Introduction
Multiple sclerosis (MS) is a lifelong demyelinating
disease of the central nervous system. The clinical onset of MS
tends to be between the second and fourth decade of life. Similarly
to other autoimmune diseases, women are affected 3-4 times more
frequently than men (1). About 10%
of MS patients experience a primary progressive MS form
characterized by the progression of neurological disability from
the onset. In about 90% of MS patients, the disease undergoes the
relapse-remitting MS course (RRMS); in most of these patients, the
condition acquires secondary progressive course (SPMS) (2).
The cause of MS is not clear. It is widely believed
to be an autoimmune disorder triggered in genetically predisposed
subjects by environmental factors such as infection, exposure to
sunlight, and vitamin D deficiency (3-5).
The vitamin deficiency and other genetic factors may also
contribute to the augmented production of proinflammatory cytokines
such as IL-12, IL-18 and macrophage migration inhibitory factor
that have been found during the active phases of the disease
(6-9).
Recent evidence indicated that these abnormal production of
proinflammatory cytokines may reside in the hyperactivation of the
inflammasome (10). Simultaneously,
MS patients showed reduced production of anti-inflammatory
cytokines such as IL-37 as well as endogenous anti-inflammatory
mediators such as IL-1 receptor antagonist, the soluble IL-1
receptor type 2 and heme oxygenase I (11-14).
MS susceptible loci have been identified in regions containing
genes with immune, co-stimulatory, signal transduction functions
and related to vitamin D function (15-21).
Ubiquitin proteasome system (UPS) plays a vital role
in immunity and its deregulation and/or modulation may influence MS
development and progression. The 20S proteasome has been identified
as a target of the humoral autoreactive immune response (22) and a major autoantigen in MS patients
(23). Proteolytic activities of
proteasomes are reduced in the brain tissue of MS patients
(24). Inhibition of proteasomes
and lysosomal proteases involved in major histocompatibility
complex (MHC) II antigen presentation has shown to improve MS
therapeutic efficacy (22,25). Disease-modifying treatments
significantly decrease the frequency of relapses and impede the
progression of disability during MS (26). Interferon-β-1b (IFNβ-1b) reduces
relapses in MS and improves magnetic resonance imaging (MRI)
outcomes. Plasma levels of ubiquitin and proteasome enzymatic
activity in MS patients before IFNβ-1b therapy have been shown to
decrease after six months of treatment (25). In a similar manner, treatment with
IFNβ augments other endogenous anti-inflammatory mediators such as
TGFβ, Heme oxygenase 1 and IL-1 receptor antagonist thus pointing
out to its pleiotropic pharmacological effects (12,14,27).
Preliminary studies have shown that the potency of IFNβ can be
increased further by simultaneous treatment with cyclophosphamide
in patients with aggressive diseases (28). In this case, studies to identify
pathways genes that predict a successful response to this treatment
combination appear promising. Similarly, on the basis of analysis
of genes and biomarkers it was possible to predict therapeutic
effects of natalizumab (6,29-31).
Genetic variations in PSMA3 and PSMB9
proteasome genes were reported to be associated with the risk of MS
in Latvians (32) and Italians
(33). Polymorphisms of the
14q11-24 proteasome genes PSMA6 and PSMC6 were
previously involved in susceptibility to juvenile idiopathic
arthritis (JIA) (34), asthma in
children (BA) (35) and obesity
(OB) (36), type 2 and type 1
diabetes mellitus (T1DM and T2DM) (37-39),
cardiovascular disorders (39,40)
and adaptation to the environment of the population (41). Taking into account that the
statistically proven phenomena of the coexistence of autoimmune
diseases (42) might happen that
some of these polymorphisms are associated with susceptibility to
MS, disease prognosis and/or response to therapy.
The aim of the current study was to evaluate the
association of genetic variations in the proteasome genes
PSMA6 (rs2277460 and rs1048990) and PSMC6 (rs2295826
and rs2295827) encoding regulatory α-subunit and ATPase subunit
respectively with MS development and response to the therapy in
Latvians. Additionally, the interaction of the PSMA6 and
PSMC6 polymorphisms with the rs2348071 locus in the
PSMA3 genes were analysed on MS susceptibility. Further on,
using publicly available SNPexp database (43) on correlations between SNP genotype
and gene expression levels in lymphoblastoid cells of individuals
included in the HapMap populations, we performed an in
silico analysis of the association between the SNPs analysed
and the expression levels of the number of previously reported MS
gene candidates and genes involved in the interferon signalling
pathway. Genotype-phenotype correlations identified in the study
may provide a new insight on MS pathogenesis and response to the
therapy.
Materials and methods
Case-control study
The case group consisted of 280 MS patients (mean
age: 41.66±12.16 years) attending the Latvian Maritime Medicine
Centre, Vecmilgravis Hospital. MS patients were diagnosed according
to the revised 2010 McDonald criteria (44) and assigned to RRMS (187 patients;
mean age: 38.88±12.81 years) and SPMS (93 patients; mean age:
51.20±9.94 years) groups. The control group of 305 individuals
representing the total Latvian population was previously described
(32,38). All controls were carefully assessed
to exclude the diagnosis of any inflammatory disorders. Written
consent was obtained from all participants of the study. The study
was performed according to the Declaration of Helsinki and the
study protocol was approved by the Central Medical Ethics Committee
of Latvia (Protocol Nr. 01-29.1/17).
Treatment
Three disease-modifying agents, including IFNβ,
glatiramer acetate and mitoxantrone, were used for the treatment of
210 MS patients. Details on therapy are given in Table I.
 | Table IMultiple sclerosis treatment and
response to therapy. |
Table I
Multiple sclerosis treatment and
response to therapy.
| | Patient number |
|---|
| Disease-modifying
agent | Commercial
name | Manufacturer | Formulation of
treatment | Total treated | Respon-ders | Non-responders |
|---|
| IFNβ total | | | | 148 | 37 | 111 |
|
IFNβ-1a |
Avonex® | Biogen Idec
Limited | Intramuscular 30 µg
once weekly | 25 | 10 | 15 |
|
IFNβ-1a |
Rebif® | Merck Serono; Merck
KGaA | Subcutaneous 44 µg
three times per week | 59 | 14 | 45 |
|
IFNβ-1b |
Betaseron® or
Extavia® | Bayer AG | Subcutaneous 250 µg
every other day | 64 | 13 | 51 |
| Glatiramer
acetate |
Copaxone® | TEVA Pharmaceutical
Industries, Ltd | Subcutaneous 20 mg
once a day | 39 | 4 | 35 |
| Mitoxan-trone |
Novantrone® | Wyeth
Laboratories | Intravenous once
every 3 months | 23 | 0 | 23 |
Response to therapy evaluation
Progress of disease and response to treatment were
evaluated in terms of ‘no evidence of disease activity’ (NEDA)
standard (45) measured by
relapses, the progression of disability and new and/or enlarging
demyelinating lesions as seen on MRI.
DNA extraction and genotyping
Genomic DNA extraction and the rs2277460, rs1048990,
rs2295826 and rs2295827 genotyping technologies (primer sequences,
necessary Polymerase Chain Reaction procedure and analysis of
amplified and digested products) were the same as in (34,41).
For quality control, 16 randomly chosen samples per each marker
were genotyped in two different experiments. Genotyping data were
verified by direct sequencing of the corresponding DNA fragments in
both directions using Applied Biosystems 3130xl Genetic
Analyzer (Fig. S1). Loci
description and nucleotide numbering are given according to the
recommended nomenclature system (http://www.genomic.unimelb.edu.au/mdi/mutnomen/recs.html).
