Introduction
According to the 2019 US Cancer Statistics report
(1), although the incidence of lung
cancer is lower compared with that of prostate and breast cancer,
lung cancer is associated with the highest rate of cancer-related
morbidity in the USA. In China, the morbidity and mortality rates
of lung cancer are the highest among all types of cancer (2). Non-small cell lung cancer (NSCLC) is a
subtype of lung cancer that accounts for ~85% of all lung cancer
cases worldwide, which is also the main cause of lung
cancer-related mortality (3). At
present, available clinical treatment options for NSCLC primarily
includes surgery and radiotherapy, combined with drug chemotherapy
(4-6).
However, NSCLC is prone to drug resistance, metastasis and
recurrence, leading to poor survival rates (7). Therefore, investigating the molecular
mechanism underlying the proliferation, migration and invasion of
NSCLC cells is crucial for prolonging the survival of patients with
NSCLC.
Etomidate (ETO) is a commonly used intravenous
anesthetic that maintains good hemodynamic stability during
anesthesia (8). It has been
reported that ETO exerts an inhibitory role in several types of
cancer. For example, it has been demonstrated that ETO could
attenuate the proliferation of human adrenocortical cancer cells
(9) and enhance the apoptosis of
N2a neuroblastoma cells (10). In
addition, ETO was found to significantly inhibit the migratory and
invasive abilities of NSCLC cells (11). However, the effect of ETO on the
apoptosis of NSCLC cells has not been previously reported.
Therefore, the present study aimed to explore the effect of ETO on
the proliferation and apoptosis of NSCLC cells.
Subsequently, the STITCH database was used to
predict the proteins interacting with ETO and to explore the
possible relationship between ETO and WW domain containing E3
ubiquitin protein ligase 2 (WWP2) in the WW domain. WWP2 is a
member of the C2-WW-HECT family (NEDD4 family) of E3 ubiquitin
ligases (E3), which act as acceptors of ubiquitin from E2 enzymes
and then transfer ubiquitin to a specific lysine residue on the
substrate (12). WWP2 has a role in
protecting cartilage from osteoarthritis through runt-related
transcription factor 2 (Runx2) polyubiquitination and degradation
to inhibit Runx2-induced disintegrin and metalloproteinase with
thrombospondin motifs 5(13). WWP2
is a novel cancer-related factor that has been reported to be
associated with the occurrence of liver cancer and lung
adenocarcinoma (14). A previous
study demonstrated that hypoxia-inducible factor-1α may promote
apoptosis and inhibit the invasion of thyroid cancer cells by
downregulating the expression of factors, such as WWP2(15). Another study showed that the
expression of WWP2 was notably upregulated in NSCLC tissues, where
WWP2 overexpression could effectively promote the proliferation of
NSCLC cells (16). Therefore, it
was hypothesized that ETO may affect the progression of NSCLC by
interacting with WWP2.
The present study aimed to uncover the role of ETO
in the proliferation, migration and apoptosis of NSCLC cells and
WWP2 expression, which could hopefully provide a theoretical basis
for a novel treatment strategy for NSCLC.
Materials and methods
Cell culture
A549 cells were purchased from the American Type
Culture Collection and maintained in RPMI-1640 medium (Thermo
Fisher Scientific, Inc.) supplemented with 10% FBS (Thermo Fisher
Scientific, Inc.) in a 5% CO2 incubator at 37˚C. BESA-2B
cells were also purchased from the American Type Culture Collection
and maintained in LHC medium (Thermo Fisher Scientific, Inc.)
supplemented with 10% FBS (Thermo Fisher Scientific, Inc.) in a 5%
CO2 incubator at 37˚C. The cells were passaged once
every 3 days, whilst only cells in the logarithmic growth phase
were used for the subsequent experiments.
Bioinformatics
The STITCH DataBase (version 5.0; http://stitch.embl.de/) is a database that can be used
to explore known and predicted interactions between chemicals and
proteins (17). Proteins that
directly interact with ETO will be selected as putative targets
(minimum required interaction score: 0.400).
