Introduction
Gastric cancer (GC) is one of the commonest
malignant tumors of the digestive system, ranking fifth in
incidence and third in mortality worldwide (1). Cell invasion and metastasis are
considered as important causes leading to postoperative recurrence
and mortality in patients with GC. Therefore, the early
identification of factors involved in cell invasion and metastasis
could improve the cure rate and prolong the survival rate of
patients with GC (2). Among tens
of thousands genes in the tumor tissues, genes associated with
tumor development are called driver genes (3). Changes in the expression of driver
genes may result in changes in the incidence of tumors. Therefore,
selecting the appropriate molecular targeted drugs for each driving
gene could improve personalized therapy for the recovery of
patients with GC.
NOD like receptor (NLR) family, CARD domain
containing 5 (NLRC5) is a member of the NLR family. It has been
reported that the short-term effects of NLRC5 are associated with
immune responses and inflammation, while the long-term ones may
lead to the dysregulation of the immune system (4). The above findings indicate that NLRC5
could be involved in the development of cancer. A previous study
demonstrated that NLRC5 is upregulated and promoted cell
proliferation, migration and invasion in clear cell renal carcinoma
by activating the Wnt/catenin signaling pathway (5). Additionally, NLRC5 can promote cell
proliferation in hepatocellular carcinoma by regulating the
AKT/VEGF-A signaling pathway (6).
NLRC5 is upregulated in GC tissues, while increased NLRC5
expression is associated with worse prognosis (7). However, the specific regulatory
mechanism of NLRC5 in GC has not been previously reported. GC is
characterized by strong heterogeneity, poor sensitivity to
chemotherapy and poor prognosis (8). Therefore, GC is considered as a tumor
with high requirements for individualized therapy (9). A previous study suggests that
regulatory NLRC5 variation can affect the survival of patients with
colorectal cancer and their response to 5-fluorouracil (5-FU)
chemotherapy (10). Therefore, the
effect of NLRC5 regulation on the sensitivity of GC cells to
chemotherapy remains to be elucidated.
Yin Yang 1 (YY1) is a transcriptional protein
involved in a variety of biological functions, including cell cycle
progression, cell proliferation, differentiation and apoptosis
(11). A number of studies have
shown that abnormal expression of YY1 can serve a regulatory role
in tumor proliferation and metastasis through interaction with
different protein cofactors (12,13).
YY1 is upregulated in GC cell lines and primary GC (14). Moreover, YY1 has extensive
carcinogenic effect in GC (15,16).
YY1 is predicted to start NLRC5 by ALGGEN-PROMO, and YY1 predicted
to bind to NLRC5 promoter by JASPAR database. Therefore, it was
hypothesized that NLRC5 could be activated by YY1 transcription
factor to participate in the malignant process of GC cells.
The present study aimed to investigate the
regulation and underlying mechanism of NLRC5 on the proliferation,
invasion and migration of GC cells and their sensitivity to
chemotherapy, thus providing a new theoretical basis for the
targeted therapy of GC.
Materials and methods
Databases
The ALGGEN-PROMO (17) and JASPAR databases (18) (hppt://jaspar.genereg.net) were used to predict the
binding capacity of YY1 on NLRC5 promoter.
Cell culture
The human gastric mucosa cell line GES-1 and the GC
cell lines AGS, MKN-45, KATO III and NCI-87, procured from Bena
Culture Collection, were cultured in DMEM supplemented with 10%
FBS, 1% penicillin and 1% streptomycin (all from Gibco; Thermo
Fisher Scientific, Inc.) at 37˚C and 5% CO2. Cells were
treated with 5 µg/ml 5-FU (Beyotime Institute of Biotechnology) at
37˚C for 48 h.
Cell viability assay
A Cell Counting Kit 8 (CCK-8; Nanjing Jiancheng
Bioengineering Institute) assay was used to evaluate cell
viability. Briefly, cells were seeded into a 96-well plate at a
density of 1x104 cells/well. Following treatment with
the appropriate compounds, cells in each well were supplemented
with 10 µl CCK-8 solution and incubated at 37˚C for an additional 4
h. Finally, the absorbance at a wavelength of 450 nm was measured
in each well using a microplate spectrophotometer reader (BioTek
Instruments, Inc.).
Reverse transcription-quantitative
(RT-q)PCR
Total RNA was extracted from cells
(1.5x106) using a TRIzol® reagent (Thermo
Fisher Scientific, Inc.) according to the manufacturer's protocols.
Subsequently, RNA was reverse transcribed into cDNA using the
PrimeScript RT Reagent kit (Takara Bio, Inc.) according to the
manufacturer's protocols. The expression levels of the target gene
were quantified with real-time PCR using the SYBR-Green PCR Master
Mix (Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's instructions. qPCR was performed on the Applied
Biosystems real time PCR system (Thermo Fisher Scientific, Inc.)
and the gene expression levels were quantified using the
2-ΔΔCq method (19).