Chromosome 14, GRCh37.p5 assembly (NCBI reference sequence:
NC_000014.8) sequence information was used for loci
description.
Data analysis
Single locus genotypes' and alleles' frequencies
were estimated in the MS group by direct gene counting. The genetic
diversity of SNPs rs2277460, rs1048990, rs2295826 and rs2295827 was
studied previously by Sjakste et al, 2016(38) in the control group. Genotyping data
on the rs2348071 variations in MS patients were described by
Kalnina et al, 2014(32).
These primary genotyping data were used in the current case/control
study to construct multi-locus genotypes, haplotypes and identify
the main effects of MS in single- and multi-locus models. Deviation
from the Hardy-Weinberg equilibrium (HWE) and the differences
between case and control groups in allele, genotype and haplotype
frequencies were evaluated by χ2 using XLSTAT software
for Windows (Data Analysis and Statistical Solution for Microsoft
Excel, https://www.xlstat.com/en, Addinsoft,
Paris, France 2017). Dominant, recessive, over dominant and
multiplicative genetic models for every individual locus were
designed according to (46), and
two- or one-tailed P-value (significant <0.05) was evaluated by
χ2 using 2x2 contingency tables. Odds ratio (OR) higher
than two (2) and lower than 0.5 was
considered to be clinically significant (46). Stratification was performed by MS
course, NEDA standard and response to the therapy.
Genotype-phenotype correlation
analysis
Data on the correlation between SNPs' genotypes and
expression levels of the number of genes potentially involved in MS
pathogenesis (15-20)
and/or response to the IFNβ therapy (PathCards, https://pathcards.genecards.org/card/immune_response_ifn_alphabeta_signaling_pathway)
were derived using SNPexp online tool [http://tinyurl.com/snpexp., Norwegian PSC Research
Center, Rikshospitalet, Oslo, Norway (43)] for lymphoblastoid cells of all
unrelated individuals of the HapMap phase II release 23 data set
which consist of 3.96 million SNP genotypes from 270 individuals
from four populations: CEU (90 Utah residents with ancestry from
northern and western Europe), HCB (45 unrelated Han Chinese in
Beijing), JPT (45 unrelated Japanese in Tokyo), YRI (90 Yoruba in
Ibadan, Nigeria). Dominant and additive genetic models were tested
for correlation analysis. The Spearman' correlation between
genotype and the gene expression level was considered to be
positive if at least one probe per transcript showed a
statistically significant association (P<0.05).
Taking into account that in the SNPexp platform
(43) one gene can be represented
by several probes, but one probe maps only one gene, the results
were manually filtered: 1) To exclude possible false calls when one
probe is found to belong to several genes; 2) to optimize repeated
data when several probes map to one gene; 3) to exclude potentially
false calls identified for the rs2295826, but not for the
rs2295827. Only Spearman correlations with P-value less than 0.05
or significant correlation found for both the rs2295826 and
rs2295827 were taken into account. The data obtained as a result of
filtration is given in Tables SI
and SII.
Results
Genotyping results and single-locus
association analysis
In the current study, we have genotyped the
rs2277460, rs1048990, rs2295826 and rs2295827 in 280 MS patients.
In Latvia there are 2035 registered MS patients in total. The study
can be considered to be a representative for the Latvian
population, as 13.7% of all patients were covered by our study.
Since significance was achieved and revealed the trend on the
association with the disease, our results appeared to predict the
common association trends for larger sample groups, with the
possibility of further studies reproduced in other populations.
Data on alleles' and genotypes' distributions and
single-locus associations are given in Table II. The Genotyping call rate was
100% and all genotyped markers were found to be in a perfect HWE
(P>0.05). The rs2295826 and rs2295827 were observed in a
complete (D'=1, r2=1) linkage disequilibrium and in
further analysis, the linkage block rs2295826-rs2295827 will be
represented by the rs2295826 locus. No linkage had been identified
between other polymorphic loci analysed (P>0.05).
 | Table IISNP genotype and MA frequencies and
data on association with MS. |
Table II
SNP genotype and MA frequencies and
data on association with MS.
| | Data on
association |
|---|
| SNP ID and MA or
genotype | MS (n=280) | Controls
(n=305) | Genetic model | P-value | OR [95% CI] |
|---|
|
rs2277460 | | | | | |
|
A | 51 (9.11) | 44 (7.21) | A vs. C | n/s | - |
|
CC | 229 (81.79) | 261 (85.57) | - | - | - |
|
CA | 51 (18.21) | 44 (14.43) | CA vs. CC | n/s | - |
|
rs1048990 | | | | | |
|
G | 50 (8.93) | 57 (9.34) | G vs. C | n/s | - |
|
CC | 233 (83.21) | 250 (81.97) | - | - | - |
|
CG | 44 (15.72) | 53 (17.38) | CG + GG vs. CC | n/s | - |
|
GG | 3 (1.07) | 2 (0.65) | - | - | - |
|
rs2295826 | | | | | |
|
G | 96 (17.14) | 69 (11.31) | G vs. A | 0.0042 | 1.62
[1.16-2.26] |
|
AA | 187 (66.79) | 243 (79.67) | - | - | - |
|
AG | 90 (32.14) | 55 (18.03) | AG + GG vs. AA | 0.0004 | 1.95
[1.34-2.83] |
|
GG | 3 (1.07) | 7 (2.30) | - | - | - |
|
rs2295827 | | | | | |
|
T | 96 (17.14) | 69 (11.31) | T vs C | 0.0042 | 1.62
[1.16-2.26] |
|
CC | 187 (66.79) | 243 (79.67) | - | - | - |
|
CT | 90 (32.14) | 55 (18.03) | CT + TT vs. CC | 0.0004 | 1.95
[1.34-2.83] |
|
TT | 3 (1.07) | 7 (2.30) | - | - | - |
The rs2277460 and rs1048990 exhibited similar
alleles' and genotypes' frequencies in the cases and controls and
appeared to be MS neutral.
The linkage block rs2295826-rs2295827 showed
statistically significant MS main effect for both rare alleles
[P<0.05; odds ratio, OR=1.62, 95% confidence interval (CI)
(1.16-2.26)] and genotypes in the dominant model [P<0.001;
OR=1.95, 95% CI (1.34-2.83)].
Identification of the risk/protective
multi-loci genotypes
Using personalized data documentation, we
reconstructed individual multi-loci genotypes and tested them on
the association with the disease. Full-spectrum and frequencies of
the rs2277460-rs1048990-rs2295826-rs2295827-rs2348071 genotypes is
presented in Table SIII. Genotype
having the homozygotes on major alleles simultaneously at the
rs2277460 and rs1048990 and MS risk genotypes at the
rs2295826-rs2295827 and rs2348071 showed a strong association with
the disease [4-LG7, P=0.0001; OR=3.01, 95% CI (1.70-5.33)].
Genotypes including risk alleles at loci rs2277460,
rs2295826-rs2295827 and rs2348071 (4-LG12) and risk genotypes at
loci rs2277460 and rs2295826-rs2295827 (4-LG11) were found about
two times more frequently in MS patients than in controls.
