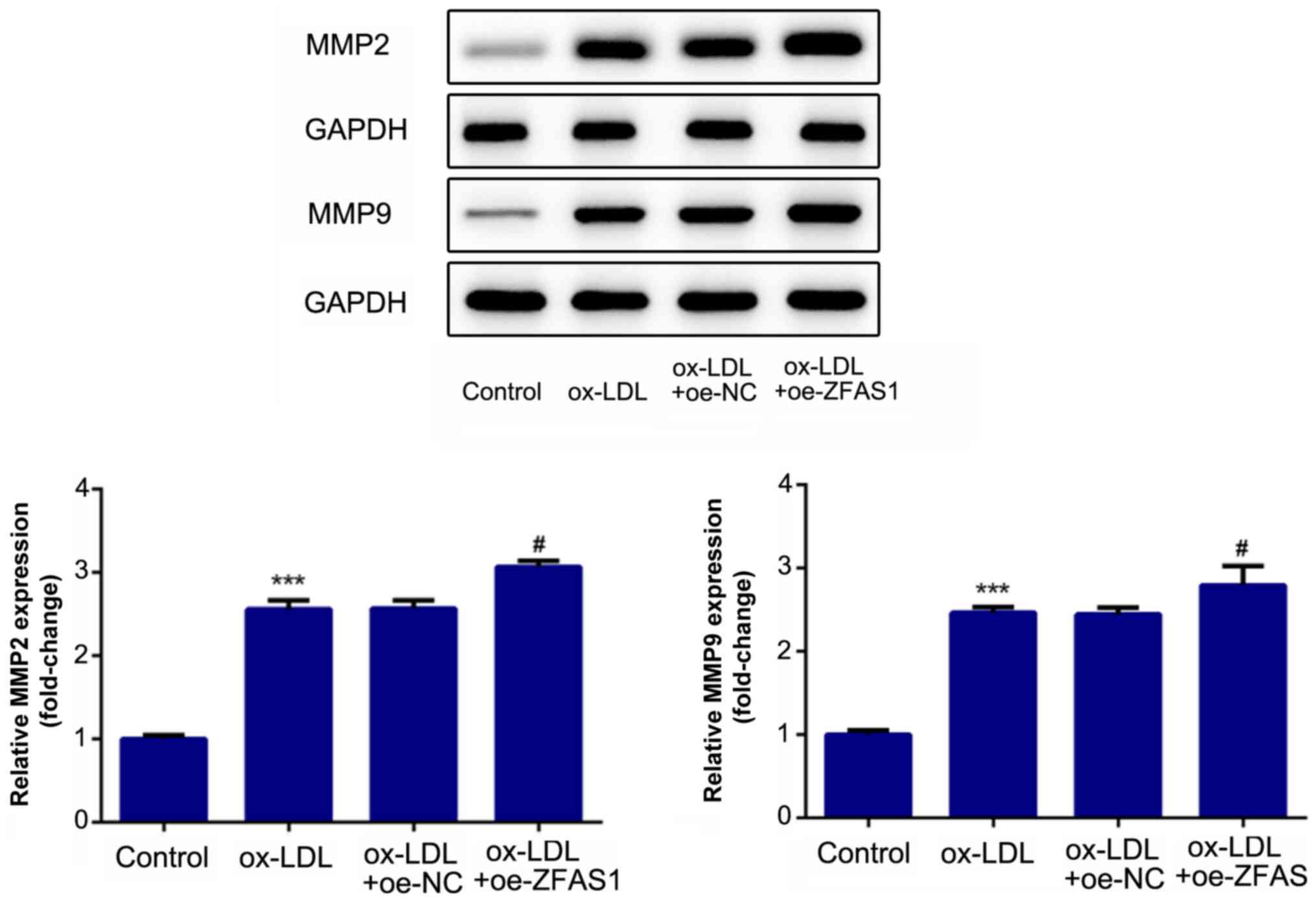
D). Additionally, the expression
levels of proteins associated with migration and invasion were
quantified. MMP2 and MMP9 are involved in cell migration and
invasion. The western blotting results revealed that the expression
levels of MMP2 and MMP9 in the ox-LDL and ox-LDL + si-NC groups
were significantly higher compared with that in the control group
(Fig. 5). Notably, ZFAS1-knockdown
significantly inhibited the expression levels of MMP2 and MMP9
following ox-LDL stimulation (Fig.
5). These results indicated that ZFAS1-knockdown suppressed the
excessive migration and invasion of VSMCs induced by ox-LDL.