Cell transfection
The WWP2 overexpression vector, pcDNA3.1-WWP2 and
empty control vector, pcDNA3.1-NC, were synthesized by Shanghai
GeneChem Co., Ltd.. Cells were seeded onto 12-well plates at a
density of 4x105 cells/well and cultured for 24 h at
37˚C. Following incubation, cells were transfected with the
aforementioned plasmids (1.5 µg per well) using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocols.
Following transfection for 48 h, the transfection efficiency was
evaluated by reverse transcription-quantitative PCR (RT-qPCR).
After transfection, 1.0, 2.0 and 3.0 µg/ml ETO (cat. no. A28229;
Beijing Wokai Biological Technology Co., Ltd.; https://www.bjoka-vip.com) were added and co-incubated
for 24, 48 and 72 h at 37˚C for subsequent experiments.
Cell counting kit-8 (CCK-8) assay
The cell viability was assessed by CCK-8 assay
(Sigma-Aldrich; Merck KGaA). Briefly, cells were seeded onto
96-well plates at a density of 2x103 cells/well and
incubated for 24, 48 and 72 h at 37˚C. Following incubation, 10 µl
CCK-8 solution was added into each well and cells were cultured for
an additional 2 h at 37˚C. The absorbance in each well was measured
at a wavelength of 450 nm using a microplate reader (Synergy 2
Multi-Mode Microplate Reader; BioTek Instruments, Inc.).
Colony formation assay
The cells with 4x102 cells/well suspended
in RPMI-1640 medium were seeded into six-well plates and cultured
in a 5% CO2 incubator at 37˚ for 14 days. Subsequently,
the cells were fixed with 70% ethanol at room temperature for 15
min and stained with 0.05% crystal violet for 20 min at 37˚C. The
number of colonies formed (>50 cells/colony) were counted under
a Olympus BX40 light microscope (magnification, x200; Olympus
Corporation).
TUNEL assay
Apoptosis was assessed using the TUNEL Apoptosis
Assay Kit (cat. no. C1088; Beyotime Institute of Biotechnology).
Briefly, the cells (1x106 cells/well) were washed with
PBS, fixed at room temperature with 4% paraformaldehyde for 20 min
and then treated with 0.1% Triton X-100 for 10 min. Subsequently,
50 µl TUNEL detection solution was added to each well, incubated at
37˚C for 60 min in dark and washed with PBS three times. A small
amount of DAPI staining solution (final concentration: 5 mg/ml) was
added (covering the sample) and placed at room temperature for 3-5
min and then washed with PBS three times. Anti-fluorescence
quenching mounting solution was used to mount the slides (Beyotime
Institute of Biotechnology). The morphological changes of apoptotic
cells were observed under the AMG EVOS fluorescence microscope
(magnification, x200; Thermo Fisher Scientific, Inc.). Three fields
of each sample were randomly selected for apoptosis analysis. Cells
with green fluorescence were considered to be apoptotic and
quantified using the following formula: Cell apoptosis (%)=Green
fluorescence area/total area x100%.
RT-qPCR analysis
Total RNA was extracted from A549 cells using a
TRIzol® reagent (Thermo Fisher Scientific, Inc.) and was
then reverse-transcribed to cDNA using the FastQuant RT kit (cat.
no. KR106; Tiangen Biotech Co., Ltd.) according to the
manufacturer's protocol. qPCR reactions were performed using the
PowerUp™ SYBR™ Green Master Mix (cat. no. A25779; Applied
Biosystems; Thermo Fisher Scientific, Inc.) on the ABI 7500 PCR
system (Applied Biosystems; Thermo Fisher Scientific, Inc.). The
thermocycling conditions used were as follows: Initial denaturation
at 94˚C for 30 sec, followed by 22 cycles at 55˚C for 30 sec and
72˚C for 30 sec. The relative expression levels of target genes
were normalized to those of the housekeeping gene GAPDH and
calculated by the 2-ΔΔCq method (18). The sequences of PCR primers were as
follows: Proliferating cell nuclear antigen (PCNA) forward,
5'-GGGTGAAGTTTTCCGCCAGT-3' and reverse,
5'-CTGTAGGAGAAAGCGGAGTGG-3'; Ki-67 forward,
5-ATCCTTACCTCCCAACCTCTGT-3 and reverse,
5'-AACTTCTGGCTCTTCCTGTAGC-3'; WWP2 forward,
5'-CGGTGTAGGCAGAGCTGATG-3' and reverse, 5'-CCACAAGGCAGAAACACCAA-3';
PTEN forward, 5'-CTCCTACTTCCACCTGCTCAC-3' and reverse,
5'-AAGGATCTCCAGGCTCGAAA-3' and GAPDH forward,
5'-GATGATGTTGAACTCGTCGC-3' and reverse,
5'-CTCTTCTGGGTTTCTCACACC-3'.