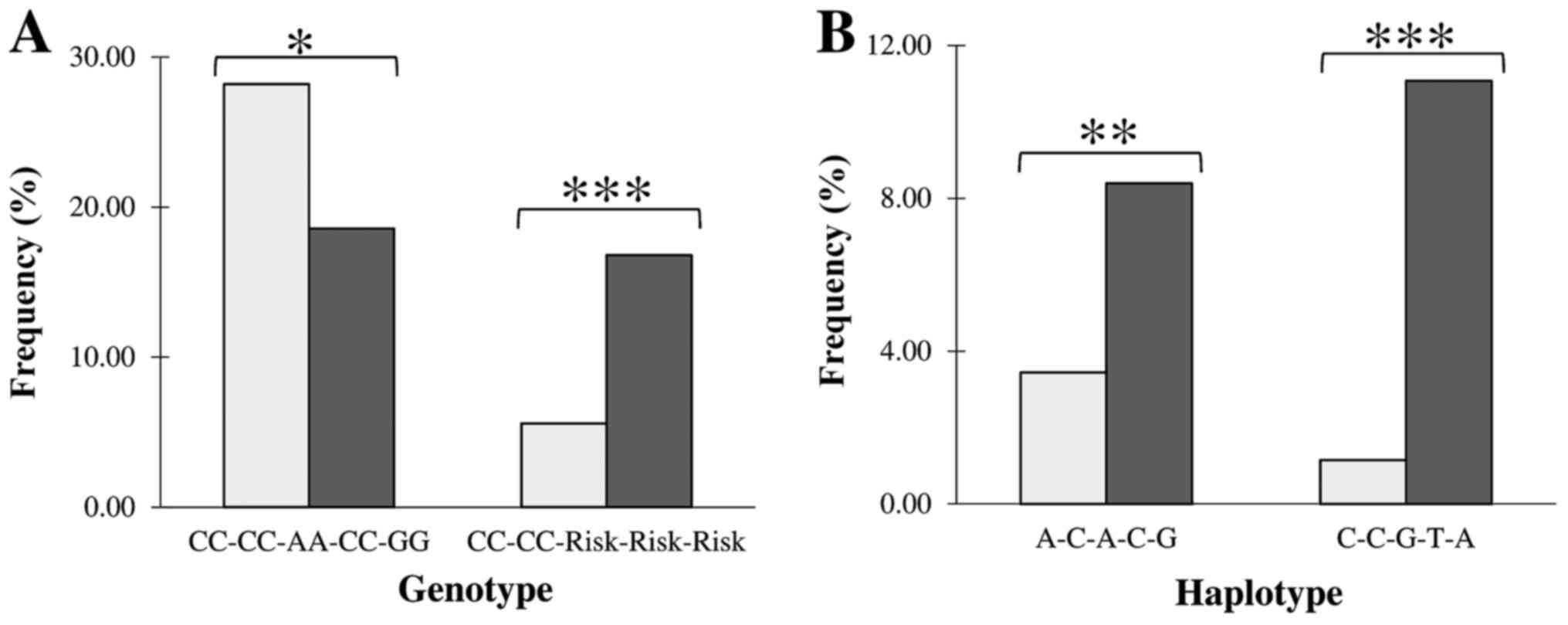
Genotype homozygous on major alleles at all five
loci was the most frequent in controls (28%) and second by
frequency in MS patients (~18%) showing significant disease
protective effect [P<0.05; OR=0.31, 95% CI (0.66-0.96), Fig. 1A, Table
SIII].
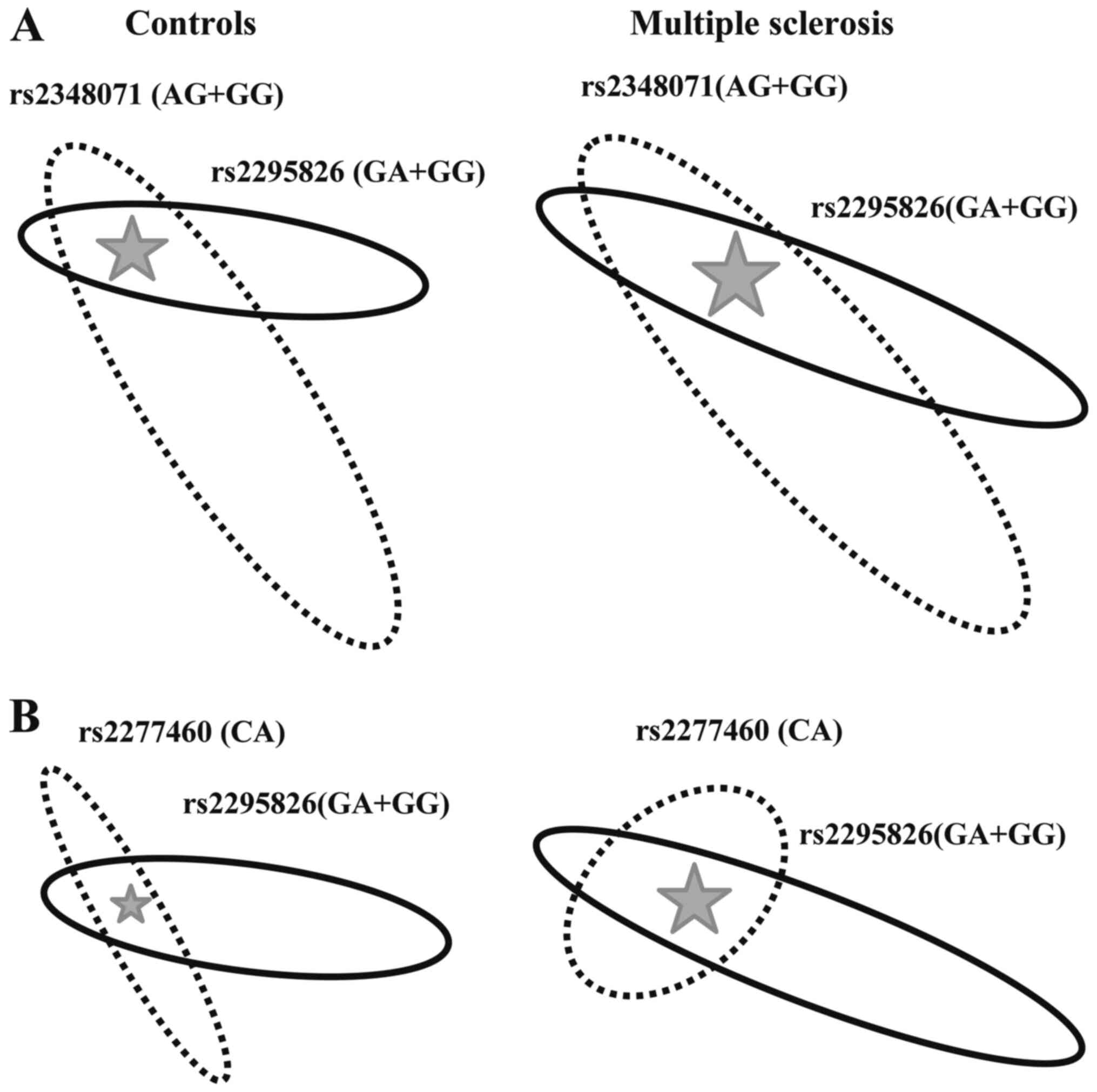
Further on, we analysed the interaction between any
two loci (the rs2295826-rs2295827 linkage block was considered as
one locus) on MS susceptibility. Data on statistically significant
interactions are illustrated in Fig.
2.