ZFAS1 overexpression promotes the
migration and invasion of VSMCs induced by ox-LDL
To investigate the effect of ZFAS1 expression on
cell migration and invasion, VSMC migration and invasion were
determined following ZFAS1 overexpression. As shown in Fig. 6, ZFAS1 overexpression significantly
promoted the migration and invasion of ox-LDL-induced VSMCs.
Moreover, ZFAS1 overexpression significantly increased MMP2 and
MMP9 expression (Fig. 7). These
results suggested that ZFAS1 overexpression promoted the migration
and invasion of VSMCs induced by ox-LDL.
Discussion
Atherosclerosis is characterized by endothelium
dysfunction, accumulation of ox-LDL and intimal hyperplasia
(20). A previous study has
reported that the excessive proliferation and migration of VSMCs in
response to vascular injury, inflammation and lipoprotein
accumulation mainly leads to the initiation of intimal hyperplasia
(21). Hence, VSMCs were treated
with ox-LDL to simulate the high blood lipid environment in the
present study, which focused on the functional role of ZFAS1 in the
behavior of ox-LDL-induced VSMCs. The current findings demonstrated
that ox-LDL treatment significantly increased ZFAS1 mRNA expression
in VSMCs in a dose- and time-dependent manner, suggesting that
ZFAS1 expression is closely associated with the pathogenesis of
atherosclerosis. The aim of the present study was to verify the
effect of ZFAS1 on the behavior of ox-LDL-induced VSMCs.
Multiple lncRNAs have recently emerged as regulators
of different processes in cardiovascular diseases (22-24).
ZFAS1 has been reported as a key lncRNA in atherosclerosis
(12). Moreover, ZFAS1 attenuates
the rate of cholesterol efflux and facilitates inflammatory
responses in atherosclerosis (25).
Additionally, ZFAS1 has been found to be dysregulated in various
types of cancer and to serve an oncogenic role in the onset and
development of malignant tumors, such as ovarian cancer and glioma,
by promoting cancer metastasis, growth and
epithelial-to-mesenchymal transition (26-28).
However, the functional effect of ZFAS1 on cell migration and the
proliferation of VSMCs has yet to be fully elucidated. PCNA and
Ki67 are identified as cell proliferation markers (29,30).
Moreover, it has been reported that MMP2 and MMP9 are associated
with cancer metastasis, angiogenesis and invasion (31,32).
In the present study, it was observed that ZFAS1 overexpression
promoted the ox-LDL-induced proliferation, migration and invasion
of VSMCs, and upregulated the expression levels of proteins
associated with cell proliferation, migration and invasion (Ki67,
PCNA, MMP2 and MMP9). Notably, ZFAS1-knockdown partly reversed the
effect of ox-LDL treatment on the proliferation, migration and
invasion of VSMCs, and the expression levels of proteins associated
with cell proliferation, migration and invasion. Previous studies
have demonstrated that the phenotype of VSMCs can be switched from
contractile to proliferative and migratory during the process of
atherosclerosis (33,34). Furthermore, inhibiting cell
proliferation, migration and invasion may serve as an effective
therapeutic strategy for preventing cardiovascular disease
(35). Thus, the present findings
indicated that ZFAS1 may serve an important regulatory role in the
phenotypic transition of ox-LDL-induced VSMCs. Overall, the
aforementioned findings indicated that ZFAS1 promoted the
ox-LDL-induced proliferation, invasion and migration of VSMCs, and
may serve as a potential biomarker for the dysfunction of VSMCs in
the pathological condition of atherosclerosis. However, there are
some limitations in the present study, since the conclusion was
only from the results of in vitro experiments. It is
necessary to further investigate the role of ZFAS1 in vivo
to confirm the findings of the present study. Hence, further
studies should be performed to confirm whether ZFAS1 may be used as
a biomarker and therapeutic target of atherosclerosis.
In conclusion, the findings of the present study
suggested that ZFAS1 expression was upregulated by ox-LDL
stimulation in VSMCs. Moreover, ZFAS1 promoted the ox-LDL-induced
proliferation, invasion and migration of VSMCs, as well as the
expression levels of Ki67, PCNA, MMP2 and MMP9, and may represent a
novel biomarker for dysfunction of VSMCs in the pathological
condition of atherosclerosis.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
HW, HH, JM, YJ and RC conceived and designed the
study, collected, analyzed and interpreted the data, and revised
the manuscript. HW wrote the manuscript. HW and RC confirmed the
authenticity of the raw data. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Liu M, Song Y and Han Z: Study on the
effect of lncRNA AK094457 on OX-LDL induced vascular smooth muscle
cells. Am J Transl Res. 11:5623–5633. 2019.PubMed/NCBI
|
|
2
|
Bennett MR, Sinha S and Owens GK: Vascular
smooth muscle cells in atherosclerosis. Circ Res. 118:692–702.