Western blot analysis
Total proteins were extracted from A549 cells using
the RIPA buffer (cat. no. P0013B; Beyotime Institute of
Biotechnology). The protein concentration was measured utilizing a
BCA protein quantitative kit (cat. no. P0012; Beyotime Institute of
Biotechnology). Subsequently, 20 µg protein extracts were separated
by 10% SDS-PAGE and transferred onto PVDF membranes (Beyotime
Institute of Biotechnology). Following blocking with 5% skimmed
milk for 30 min at room temperature, the membranes were incubated
with primary antibodies (dilution, 1:1,000; all from Abcam) against
PCNA (cat. no. ab92552), Ki67 (cat. no. ab15580), Bcl-2 (cat. no.
ab182858), Bax (cat. no. ab182733), caspase 3 (cat. no. ab32150),
cleaved caspase 3 (cat. no. ab2302), WWP2 (cat. no. ab103527), PTEN
(cat. no. ab267787), PI3K (cat. no. ab32089), AKT (cat. no.
ab18785), phosphorylated (p)-AKT (cat. no. ab38449) and GAPDH (cat.
no. ab9485) overnight at 4˚C. The next day, the membranes were
incubated with the corresponding HRP-conjugated secondary
antibodies (cat. no. ab97190; dilution, 1:1,000; Abcam) at 37˚C for
2 h. The ECL Plus kit (cat. no. P0018; Beyotime Institute of
Biotechnology) was utilized to visualize the protein bands (Image
J; version number: 1.4.3.67; National Institutes of Health).
Statistical analysis
All data were analyzed with the GraphPad Prism 7
software (GraphPad Software, Inc.). All data are expressed as the
mean ± SD (n=3). Differences between two groups were compared using
an unpaired Student's t-test, whilst those among multiple groups by
one-way ANOVA followed by Tukey's post hoc test. P<0.05 was
considered to indicate a statistically significant difference.
Results
ETO attenuates A549 cell proliferation
and induces apoptosis
First, CCK-8, colony formation and TUNEL assays were
performed to evaluate the viability, proliferation and apoptosis of
A549 cells, respectively, following treatment with different
concentrations of ETO (0, 1, 2 or 3 µg/ml). The effect of different
concentrations of ETO on the viability of normal lung epithelial
BESA-2B cells was tested first, which yielded no difference
(Fig. 1A). As shown in Fig. 1B, CCK-8 assay results showed that
ETO significantly reduced the viability of A549 cells in a
dose-dependent manner. In addition, the inhibitory effects of ETO
on the expression of the proliferation-related genes, Ki67 and PCNA
(19) was stronger with increasing
concentrations of ETO (Fig. 1C and
D). The results of colony formation
assays showed that ETO also significantly decreased the number of
colonies formed in a dose-dependent manner (Fig. 1E).