The rs2295826-rs2348071 Risk-Risk genotype, was more
than twice more frequent in MS patients (22.14%) than in controls
(8.54%) and showed strong (P<0.0001) association with the
disease [OR=3.60, 95% CI (1.60-4.22); Fig. 2A]. The rs2277460-rs2295826 genotype
of the CA-Risk structure rarely observed in controls (2.63%) was
more frequent in MS patients (6.43%) showing nominal (P<0.05)
association with the disease [OR=2.45, 95% CI (1.05-5.73); Fig. 2B].
Haplotype analysis
Results on haplotype analysis are given in the
Table SIV and illustrated in
Fig. 1B.
Haplotype diversity was similar in both groups of MS
patients and healthy individuals. Although, the rs2277460 locus did
not show MS susceptibility in a single-locus analysis, the
haplotype of the A-C-A-C-G structure (Hap6) showed a significant
association with the disease [P<0.001; OR=2.44 95% CI
(1.45-4.11), Fig. 1B].
The
rs2277460-rs1048990-rs2295826-rs2295827-rs2348071 haplotype of the
C-C-G-T-A composition (Hap8) having MS risk alleles at the
rs2295826-rs2295827 and rs2348071 loci simultaneously, showed
strong (P<0.0001) association with the disease [OR=9.65 95% CI
(4.49-20.76), Fig. 1B] supporting
the above presented data on the single- and multi-loci genotype
susceptibility.
Polymorphism association with the
response to therapy
Patients did not respond to mitoxantrone therapy.
Among 39 patients treated by glatiramer acetate, only four were
susceptible to this treatment. In the group of 148 patients treated
with IFNβ, 37 responders were detected (Table I).
Genotyping data were further analysed for the
association with response to IFNβ therapy (Table III). Alleles' and genotypes'
distribution were found to be similar between responders and
non-responders at the rs2277460, rs2295826-rs2295827 and rs2348071.
However, the G allele at the rs1048990 was approximately twice more
frequent in non-responders than in responders and the association
of the G allele with no IFNβ therapy response was found to be close
to statistical significance [Table
III; OR=2.53 95% CI (0.90 7.08) in the multiplicative
model].
 | Table IIIThe rs1048990 alleles and genotypes
presentation in responders and non-responders to IFNβ therapy. |
Table III
The rs1048990 alleles and genotypes
presentation in responders and non-responders to IFNβ therapy.
| | IFN-β (n) | Statistics |
|---|
| Allele or
genotype | R | NR | Model | P-value | OR [95% CI] |
|---|
| C | 70 (94.59) | 194 (87.39) | G vs. C | 0.059 | 2.53
[0.90-7.08] |
| G | 4 (5.41) | 28 (12.61) | G vs. CC | 0.056 | 2.69
[0.92-7.84] |
| CC | 33 (89.19) | 86 (77.48) | - | - | - |
| CG | 4 (10.81) | 22 (19.82) | CG + GG vs. CC | 0.090 | 2.40
[0.78-7.42] |
| GG | - | 3 (2.70) | - | - | - |
Correlation between SNPs' genotype and
gene expression
Suggesting the potential for genetic variations to
produce a local or distant functional effect in the genome, we
analysed the further correlation between genotypes of the
rs1048990, rs2295826-rs2295827 and rs2348071, identified in the
current and previous (32) studies
as MS susceptible and expression levels of 13 well-characterized
genes associated with the susceptibility to MS (15-20)
and 21 genes involved in IFNα/β signalling pathway (PathCards,
https://pathcards.genecards.org/card/immune_response_ifn_alphabeta_signaling_pathway).
The name of the CD25 gene was not found on the expression
chips. A statistically significant correlation between mRNA
transcript level and genotypes of any SNP was found for six genes
implicated in MS pathogenesis and nine genes involved in IFN
pathways (Tables IV and SII).
 | Table IVData on correlation between mRNA
expression and the rs1048990, rs2295826-rs2295827 and rs2348071
genotypes. |
Table IV
Data on correlation between mRNA
expression and the rs1048990, rs2295826-rs2295827 and rs2348071
genotypes.
| Gene group | In correlation | Without
correlation | Not found on the
expression chip | Total |
|---|
| MS |
CD58a,d, HLA-Aa, HLA-Bb,c, IL7Ra,d, STAT3a,d, TNFRSF5
(CD40)a,d | CD6,
HLA-DRB1, IL2RA, IL12A, IRF8 (ICSBP),
TNFRSF1A | CD25 | 13 |
| IFN | IFI6
(G1P3)d, IFNA1
(IFNα)a,
IRF1a,
IRF2d, PRMT1
(HMRT1L2)d,PTP-1B
(PTPN1)a,d, SHP-2
(PTPN11)d,
TYK2a,
USP18a,d | IFNAR1,
IFNAR2, IFNB1, ISG15 (G1P2), ISG54
(IFIT2), ISGF3 (IRF9), JAK1, PML
(MYL), SHP-1 (PTPN6), SOCS1, STAT1,
STAT2 | - | 21 |
| Total | 15 | 19 | 1 | 34 |
Examples of correlations revealed and details on the
correlated genes are given in Fig.
3 and Table SII, respectively.
Risk genotypes of the rs1048990 and rs2348071 were found in a
simultaneous correlation with expression levels of the IL7R
(Fig. 3B), STAT3 (Fig. 3C), TNFRSF5 (Fig. 3D), PTP-1B (Fig. 3E) and USP18 (Fig. 3F). Fig.
3A illustrated the situation when the HLA-B expression
level correlates with the rs2295826 and rs2295827.
Discussion
The main finding of this study is that the
PSMC6-rs2295826-rs2295827 and PSMA3-rs2348071 loci
showed MS susceptibility, and data on single-locus associations
were confirmed by haplotype and multi-loci genotype analysis.
Remarkably, MS susceptible to
rs2277460-rs1048990-rs2295826-rs2348071 haplotypes of the A-C-A-C-G
and C-C-G-T-A configuration is identified in this study (Fig. 1B) were found to be linked with
poly-JIA and oligo-JIA, respectively (41), and T1DM (38). Similarly, multi-loci genotype
CC-CC-Risk-Risk-Risk, associated with MS risk effect in this study
(Fig. 1A), is also a risk factor
for T1DM (38), BA (35) and JIA (41). We did not find any data about the
association of the polymorphisms studied by us with MS in the
literature. However, haplotypes encompassing the KIAA0391
localized in 14q13.2 and PSMA6 gene cluster confer a genetic
link for myocardial infarction and coronary artery disease
(47). Additionally, in a GWAS
study performed by the International Multiple Sclerosis Consortium
(48) on association of SNP
rs12436216 of the gene KIAA0391, it was reported that
14q13.2 could be a MS susceptibility locus.
Some loci of susceptibility may be shared among many
autoimmune and other immune-mediated diseases (49,50).
Earlier, the pleiotropic genetic effect as a frequent phenomenon in
human complex traits and diseases (51) has been reported for BA and OB
(52,53) and for BA and juvenile rheumatoid
arthritis (54,55). Similarly, rs2277460 associated with
BA in our previous study (35) has
been found to determine susceptibility in Latvians with JIA
(34) and T1DM (38). This locus turned out not to be
associated with MS primary cohort in a single-locus analysis in
Latvians, however, it was found in the block of interaction of risk
loci PSMA6-rs2277460-PSMC6-rs2295826, which was more
frequent in disease's group (Fig.
2B). This data was confirmed by haplotype analysis: Haplotype
A-C-A-C-G included only the rs2277460 risk allele A and showed a
significant association with MS [P<0.001; OR=2.44 95% CI
(1.45-4.11), Fig. 1B].