2016.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Wang J, Uryga AK, Reinhold J, Figg N,
Baker L, Finigan A, Gray K, Kumar S, Clarke M and Bennett M:
Vascular smooth muscle cell senescence promotes atherosclerosis and
features of plaque vulnerability. Circulation. 132:1909–1919.
2015.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Misra A, Feng Z, Chandran RR, Kabir I,
Rotllan N, Aryal B, Sheikh AQ, Ding L, Qin L, Fernández-Hernando C,
et al: Integrin beta3 regulates clonality and fate of smooth
muscle-derived atherosclerotic plaque cells. Nat Commun.
9(2073)2018.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Hu D, Yin C, Luo S, Habenicht AJR and
Mohanta SK: Vascular smooth muscle cells contribute to
atherosclerosis immunity. Front Immunol. 10(1101)2019.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Xu L, Hao H, Hao Y, Wei G, Li G, Ma P,
Ding N, Ma S, Chen AF and Jiang Y: Aberrant MFN2 transcription
facilitates homocysteine-induced VSMCs proliferation via the
increased binding of c-Myc to DNMT1 in atherosclerosis. J Cell Mol
Med. 23:4611–4626. 2019.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Harman JL and Jørgensen HF: The role of
smooth muscle cells in plaque stability: Therapeutic targeting
potential. Br J Pharmacol. 176:3741–3753. 2019.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Kattoor AJ, Kanuri SH and Mehta JL: Role
of Ox-LDL and LOX-1 in atherogenesis. Curr Med Chem. 26:1693–1700.
2019.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Huang M, Zhong Z, Lv M, Shu J, Tian Q and
Chen J: Comprehensive analysis of differentially expressed profiles
of lncRNAs and circRNAs with associated co-expression and ceRNA
networks in bladder carcinoma. Oncotarget. 7:47186–47200.
2016.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Huang X, Zhi X, Gao Y, Ta N, Jiang h and
Zheng J: lncRNAs in pancreatic cancer. Oncotarget. 7:57379–57390.
2016.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Mathy NW and Chen XM: Long non-coding RNAs
(lncRNAs) and their transcriptional control of inflammatory
responses. J Biol Chem. 292:12375–12382. 2017.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Wang CH, Shi HH, Chen LH, Li XL, Cao GL
and Hu XF: Identification of key lncRNAs associated with
atherosclerosis progression based on public datasets. Front Genet.
10(123)2019.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Chen L, Yao H, Hui JY, Ding SH, Fan YL,
Pan YH, Chen KH, Wan JQ and Jiang JY: Global transcriptomic study
of atherosclerosis development in rats. Gene. 592:43–48.
2016.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Zhang Y, Jiao L, Sun L, Li Y, Gao Y, Xu C,
Shao Y, Li M, Li C, Lu Y, et al: lncRNA ZFAS1 as a SERCA2a
inhibitor to cause intracellular Ca2+ overload and
contractile dysfunction in a mouse model of myocardial infarction.
Circ Res. 122:1354–1368. 2018.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Wang X, Jin Q, Chen W and Cai Z: lncRNA
ZFAS1 promotes proliferation and migration and inhibits apoptosis
in nasopharyngeal carcinoma via the PI3K/AKT pathway in vitro.
Cancer Biomark. 26:171–182. 2019.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Meng Q, Zhang R, Ding W and Mao B: Long
noncoding RNA ZFAS1 promotes cell proliferation and tumor growth by
upregulating LIN28 in cervical carcinoma. Minerva Med. 111:511–514.
2019.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Xie S, Ge Q, Wang X, Sun X and Kang Y:
Long non-coding RNA ZFAS1 sponges miR-484 to promote cell
proliferation and invasion in colorectal cancer. Cell Cycle.
17:154–161. 2018.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Ye D, Jian W, Feng J and Liao X: Role of
long noncoding RNA ZFAS1 in proliferation, apoptosis and migration
of chondrocytes in osteoarthritis. Biomed Pharmacother.
104:825–831. 2018.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Spartalis M, Spartalis E, Athanasiou A,
Paschou SA, Kontogiannis C, Georgiopoulos G, Iliopoulos DC and
Voudris V: The role of the endothelium in premature
atherosclerosis: Molecular mechanisms. Curr Med Chem. 27:1041–1051.