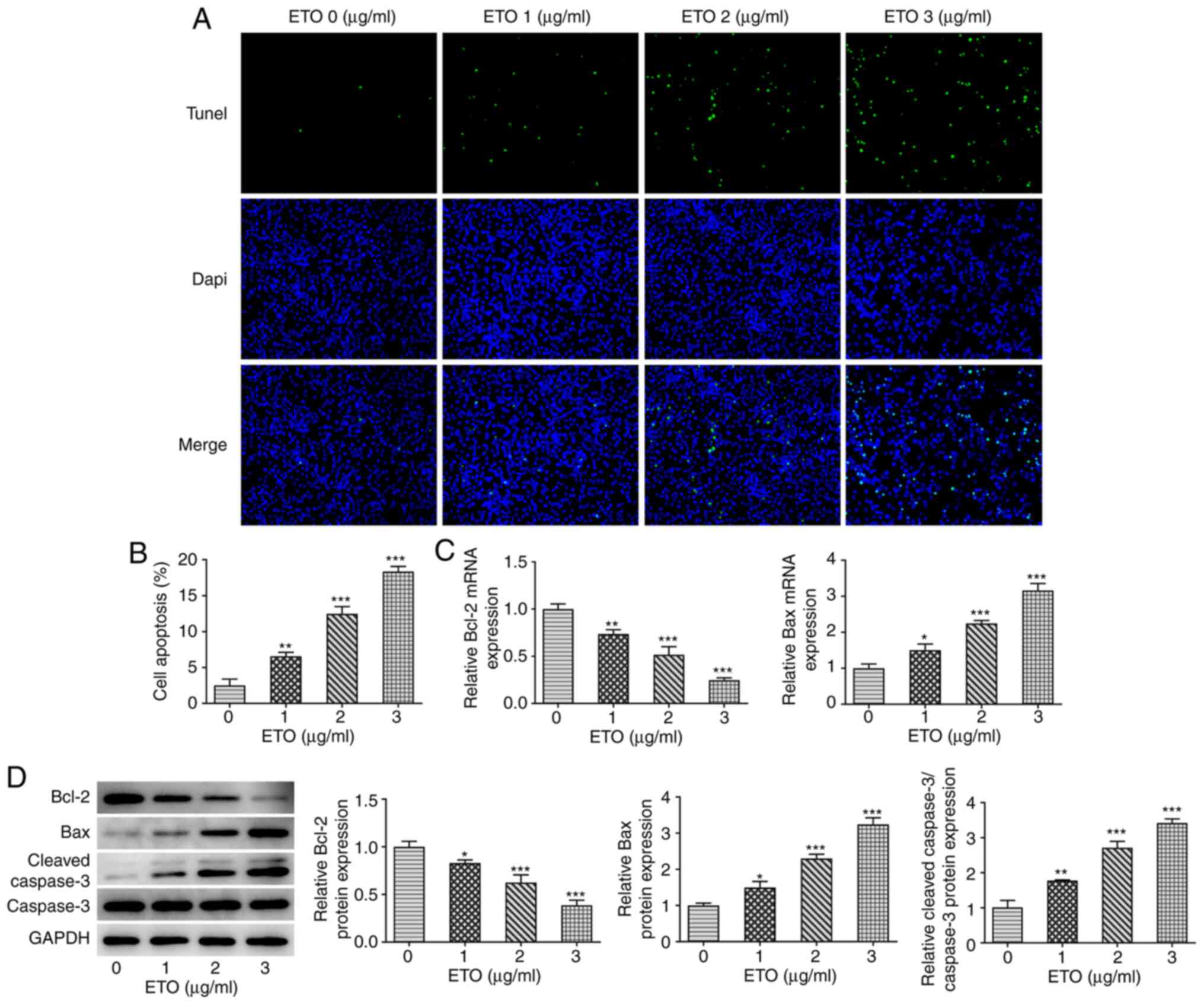
Subsequently, results from TUNEL assay revealed
that, compared with that in the control group, ETO significantly
promoted the apoptosis of A549 cells in a dose-dependent manner
(Fig. 2A and B). Similarity, the RT-qPCR and western
blot analyzes showed that ETO significantly reduced the expression
of the anti-apoptotic protein Bcl-2, and increased that of Bax and
cleaved caspase 3 in a dose-dependent manner (Fig. 2C and D).
ETO negatively regulates the
expression of WWP2 in A549 cells
Subsequently, the present study further investigated
the mechanism underlying the effects of ETO on NSCLC.
Bioinformatics analysis using the STITCH database predicted that
ETO could interact with WWP2 and PTEN by upregulating the protein
expression of WWP2 and downregulating the protein expression of
PTEN. (Fig. 3A). Data from RT-qPCR
and western blot analyzes demonstrated that, compared with that in
the control group, treatment of A549 cells with 3 µg/ml ETO
significantly downregulated WWP2 expression (Fig. 3B and C).