In silico analysis revealed (34,41)
that the nucleotide substitutions we have studied, modified
transcription factor (TFs) binding sites and miRNAs. This can
significantly modulate the transcription of related genes and gene
networks in response to the inflammation and other environmental
stimuli.
The PSMA6-rs2277460 and
PSMA3-rs2348071 rare alleles and the PSMC6-rs2295826
major allele assisted the cytoplasmic ribonucleoprotein hnRNPA1
targeting. This multifunctional protein is involved in the
pathogenesis of many neurodegenerative diseases, including MS
(45,56,57).
It can influence protein-protein interactions, including those
involving NF-κB, which are known to play a critical role in the
initiation and progression of MS (58,59)
and participate in the crosstalk with UPS at different levels of
the NF-κB signalling pathways (60). This splicing signal, affecting
splicing and post-transcriptional modification of the majority of
expressed genes in mammals, was shown to directly interact with
PSMA3 proteins (61) and may be
involved in the regulation of the PSMA3 gene expression
through the rs2348071 allele-specific targeting.
PSMA6-rs1048990 rare allele G generates
binding sites for multifunctional p53 protein, overexpression of
which can induce apoptosis of oligodendrocytes in MS (62). The PSMA6-rs2277460
substitution to A, produces binding sites for BARBIE box proteins,
which are shown to be involved in innate immunity modulation
(63). The major allele of
PSMC6-rs2295826 generates sequence affinity to transcription
factors of MYT1 and PARF families known to be involved in primary
neurogenesis; substitution at the PSMC6-rs2295827 can bind
the BRN5 and LHXF factors and replacement at the
PSMA3-rs2348071 may significantly change patterns of the
local targets for regulatory proteins of MEF2 and HBPX families
known to mediate transcriptional control of neuronal
differentiation (64-67);
HOXF family NANOG.01 is involved in signal transduction pathways
during development (68).
Previously, PSMA6-rs1048990 had shown to
affect PSMA6 gene expression in vivo (69) and in vitro (40). However, it should be taken into
account that the data on the possibility of a particular genetic
variant to have multiple transcription effects are limited.
To explain the above speculation it should be kept
in mind also, that changes in the expression of different
proteasome subunits in response to outer factors is not
co-ordinated. For example, cocaine mainly upregulates PSMB1
and PSMA5 subunits, decreases PSMA6 subunit, but does
not affect PSMB2 and PSMB5 subunit expression
(70). Similar ‘chaotic’ changes in
proteasome gene expression can happen in pathogenesis of the
diseases, including MS. General expression of the proteasomes is
increased in MS and, decreased by the interferon therapy (25). At the first stage of this process,
the expression of the PSMA6 gene could increase, which can further
promote the formation of more proteasomes with a subsequent
increase in their activity. The same might happen with immune
proteasome expression. The composition of α subunits in
‘conventional’ and immune proteasomes is identical. Thus, higher
accessibility of PSMA6 subunits can favour initial increase of the
formation of ‘conventional’ proteasomes with the transformation to
immune proteasomes.
Based on the present and previously published
results (38), we propose a concept
for correlations between genotypes involved in MS risk and the
level of expression of MS gene candidates. We should keep in mind
that the expression studies in silico using data obtained
for the lymphoblastoid cell line is a standard approach used for
the MS studies (16).
A group of well-known MS gene candidates (Tables IV and SII) is represented by genes encoding the
lymphocyte function-associated antigen 3 (CD58), MHC
antigens (HLA-A and HLA-B), interleukin-7 receptor
subunit α (IL7R), signal transducer and activator of
transcription 3STAT3, tumor necrosis factor receptor
superfamily member 5 [TNFRSF5 (CD40)].
The gene group involved in IFNα/β signalling pathway
(Tables IV and SII) includes: Genes encoding interferon-α
inducible protein 6 and interferon-α1 (IFI6 (G1P3) and
IFNA1 (IFNα), respectively), representatives of interferon
regulatory factors (IRF1 and IRF2), proteins related
to enzyme binding and protein domain specific binding (PRMT1
(HMRT1L2), PTP-1B (PTPN1), SHP-2 (PTPN11, TYK2 and
USP18)).
So, the expression of MS susceptible genes coding
proteins of immune function (CD58, IL7R, STAT3, TNFRSF5
(CD40)) and genes involved in the IFN pathway (PTP-1B
(PTPN1) and USP18) was found to be in a statistically
significant correlation with the rs1048990 and rs2348071 risk
genotypes simultaneously (Fig.
3B-F). Besides, among genes involved in IFNα/β signalling
pathway the expression levels of IFI6 (G1P3), IRF2, PRMT1
(HMRT1L2) and SHP-2 (PTPN11) were found to be in a
correlation only with PSMA3-rs2348071, but IFNA1
(IFN-α), IRF1 and TYK2 genes correlate only with
the PSMA6-rs1048990 (Table
IV). Loci PCMC6-rs2295826 and rs2295827, were identified
as susceptible to MS cohort in this study, are in a correlation
with HLA-B encoding the MHC antigen expression level
(Fig. 3A).
These results prove that the possibility of the
combined functional effect of genetic risk factors on various
metabolic aspects of MS pathogenesis is parallel with the potential
for each SNP to have its own transcriptional effect on the genes
involved in this study.
UPS components possess the potential of being a
therapeutic target for the treatment of several diseases (71). IFNβ is a pleiotropic cytokine
belonging to the type I interferons; it is involved in the
regulation of innate and adaptive immune responses and is widely
used as the first-line disease-modifying treatment for multiple
sclerosis (72). Interestingly,
among the studied loci, the association of the G allele at the
PSMA6-rs1048990 with no IFNβ therapy response in the Latvian
population in the current study was found to be close to
statistical significance (Table
III). It might be that with the increase in the number of
patients in the studied cohorts, the significance of statistical
reliability in this associative study can increase. The above data
provided evidence of the cumulative effect of immune-response genes
on IFNβ treatment efficacy.
The PSMA6-rs1048990-rs2277460,
PSMC6-rs2295826-rs2295827 and PSMA3-rs2348071 single-
and multi-loci variations may contribute to the risk of MS in
Latvians. The nucleotide substitutions may either have their own
functional effect on transcription of numerous genes involved in MS
pathogenesis or may be linked to other MS-susceptible genetic
variations.
Our results suggested the potential for the
rs1048990 of the PSMA6 gene to be an independent marker for
the prognosis of the IFNβ therapy response.
Supplementary Material
Examples of the DNA sequencing
chromatograms for PSMA6: rs2277460 (A1: genotype CC; A2:
genotype CA) and rs1048990 (B1: CC; B2: GG), PSMC6:
rs2295826 (C1: AA; C2: GG) and rs2295827 (D1: CC; D2: TT) and
PSMA3 rs2348071 (E1: GG; E2: AA) described in the present
study. On chromatograms, the sites of localization of the studied
SNPs are marked in grey. Allelic variants in the sequence are in
bold and underlined.
14q SNP gentoype polymorphisms and
correlation with gene expression level (profile) of related genes,
including: (1) genes-candidates for
MS and (2) IFN α/β signalling
pathway genes, for HapMap unrelated subjects of CEU, YRI, HCB and
JPT HapMap populations grouped in one HapMap unrelated subjects
cohort.
Summary on the genotype - phenotype
correlation used in the genes whose expression levels in
lymphoblastic cell lines are correlated with SNP genotype in the
dominant model for the rs1048990, rs2295826 and rs2295827 and in
the additive model for the rs2348071.
Spectrum and frequency of the 4 loci
genotypes in patients with multiple sclerosis and healthy
individuals and data on the association with disease.