2020.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Zhao XS, Zheng B, Wen Y, Sun Y, Wen JK and
Zhang XH: Salvianolic acid B inhibits Ang II-induced VSMC
proliferation in vitro and intimal hyperplasia in vivo by
downregulating miR-146a expression. Phytomedicine.
58(152754)2019.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Guo FX, Wu Q, Li P, Zheng L, Ye S, Dai XY,
Kang CM, Lu JB, Xu BM, Xu YJ, et al: The role of the
lncRNA-FA2H-2-MLKL pathway in atherosclerosis by regulation of
autophagy flux and inflammation through mTOR-dependent signaling.
Cell Death Differ. 26:1670–1687. 2019.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Li FP, Lin DQ and Gao LY: lncRNA TUG1
promotes proliferation of vascular smooth muscle cell and
atherosclerosis through regulating miRNA-21/PTEN axis. Eur Rev Med
Pharmacol Sci. 22:7439–7447. 2018.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Ye ZM, Yang S, Xia YP, Hu RT, Chen S, Li
BW, Chen SL, Luo XY, Mao L, Li Y, et al: lncRNA MIAT sponges
miR-149-5p to inhibit efferocytosis in advanced atherosclerosis
through CD47 upregulation. Cell Death Dis. 10(138)2019.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Tang X, Yin R, Shi H, Wang X, Shen D and
Pan C: lncRNA ZFAS1 confers inflammatory responses and reduces
cholesterol efflux in atherosclerosis through regulating
miR-654-3p-ADAM10/RAB22A axis. Int J Cardiol. 315:72–80.
2020.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Han S, Li DZ and Xiao MF: lncRNA ZFAS1
serves as a prognostic biomarker to predict the survival of
patients with ovarian cancer. Exp Ther Med. 18:4673–4681.
2019.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Li X, Luo Y, Liu L, Cui S, Chen W, Zeng A,
Shi Y and Luo L: The long noncoding RNA ZFAS1 promotes the
progression of glioma by regulating the miR-150-5p/PLP2 axis. J
Cell Physiol. 235:2937–2946. 2020.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Zeng Z, Zhao G, Rao C, Hua G, Yang M, Miao
X, Ying J and Nie L: Knockdown of lncRNA ZFAS1-suppressed non-small
cell lung cancer progression via targeting the miR-150-5p/HMGA2
signaling. J Cell Biochem: Nov 6, 2019 (Epub ahead of print). doi:
10.1002/jcb.29542.
|
|
29
|
Ben-Izhak O, Bar-Chana M, Sussman L,
Dobiner V, Sandbank J, Cagnano M, Cohen h and Sabo E: Ki67 antigen
and PCNA proliferation markers predict survival in anorectal
malignant melanoma. Histopathology. 41:519–525. 2002.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Li N, Deng W, Ma J, Wei B, Guo K, Shen W,
Zhang Y and Luo S: Prognostic evaluation of Nanog, Oct4, Sox2,
PCNA, Ki67 and E-cadherin expression in gastric cancer. Med Oncol.
32(433)2015.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Farina P, Tabouret E, Lehmann P, Barrie M,
Petrirena G, Campello C, Boucard C, Graillon T, Girard N and Chinot
O: Relationship between magnetic resonance imaging characteristics
and plasmatic levels of MMP2 and MMP9 in patients with recurrent
high-grade gliomas treated by Bevacizumab and Irinotecan. J
Neurooncol. 132:433–437. 2017.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Struckmann K, Mertz K, Steu S, Storz M,
Staller P, Krek W, Schraml P and Moch H: pVHL co-ordinately
regulates CXCR4/CXCL12 and MMP2/MMP9 expression in human clear-cell
renal cell carcinoma. J Pathol. 214:464–471. 2008.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Gomez D and Owens GK: Smooth muscle cell
phenotypic switching in atherosclerosis. Cardiovasc Res.
95:156–164. 2012.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Wei M, Liu Y, Zheng M, Wang L, Ma F, Qi Y
and Liu G: Upregulation of protease-activated receptor 2 promotes
proliferation and migration of human vascular smooth muscle cells
(VSMCs). Med Sci Monit. 25:8854–8862. 2019.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Luo Z, Deng H, Fang Z, Zeng A, Chen Y,
Zhang W and Lu Q: Ligustilide inhibited rat vascular smooth muscle
cells migration via c-Myc/MMP2 and ROCK/JNK signaling pathway. J
Food Sci. 84:3573–3583. 2019.PubMed/NCBI View Article : Google Scholar
|