ETO attenuates A549 cell proliferation
and induces apoptosis via downregulating WWP2
A549 cells were then transected with the
pcDNA3.1-WWP2 plasmid to overexpress WWP2 (ov-WWP2). As shown in
Fig. 3D and E, the expression of WWP2 in the ov-WWP2
group was significantly increased compared with that in the cell
group transfected with the empty plasmid (ov-NC). Additionally,
WWP2 overexpression significantly reversed the inhibitory effects
of ETO on A549 cell viability, colony formation, Ki67 and PCNA
expression (Fig. 3F-I). Results
from TUNEL assay revealed that WWP2 overexpression significantly
reversed the potentiating effects of ETO on A549 cell apoptosis
(Fig. 4A and B). In addition, the mRNA levels of Bcl-2
and Bax, in addition to the protein expression levels of Bcl-2,
Bax, cleaved caspase 3 and caspase 3 were detected by RT-qPCR and
western blotting. The results showed that compared with that in the
ETO + OV-NC group, the expression levels of Bcl-2 protein and
relative Bcl-2 mRNA in ETO + OV-WWP2 group were significantly
increased, whilst the expression levels of Bax protein and relative
Bax mRNA were downregulated, indicating that WWP2 overexpression
significantly reversed the potentiating effects of ETO on A549 cell
apoptosis. (Fig. 4C and D).
ETO attenuates the physiology of A549
cells by PTEN downregulation through targeting WWP2
Subsequently, the mRNA and protein expression levels
of PTEN were evaluated by RT-qPCR and western blot analyzes,
respectively. As shown in Fig. 5A
and B, ETO significantly increased
PTEN expression compared with that in the control group, which was
significantly reversed by WWP2 overexpression. In addition, the
significantly decreased AKT phosphorylation and PI3K expression
induced by ETO were also in turn significantly reversed by WWP2
overexpression (Fig. 5C).
Discussion
Lung cancer is one of the most common malignancies
worldwide, of which NSCLC is the most prevalent type of lung
cancer, accounting for ~80% of all lung cancer cases (20). Due to the lack of effective
long-term treatment strategies and difficulties in early-stage
diagnosis, the postoperative survival rate of patients with NSCLC
remains low. Previous studies have shown that among patients with
advanced NSCLC who have previously received operative, chemotherapy
or radiotherapy treatment, the 5-year overall survival rate of all
treated patients (n=129) was estimated to be 16% (21,22).
Therefore, identifying novel treatment approaches is crucial for
improving the prognosis of patients with NSCLC. In the present
study, the results demonstrated that ETO could attenuate
proliferation whilst inducing apoptosis in A549 cells in a
dose-dependent manner. In addition, the interaction between WWP2
and ETO was predicted using the STITCH database, where WWP2
overexpression was subsequently found to reverse the inhibitory
effects of ETO on A549 cell activity.
ETO is a commonly used hypnotic and intravenous
anesthetic (23). Previous studies
have shown that ETO exerts antioxidant, anti-inflammatory,
antitumor and antiplatelet aggregation effects (24,25).
For example, ETO could reduce the proliferation, migration and
invasion of human adrenocortical cancer cells (9) and induce the apoptosis of N2a brain
tumor cells (10). In lung cancer,
one previous study demonstrated that ETO can effectively attenuate
the proliferation and migration of A549 cells, supporting the
notion of antitumor effects of ETO on NSCLC (11). However, the specific role and
mechanism of action of ETO in NSCLC remain elusive. ETO treatment
conferred no effects on the immune system of patients with lung
cancer (26). Therefore, the effect
of ETO on NSCLC is worthy of further investigation. In the present
study, ETO significantly attenuated the cell viability and
proliferation of A549 cells, whilst promoting apoptosis in a
dose-dependent manner. Therefore, the results of the present study
further supported the potential antitumor and therapeutic value of
ETO in NSCLC.