Allelic composition frequency of the 5
loci haplotypes in patients with multiple sclerosis and healthy
individuals.
Acknowledgements
The authors would like to thank Ms Natalija
Mjagkova (Latvian Maritime Medicine Centre, Riga, Latvia) for
technical assistance.
Funding
The current study was funded by the European Regional
Development Fund (project no. 1.1.1.1/16/A/016; 'Identification of
proteasome related genetic, epigenetic and clinical markers for
multiple sclerosis.').
Availability of data and materials
The datasets used and/or analysed during the
current study are available from the corresponding author on
reasonable request.
Authors' contributions
TS conceived of the study and acquired the funding.
KDo and KDi analyzed and interpreted the data. IT performed the
formal analysis. NP, KDo and KDi performed the experiments. IT and
TS conceived of the methodology. NS and NP obtained resources and
were associated with the project administration. KDo and KDi ran
the software. JK performed data validation. NP, JK and NS confirmed
the authenticity of all the raw data. NP and TS wrote the original
draft of the manuscript. NP, KD, IT and NS wrote, reviewed and
edited the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was performed according to the
Declaration of Helsinki and the study protocol was approved by the
Central Medical Ethics Committee of Latvia (protocol no.
01-29.1/17). Informed consent was obtained from all individual
participants included in the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Compston A and Confavreux C: The
distribution of multiple sclerosis. In: McAlPine's multiPle
Sclerosis. 4th edition. Pioli S (ed). Churchill
Livingstone/Elsevier, Philadelphia, pp69-180, 2006.
|
|
2
|
Bellavista E, Santoro A, Galimberti D,
Comi C, Luciani F and Mishto M: Current understanding on the role
of standard and immunoproteasomes in inflammatory/immunological
pathways of multiple sclerosis. Autoimmune Dis.
2014(739705)2014.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Miller AE: Multiple sclerosis: Where will
we be in 2020? Mt Sinai J Med. 78:268–279. 2011.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Noseworthy JH, Lucchinetti C, Rodriguez M
and Weinshenker BG: Multiple sclerosis. N Engl J Med. 343:938–952.
2000.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Song GG, Choi SJ, Ji JD and Lee YH:
Genome-wide pathway analysis of a genome-wide association study on
multiple sclerosis. Mol Biol Rep. 40:2557–2564. 2013.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Nicoletti F, Patti F, Cocuzza C, Zaccone
P, Nicoletti A, Di Marco R and Reggio A: Elevated serum levels of
interleukin-12 in chronic progressive multiple sclerosis. J
Neuroimmunol. 70:87–90. 1996.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Nicoletti F, Di Marco R, Mangano K, Patti
F, Reggio E, Nicoletti A, Bendtzen K and Reggio A: Increased serum
levels of interleukin-18 in patients with multiple sclerosis.
Neurology. 57:342–244. 2001.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Cavalli E, Mazzon E, Basile MS, Mangano K,
Di Marco R, Bramanti P, Nicoletti F, Fagone P and Petralia MC:
Upregulated expression of macrophage migration inhibitory factor,
its analogue D-dopachrome tautomerase, and the CD44 receptor in
peripheral CD4 T cells from clinically isolated syndrome patients
with rapid conversion to clinical defined multiple sclerosis.
Medicina (Kaunas). 55(667)2019.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Fagone P, Mazzon E, Cavalli E, Bramanti A,
Petralia MC, Mangano K, Al-Abed Y, Bramati P and Nicoletti F:
Contribution of the macrophage migration inhibitory factor
superfamily of cytokines in the pathogenesis of preclinical and
human multiple sclerosis: In silico and in vivo evidences. J
Neuroimmunol. 322:46–56. 2018.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Govindarajan V, de Rivero Vaccari JP and
Keane RW: Role of inflammasomes in multiple sclerosis and their
potential as therapeutic targets. J Neuroinflammation.
17(260)2020.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Cavalli E, Mazzon E, Basile MS, Mammana S,
Pennisi M, Fagone P, Kalfin R, Martinovic V, Ivanovic J, Andabaka
M, et al: In silico and in vivo analysis of IL37 in multiple
sclerosis reveals its probable homeostatic role on the clinical
activity, disability, and treatment with fingolimod. Molecules.
25(20)2019.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Nicoletti F, Patti F, DiMarco R, Zaccone
P, Nicoletti A, Meroni P and Reggio A: Circulating serum levels of
IL-1ra in patients with relapsing remitting multiple sclerosis are
normal during remission phases but significantly increased either
during exacerbations or in response to IFN-beta treatment.
Cytokine. 8:395–400. 1996.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Dujmovic I, Mangano K, Pekmezovic T,
Quattrocchi C, Mesaros S, Stojsavljevic N, Nicoletti F and Drulovic
J: The analysis of IL-1 beta and its naturally occurring inhibitors
in multiple sclerosis: The elevation of IL-1 receptor antagonist
and IL-1 receptor type II after steroid therapy. J Neuroimmunol.
207:101–106. 2009.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Fagone P, Patti F, Mangano K, Mammana S,
Coco M, Touil-Boukoffa C, Chikovani T, Di Marco R and Nicoletti F:
Heme oxygenase-1 expression in peripheral blood mononuclear cells
correlates with disease activity in multiple sclerosis. J
Neuroimmunol. 261:82–86. 2013.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Australia and New Zealand Multiple
Sclerosis Genetics Consortium (ANZgene) Genome-wide association
study identifies new multiple sclerosis susceptibility loci on
chromosomes 12 and 20. Nat Genet. 41:824–828. 2009.PubMed/NCBI View Article : Google Scholar
|
|
16
|
International Multiple Sclerosis Genetics
Conssortium (IMSGC). IL12A, MPHOSPH9/CDK2AP1 and RGS1 are novel
multiple sclerosis susceptibility loci. Genes Immun. 11:397–405.
2010.PubMed/NCBI View Article : Google Scholar
|
|
17
|
International Multiple Sclerosis Genetics
Consortium. Hafler DA, Compston A, Sawcer S, Lander ES, Daly MJ, de
Jager PL, de Bakker PI, Gabriel SB, Mirel DB, et al: Risk allels
for multiple sclerosis identified by a genome wide study. N Engl J
Med. 357:851–862. 2007.PubMed/NCBI View Article : Google Scholar
|
|
18
|
De Jager PL, Jia X, Wang J, de Bakker PI,
Ottoboni L, Aggarwal NT, Piccio L, Raychaudhuri S, Tran D, Aubin C,
et al: Meta-analysis of genome scans and replication identify CD6,
IRF8 and TNFRSF1A as new multiple sclerosis susceptibility loci.
Nat Genet. 41:776–782. 2009.PubMed/NCBI View
Article : Google Scholar
|
|
19
|
Kofler DM, Severson CA, Mousissian N, De
Jager PL and Hafler DA: The CD6 multiple sclerosis susceptibility
allele is associated with alterations in CD4+ T cell
proliferation. J Immunol. 187:3286–3291. 2011.PubMed/NCBI View Article : Google Scholar
|
|
20
|
International Multiple Sclerosis Genetics
Consortium; Wellcome Trust Case Control Consortium 2. Sawcer S,
Hellenthal G, Pirinen M, Spencer CC, Patsopoulos NA, Moutsianas L,
Dilthey A, Su Z, et al: Genetic risk and a primary role for
cell-mediated immune mechanisms in multiple sclerosis. Nature.
476:214–219. 2011.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Zhang K, Chang S, Cui S, Guo L, Zhang L
and Wang J: ICSNPathway: Identify candidate causal SNPs and
pathways from genome-wide association study by one analytical
framework. Nucleic Acids Res. 39:W437–W443. 2011.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Fissolo N, Kraus M, Reich M, Ayturan M,
Overkleeft H, Driessen C and Weissert R: Dual inhibition of
proteasomal and lysosomal proteolysis ameliorates autoimmune
central nervous system inflammation. Eur J Immunol. 38:2401–2411.
2008.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Mayo I, Arribas J, Villoslada P, Alvarez
DoForno R, Rodríguez-Vilariño S, Montalban X, De Sagarra MR and
Castaño JG: The proteasome is a major autoantigen in multiple
sclerosis. Brain. 125:2658–2667. 2002.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Zheng J and Bizzozero OA: Decreased
activity of the 20S proteasome in the brain white matter and gray
matter of patients with multiple sclerosis. J Neurochem.
117:143–153. 2011.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Minagar A, Ma W, Zhang X, Wang X, Zhang K,
Alexander JS, Gonzalez-Toledo E and Albitar M: Plasma
ubiquitin-proteasome system profile in patients with multiple
sclerosis: Correlation with clinical features, neuroimaging, and
treatment with interferon-beta-1b. Neurol Res. 34:611–618.
2012.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Pardo G and Jones DE: The sequence of
disease-modifying therapies in relapsing multiple sclerosis: Safety
and immunologic considerations. J Neurol. 264:2351–2374.
2017.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Nicoletti F, Di Marco R, Patti F, Reggio
E, Nicoletti A, Zaccone P, Stivala F, Meroni PL and Reggio A: Blood
levels of transforming growth factor-beta 1 (TGF-beta1) are
elevated in both relapsing remitting and chronic progressive
multiple sclerosis (MS) patients and are further augmented by
treatment with interferon-beta 1b (IFN-beta1b). Clin Exp Immunol.
113:96–99. 1998.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Patti F, Cataldi ML, Nicoletti F, Reggio
E, Nicoletti A and Reggio A: Combination of cyclophosphamide and
interferon-beta halts progression in patients with rapidly
transitional multiple sclerosis. J Neurol Neurosurg Psychiatry.
71:404–407. 2001.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Fagone P, Mazzon E, Mammana S, Di Marco R,
Spinasanta F, Basile MS, Petralia MC, Bramanti P, Nicoletti F and
Mangano K: Identification of CD4+ T cell biomarkers for
predicting the response of patients with relapsing-remitting
multiple sclerosis to natalizumab treatment. Mol Med Rep.
20:678–684. 2019.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Derfuss T, Mehling M, Papadopoulou A,
Bar-Or A, Cohen JA and Kappos L: Advances in oral immunomodulating
therapies in relapsing multiple sclerosis. Lancet Neurol.
19:336–347. 2020.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Kulakova OG, Tsareva EY, Boyko AN, Shchur
SG, Gusev EI, Lvovs D, Favorov AV, Vandenbroeck K and Favorova OO:
Allelic combinations of immune-response genes as possible composite
markers of IFN-β efficacy in multiple sclerosis patients.
Pharmacogenomics. 13:1689–1700. 2012.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Kalnina J, Paramonova N, Sjakste N and
Sjakste T: Study of association between polymorphisms in the PSMB5
(rs11543947) and PSMA3 (rs2348071) genes and multiple sclerosis in
Latvians. Biopolymers Cell. 30:305–309. 2014.
|
|
33
|
Mishto M, Bellavista E, Ligorio C,
Textoris-Taube K, Santoro A, Giordano M, D'Alfonso S, Listì F,
Nacmias B, Cellini E, et al: Immunoproteasome LMP2 60HH variant
alters MBP epitope generation and reduces the risk to develop
multiple sclerosis in Italian female population. PLoS One.
5(e9287)2010.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Sjakste T, Paramonova N, Rumba-Rozenfelde
I, Trapina I, Sugoka O and Sjakste N: Juvenile idiopathic arthritis
subtype- and sex-specific associations with genetic variants in the
PSMA6/PSMC6/PSMA3 gene cluster. Pediatr Neonatol. 55:393–403.
2014.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Paramonova N, Wu LS, Rumba-Rozenfelde I,
Wang JY, Sjakste N and Sjakste T: Genetic variants in the PSMA6,
PSMC6 and PSMA3 genes associated with childhood asthma in Latvian
and Taiwanese populations. Biopolymers Cell. 30:377–387. 2014.
|
|
36
|
Paramonova N, Kupca S, Rumba-Rozenfelde I,
Sjakste N and Sjakste T: Association between the PSMB5 and PSMC6
genetic variations and children obesity in the Latvian population.
Biopolymers Cell. 30:477–480. 2014.
|
|
37
|
Sjakste T, Kalis M, Poudziunas I, Pirags
V, Lazdins M, Groop L and Sjakste N: Association of microsatellite
polymorphisms of the human 14q13.2 region with type 2 diabetes
mellitus in Latvian and Finnish populations. Ann Hum Genet.
71:772–776. 2007.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Sjakste T, Paramonova N, Osina K, Dokane
K, Sokolovska J and Sjakste N: Genetic variations in the PSMA3,
PSMA6 and PSMC6 genes are associated with type 1 diabetes in
Latvians and with expression level of number of UPS-related and
T1DM-susceptible genes in HapMap individuals. Mol Genet Genomics.
291:891–903. 2016.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Sjakste T, Poudziunas I, Ninio E, Perret
C, Pirags V, Nicaud V, Lazdins M, Evanss A, Morrison C, Cambien F
and Sjakste N: SNPs of PSMA6 gene-investigation of possible
association with myocardial infarction and type 2 diabetes
mellitus. Genetika. 43:553–559. 2007.PubMed/NCBI
|
|
40
|
Wang H, Jiang M, Zhu H, Chen Q, Gong P,
Lin J, Lu J and Qiu J: Quantitative assessment of the influence of
PSMA6 variant (rs1048990) on coronary artery disease risk. Mol Biol
Rep. 40:1035–1041. 2013.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Sjakste T, Paramonova N, Wu LS, Zemeckiene
Z, Sitkauskiene B, Sakalauskas R, Wang JY and Sjakste N: PSMA6
(rs2277460, rs1048990), PSMC6 (rs2295826, rs2295827) and PSMA3
(rs2348071) genetic diversity in Latvians, lithuanians and
taiwanese. Meta Gene. 2:283–298. 2014.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Rojas-Villarraga A, Amaya-Amaya J,
Rodriguez-Rodriguez A, Mantilla RD and Anaya JM: Introducing
polyautoimmunity: Secondary autoimmune diseases no longer exist.
Autoimmune Dis. 2012(254319)2012.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Holm K, Melum E, Franke A and Karlsen TH:
SNPexp-A web tool for calculating and visualizing correlation
between HapMap genotypes and gene expression levels. BMC
Bioinformatics. 11(600)2010.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Polman CH, Reingold SC, Banwell B, Clanet
M, Cohen JA, Filippi M, Fujihara K, Havrdova E, Hutchinson M,
Kappos L, et al: Diagnostic criteria for multiple sclerosis: 2010
Revisions to the McDonald criteria. Ann Neurol. 69:292–302.
2011.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Stangel M, Penner IK, Kallmann BA, Lukas C
and Kieseier BC: Towards the implementation of ‘no evidence of
disease activity’ in multiple sclerosis treatment: The multiple
sclerosis decision model. Ther Adv Neurol Disord. 8:3–13.