Furthermore, the present study further investigated
the mechanism underlying the effect of ETO on NSCLC. Bioinformatics
analysis by the STITCH database revealed that WWP2 could interact
with ETO. WWP2 belongs to the ubiquitin ligase protein family and
has been reported to serve an important role in liver cancer and
lung adenocarcinoma (27). Previous
studies in prostate cancer models have shown that WWP2 served as an
oncogene, which mainly operated through the PTEN/Akt signaling
pathway to promote carcinogenesis (14,28),
In gastric cancer, overexpression of WWP2 enhanced cell
proliferation by silencing PTEN protein expression and upregulating
of Akt phosphorylation (29). Loss
of PTEN protein expression has been widely reported in several
types of malignant tumors, including gastric cancer, liver cancer
and lung adenocarcinoma, where they are closely associated with
histological grade, metastasis and prognosis (30-32).
PTEN lie upstream of the PI3K/AKT signaling pathway and functions
as an important regulator of non-small cell lung cancer (33). A previous study showed that PTEN
played an inhibitory role on Human cervical cancer cells (HeLa),
human prostate cancer cells (DU145) and human prostatic hyperplasia
cells (BPH1) by negatively regulating the PI3K/Akt signaling
pathway (28). Downstream, the
PI3K/AKT pathway regulates various cellular functions during
tumorigenesis and development, including cell proliferation,
migration and apoptosis, thereby serving a key role in promoting
cancer progression (29). It has
been suggested that ETO can reduce PI3K/AKT activation in A549
cells (11). Therefore, in the
present study it was hypothesized that ETO may act through this
pathway. It was found that PTEN and WWP2 could interact with each
other. WWP2 was previously found to promote the proliferation of
gastric cancer cells in a PTEN-dependent manner in gastric cancer
(29). WWP2 was also found to be
highly expressed in NSCLC, suggesting that it may function as a
tumor-promoting factor (16).
Therefore, the present study investigated the effects of WWP2 on
the proliferation of NSCLC cells and the PTEN/PI3K/AKT axis.
Treatment of A549 cells with ETO inhibited the PI3K/AKT signaling
pathway by downregulating WWP2 and upregulating PTEN, which also
attenuated A549 cell proliferation and enhancing apoptosis.
However, it should be noted that there are
limitations in the present study. Only one cell line was used for
present study. In future studies, multiple NSCLC cell lines must be
used for in vitro experiments for more comprehensive and
in-depth validation. A549 cells are also of the wild-type p53
genotype, whilst most other lung cancer cell lines contain a
mutated p53 genotype. Since p53 is one of the key mediators of
apoptosis (34), the role of ETO in
cell lines with mutant p53 should be explored. In addition, ETO was
not only found to interact with WWP2, but also with eight other
proteins, namely cytochrome P450, family 11, subfamily B,
polypeptide 2, cytochrome P450, family 11, subfamily B, polypeptide
1, γ-aminobutyric acid (GABA) A receptor α1, ADRA2B: adrenoceptor
α2B, sulfotransferase family, cytosolic, 2A,
dehydroepiandrosterone-preferring, member 1, GABA A receptor γ2,
unc-13 homolog B and GABA A receptor γ1, which should be further
explored in future studies. The molecular mechanism of ETO and
WWP2/PTEN on NSCLC cell function has not been fully investigated in
the present study. These issues require further in-depth analysis
and should be addressed in future studies.
Overall, results of the present study demonstrated
that ETO reduced the prolfieration of NSCLC cells in a
dose-dependent manner. The mechanism underlying the effects of ETO
on NSCLC may be associated with the downregulation of WWP2 and
activation of PTEN. These findings may provide a theoretical basis
for the clinical treatment of NSCLC using ETO.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XM and DL contributed to conception and design of
the study. DL, JZ and LY contributed to the experiments and data
collection. ZJ and XC contributed to analysis and interpretation of
data. XM revised the manuscript critically for important
intellectual content. XM and DL confirmed the authenticity of all
the raw data. All authors read and approved the final version of
the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Miller KD, Nogueira L, Mariotto AB,
Rowland JH, Yabroff KR, Alfano CM, Jemal A, Kramer JL and Siegel
RL: Cancer treatment and survivorship statistics, 2019. CA Cancer J
Clin. 69:363–385. 2019.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2019. CA Cancer J Clin. 69:7–34. 2019.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Hoffman PC, Mauer AM and Vokes EE: Lung
cancer. Lancet. 355:479–485. 2000.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Vinod SK: International patterns of
radiotherapy practice for non-small cell lung cancer. Semin Radiat
Oncol. 25:143–150. 2015.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Gandhi L, Rodríguez-Abreu D, Gadgeel S,
Esteban E, Felip E, De Angelis F, Domine M, Clingan P, Hochmair MJ,
Powell SF, et al: Pembrolizumab plus chemotherapy in metastatic
non-small-cell lung cancer. N Engl J Med. 378:2078–2092.
2018.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Groth SS, Rueth NM, Hodges JS, Habermann
EB, Andrade RS, D'Cunha J and Maddaus MA: Conditional
cancer-specific versus cardiovascular-specific survival after
lobectomy for stage I non-small cell lung cancer. Ann Thorac Surg.
90:375–382. 2010.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Herbst RS, Heymach JV and Lippman SM: Lung
cancer. N Engl J Med. 359:1367–1380. 2008.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Erdoes G, Basciani RM and Eberle B:
Etomidate-a review of robust evidence for its use in various
clinical scenarios. Acta Anaesthesiol Scand. 58:380–389.
2014.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Fassnacht M, Hahner S, Beuschlein F, Klink
A, Reincke M and Allolio B: New mechanisms of adrenostatic
compounds in a human adrenocortical cancer cell line. Eur J Clin
Invest. 30 (Suppl 3):S76–S82. 2000.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Chen HT, Zhou J, Fan YL, Lei CL, Li BJ,
Fan LX, Xu L, Xu M, Hu XQ and Yu ZY: Anesthetic agent etiomidate
induces apoptosis in N2a brain tumor cell line. Mol Med Rep.
18:3137–3142. 2018.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Chu CN, Wu KC, Chung WS, Zheng LC, Juan
TK, Hsiao YT, Peng SF, Yang JL, Ma YS, Wu RS and Chung JG:
Etomidate suppresses invasion and migration of human A549 lung
adenocarcinoma cells. Anticancer Res. 39:215–223. 2019.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Bernassola F, Karin M, Ciechanover A and
Melino G: The HECT family of E3 ubiquitin ligases: Multiple players
in cancer development. Cancer Cell. 14:10–21. 2008.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Mokuda S, Nakamichi R, Matsuzaki T, Ito Y,
Sato T, Miyata K, Inui M, Olmer M, Sugiyama E, Lotz M and Asahara
H: Wwp2 maintains cartilage homeostasis through regulation of
Adamts5. Nat Commun. 10(2429)2019.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Zhang R, Zhang J, Luo W, Luo Z and Shi S:
WWP2 is one promising novel oncogene. Pathol Oncol Res. 25:443–446.
2019.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Ding ZY, Huang YJ, Tang JD, Li G, Jiang PQ
and Wu HT: Silencing of hypoxia-inducible factor-1α promotes
thyroid cancer cell apoptosis and inhibits invasion by
downregulating WWP2, WWP9, VEGF and VEGFR2. Exp Ther Med.
12:3735–3741. 2016.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Yang R, He Y, Chen S, Lu X, Huang C and
Zhang G: Elevated expression of WWP2 in human lung adenocarcinoma
and its effect on migration and invasion. Biochem Biophys Res
Commun. 479:146–151. 2016.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Kuhn M, von Mering C, Campillos M, Jensen
LJ and Bork P: STITCH: Interaction networks of chemicals and
proteins. Nucleic Acids Res. 36 (Database Issue):D684–D688.
2008.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Liao XH, Lu DL, Wang N, Liu LY, Wang Y, Li
YQ, Yan TB, Sun XG, Hu P and Zhang TC: Estrogen receptor α mediates
proliferation of breast cancer MCF-7 cells via a
p21/PCNA/E2F1-dependent pathway. FEBS J. 281:927–942.
2014.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Dey N, De P and Leyland-Jones B:
PI3K-AKT-mTOR inhibitors in breast cancers: From tumor cell
signaling to clinical trials. Pharmacol Ther. 175:91–106.