2015.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Lewis CM: Genetic association studies:
Design, analysis and interpretation. Brief Bioinform. 3:146–153.
2002.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Alsmadi O, Muiya P, Khalak H, Al-Saud H,
Meyer BF, Al-Mohanna F, Alshahid M and Dzimiri N: Haplotypes
encompassing the KIAA0391 and PSMA6 gene cluster confer a genetic
link for myocardial infarction and coronary artery disease. Ann Hum
Genet. 73:475–483. 2009.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Andreassen OA, Harbo HF, Wang Y, Thompson
WK, Schork AJ, Mattingsdal M, Zuber V, Bettella F, Ripke S, Kelsoe
JR, et al: Genetic pleiotropy between multiple sclerosis and
schizophrenia but not bipolar disorder: Differential involvement of
immune-related gene loci. Mol Psychiatry. 20:207–214.
2015.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Lee S, Xu L, Shin Y, Gardner L, Hartzes A,
Dohan FC, Raine C, Homayouni R and Levin MC: A potential link
between autoimmunity and neurodegeneration in immune-mediated
neurological disease. J Neuroimmunol. 235:56–69. 2011.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Thompson SD, Barnes MG, Griffin TA, Grom
AA and Glass DN: Heterogeneity in juvenile idiopathic arthritis:
Impact of molecular profiling based on DNA polymorphism and gene
expression patterns. Arthritis Rheum. 62:2611–2615. 2010.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Sivakumaran S, Agakov F, Theodoratou E,
Prendergast JG, Zgaga L, Manolio T, Rudan I, McKeigue P, Wilson JF
and Campbell H: Abundant pleiotropy in human complex diseases and
traits. Am J Hum Genet. 89:607–618. 2011.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Hallstrand TS, Fischer ME, Wurfel MM,
Afari N, Buchwald D and Goldberg J: Genetic pleiotropy between
asthma and obesity in a community-based sample of twins. J Allergy
Clin Immunol. 116:1235–1241. 2005.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Murphy A, Tantisira KG, Soto-Quirós ME,
Avila L, Klanderman BJ, Lake S, Weiss ST and Celedón JC: PRKCA: A
positional candidate gene for body mass index and asthma. Am J Hum
Genet. 85:87–96. 2009.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Lee SH, Lee EB, Shin ES, Lee JE, Cho SH,
Min KU and Park HW: The interaction between allelic variants of
CD86 and CD40LG: A common risk factor of allergic asthma and
rheumatoid arthritis. Allergy Asthma Immunol Res. 6:137–141.
2014.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Ramírez-Bello J, Jiménez-Morales S,
Espinosa-Rosales F, Gómez-Vera J, Gutiérrez A, Velázquez Cruz R,
Baca V and Orozco L: Juvenile rheumatoid arthritis and asthma, but
not childhood-onset systemic lupus erythematosus are associated
with FCRL3 polymorphisms in Mexicans. Mol Immunol. 53:374–378.
2013.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Johnson DA, Amirahmadi S, Ward C, Fabry Z
and Johnson JA: The absence of the pro-antioxidant transcription
factor Nrf2 exacerbates experimental autoimmune encephalomyelitis.
Toxicol Sci. 114:237–246. 2010.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Bekenstein U and Soreq H: Heterogeneous
nuclear ribonucleoprotein A1 in health and neurodegenerative
disease: From structural insights to post-transcriptional
regulatory roles. Mol Cell Neurosci. 56:436–446. 2013.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Leibowitz SM and Yan J: NF-κB pathways in
the pathogenesis of multiple sclerosis and the therapeutic
implications. Front Mol Neurosci. 9(84)2016.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Yue Y, Stone S and Lin W: Role of nuclear
factor κB in multiple sclerosis and experimental autoimmune
encephalomyelitis. Neural Regen Res. 13:1507–1515. 2018.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Wu ZH and Shi Y: When ubiquitin meets
NF-κB: A trove for anti-cancer drug development. Curr Pharm Des.
19:3263–3275. 2013.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Fedorova OA, Moiseeva TN, Nikiforov AA,
Tsimokha AS, Livinskaya VA, Hodson M, Bottrill A, Evteeva IN,
Ermolayeva JB, Kuznetzova IM, et al: Proteomic analysis of the 20S
proteasome (PSMA3)-interacting proteins reveals a functional link
between the proteasome and mRNA metabolism. Biochem Biophys Res
Commun. 416:258–265. 2011.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Wosik K, Antel J, Kuhlmann T, Brück W,
Massie B and Nalbantoglu J: Oligodendrocyte injury in multiple
sclerosis: A role for p53. J Neurochem. 85:635–644. 2003.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Dozmorov M, Wu W, Chakrabarty K, Booth JL,
Hurst RE, Coggeshall KM and Metcalf JP: Gene expression profiling
of human alveolar macrophages infected by B. anthracis spores
demonstrates TNF-alpha and NF-kappab are key components of the
innate immune response to the pathogen. BMC Infect Dis.
9(152)2009.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Gill GN: Decoding the LIM development
code. Trans Am Clin Climatol Assoc. 114:179–189. 2003.PubMed/NCBI
|
|
65
|
Phillips K and Luisi B: The virtuoso of
versatility: POU proteins that flex to fit. J Mol Biol.
302:1023–1039. 2000.PubMed/NCBI View Article : Google Scholar
|
|
66
|
She H and Mao Z: Regulation of myocyte
enhancer factor-2 transcription factors by neurotoxins.
Neurotoxicology. 32:563–566. 2011.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Uzumcu A, Karaman B, Toksoy G, Uyguner ZO,
Candan S, Eris H, Tatli B, Geckinli B, Yuksel A, Kayserili H and
Basaran S: Molecular genetic screening of MBS1 locus on chromosome
13 for microdeletions and exclusion of FGF9, GSH1 and CDX2 as
causative genes in patients with Moebius syndrome. Eur J Med Genet.
52:315–320. 2009.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Ho B, Olson G, Figel S, Gelman I, Cance WG
and Golubovskaya VM: Nanog increases focal adhesion kinase (FAK)
promoter activity and expression and directly binds to FAK protein
to be phosphorylated. J Biol Chem. 287:18656–18673. 2012.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Ozaki K, Sato H, Iida A, Mizuno H,
Nakamura T, Miyamoto Y, Takahashi A, Tsunoda T, Ikegawa S, Kamatani
N, et al: A functional SNP in PSMA6 confers risk of myocardial
infarction in the Japanese population. Nat Genet. 38:921–925.
2006.PubMed/NCBI View
Article : Google Scholar
|
|
70
|
Caputi FF, Carboni L, Mazza D, Candeletti
S and Romualdi P: Cocaine and ethanol target 26S proteasome
activity and gene expression in neuroblastoma cells. Drug Alcohol
Depend. 161:265–275. 2016.PubMed/NCBI View Article : Google Scholar
|
|
71
|
Bedford L, Lowe J, Dick LR, Mayer RJ and
Brownell JE: Ubiquitin-like protein conjugation and the
ubiquitin-proteasome system as drug targets. Nat Rev Drug Discov.
10:29–46. 2011.PubMed/NCBI View Article : Google Scholar
|
|
72
|
Torkildsen Ø, Myhr KM and Bø L:
Disease-modifying treatments for multiple sclerosis-a review of
approved medications. Eur J Neurol. 23 (Suppl 1):S18–S27.
2016.PubMed/NCBI View Article : Google Scholar
|