2017.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Gettinger S, Horn L, Jackman D, Spigel D,
Antonia S, Hellmann M, Powderly J, Heist R, Sequist LV, Smith DC,
et al: Five-year follow-up of nivolumab in previously treated
advanced non-small-cell lung cancer: Results from the CA209-003
study. J Clin Oncol. 36:1675–1684. 2018.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Garon EB, Hellmann MD, Rizvi NA, Carcereny
E, Leighl NB, Ahn MJ, Eder JP, Balmanoukian AS, Aggarwal C, Horn L,
et al: Five-year overall survival for patients with advanced
non-small-cell lung cancer treated with pembrolizumab: Results from
the phase I KEYNOTE-001 study. J Clin Oncol. 37:2518–2527.
2019.PubMed/NCBI View Article : Google Scholar
|
|
23
|
McGrath M, Ma C and Raines DE:
Dimethoxy-etomidate: A nonhypnotic etomidate analog that potently
inhibits steroidogenesis. J Pharmacol Exp Ther. 364:229–237.
2018.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Forman SA: Clinical and molecular
pharmacology of etomidate. Anesthesiology. 114:695–707.
2011.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Av Sá LGD, Silva CRD, de A Neto JB,
Cândido TM, de Oliveira LC, do Nascimento FB, Barroso FD, da Silva
LJ, de Mesquita JR, de Moraes MO, et al: Etomidate inhibits the
growth of MRSA and exhibits synergism with oxacillin. Future
Microbiol. 15:1611–1619. 2020.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Liu J, Dong W, Wang T, Liu L, Zhan L, Shi
Y and Han J: Effects of etomidate and propofol on immune function
in patients with lung adenocarcinoma. Am J Transl Res. 8:5748–5755.
2016.PubMed/NCBI
|
|
27
|
Chantry A: WWP2 ubiquitin ligase and its
isoforms: New biological insight and promising disease targets.
Cell Cycle. 10:2437–2439. 2011.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Maddika S, Kavela S, Rani N, Palicharla
VR, Pokorny JL, Sarkaria JN and Chen J: WWP2 is an E3 ubiquitin
ligase for PTEN. Nat Cell Biol. 13:728–733. 2011.PubMed/NCBI View
Article : Google Scholar
|
|
29
|
Wang K, Liu J, Zhao X, Li H, Luo G, Yu Y,
Guo Y, Zhang L, Zhu J, Wang S, et al: WWP2 regulates proliferation
of gastric cancer cells in a PTEN-dependent manner. Biochem Biophys
Res Commun. 521:652–659. 2020.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Dillon LM and Miller TW: Therapeutic
targeting of cancers with loss of PTEN function. Curr Drug Targets.
15:65–79. 2014.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Trigka EA, Levidou G, Saetta AA,
Chatziandreou I, Tomos P, Thalassinos N, Anastasiou N, Spartalis E,
Kavantzas N, Patsouris E and Korkolopoulou P: A detailed
immunohistochemical analysis of the PI3K/AKT/mTOR pathway in lung
cancer: Correlation with PIK3CA, AKT1, K-RAS or PTEN mutational
status and clinicopathological features. Oncol Rep. 30:623–636.
2013.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Daniilidou K, Frangou-Plemenou M,
Grammatikakis J, Grigoriou O, Vitoratos N and Kondi-Pafiti A:
Prognostic significance and diagnostic value of PTEN and p53
expression in endometrial carcinoma. A retrospective
clinicopathological and immunohistochemical study. J BUON.
18:195–201. 2013.PubMed/NCBI
|
|
33
|
Pérez-Ramírez C, Cañadas-Garre M, Molina
MÁ, Faus-Dáder MJ and Calleja-Hernández MÁ: PTEN and PI3K/AKT in
non-small-cell lung cancer. Pharmacogenomics. 16:1843–1862.
2015.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Soond SM, Savvateeva LV, Makarov VA,
Gorokhovets NV, Townsend PA and Zamyatnin AA Jr: Making
connections: p53 and the cathepsin proteases as co-regulators of
cancer and apoptosis. Cancers (Basel). 12(3476)2020.PubMed/NCBI View Article : Google Scholar
|