The following thermocycling conditions for qPCR were used: Initial
denaturation at 95˚C for 5 min; 40 cycles of denaturation at 95˚C
for 10 sec, and annealing and extension at 60˚C for 45 sec. The
following primers were used: NLRC5 forward,
5'-TGAGGGAGTCTGCACTATGGA-3' and reverse,
5'-TCCGATTCAGGGCTCAGGTA-3'; YY1 forward, 5'-TCAGACAAGTCACGTCAGGC-3'
and reverse, 5'-CTCCATGTGTCACCTCCCAC-3'; and GAPDH forward,
5'-CGGAGTCAACGGATTTGGTCGTAT-3' and reverse,
5'-AGCCTTCTCCATGGTGGTGAAGAC-3'.
Western blot analysis
AGS cells were lysed using a protein extraction
solution (Beyotime Institute of Biotechnology) and the protein
concentration was measured using a BCA kit (SinoBio Biotech).
Proteins (50 µg) were uploaded and were then transferred onto a
PVDF membrane (MilliporeSigma). Following blocking with 5% skimmed
milk powder in Tris-buffered saline containing 0.05% Tween-20
(TBST) at room temperature for 1 h, the membrane was incubated with
primary antibodies (dilution, 1:1,000) at 4˚C overnight.
Subsequently, the membrane was incubated with the corresponding
HRP-conjugated secondary antibodies (dilution, 1:5,000; Abcam) at
37˚C for 1 h. The protein bands were visualized using an ECL
reagent (ECL-plus; Thermo Fisher Scientific, Inc.) and analyzed
using ImageJ software (version 1.8.0; National Institutes of
Health). Primary antibodies used in this study were as follows:
NLRC5 (cat. no. GTX85160; GeneTex, Inc.), YY1 (cat. no. ab109237;
Abcam), MMP2 (cat. no. ab92536; Abcam), MMP9 (cat. no. ab76003;
Abcam), cleaved caspase 3 (cat. no. ab32042; Abcam), p53 (cat. no.
ab32389; Abcam) and GAPDH (cat. no. ab9485; Abcam).
Cell transfection
Small interfering (si)RNAs targeting NLRC5
(si-NLRC5-1 and si-NLRC5-2) and YY1 (si-YY1), their corresponding
blank controls (si-NC), YY1 overexpression (ov) plasmid (ov-YY1)
and ov-NC were synthesized by Guangzhou RiboBio Co., Ltd. Cell
transfection was performed using Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) at 37˚C for 24 h
according to the manufacturer's instructions. siRNA-NLRC5-1-sense:
5'-AAGAACGAGAGACUCUGCCAACUGCdTdT-3', siRNA-NLRC5-1-antisense:
5'-GCAGUUGGCAGAGUCUCUCGUUCUUdTdT-3', siRNA-NLRC5-2-sense:
5'-GGGACTGAGAGCTTTGTAT-3', siRNA-NLRC5-2-antisense:
5'-CGCACCCTAGACTGAAA-3', scrambled-RNAi-sense:
5'-UUCUCCGAACGUGUCACGUTT-3', and scrambled-RNAi-antisense:
5'-ACGUGACACGUUCGGAGAATT-3'. At 48 h post-transfection, subsequent
experiments were conducted.
Experimental groups
To determine the impact of NLRC5 on the sensitivity
of GC cells to 5-FU, cells were divided into the control, 5-FU,
5-FU + si-NC and 5-FU + si-NLRC5 groups. To substantiate the impact
of the YY1/NLRC5 axis on the sensitivity of GC cells to 5-FU, GC
cells were divided into the following five groups: The control
group, the ov-NC group, the ov-YY1 group, the ov-YY1 + si-NC group
and the ov-YY1 + si-NLRC5 group.
Colony formation assay
Cells were seeded into 6-well plates at a density of
103 cells/well and incubated at 37˚C in humidified 5%
CO2 incubator for 15 days. Subsequently, the formed
colonies (>50 cells/colony) were stained with 0.5% crystal
violet for 30 min at room temperature. Images were captured and the
number of the colonies (>50 cells/colony) in three fields of
view was counted with the naked eye under a light microscope
(magnification, x10).
Wound healing assay
For wound healing assays, AGS cells were seeded into
a 6-well plate at a density of 105 cells/well overnight
at 37˚C. When cells grown in complete medium reached 75%
confluency, a wound was created using a sterile pipette tip.
Subsequently, cells were washed with PBS several times to remove
cell debris and incubated at 37˚C for an additional 48 h in
serum-free medium. Images of the wound were captured under an
inverted fluorescence microscope and the wound closure rate was
then assessed.
Transwell assay
Transwell chambers (Corning Life Sciences) with 8-µm
pore inserts coated with Matrigel at 37˚C for 30 min were used to
evaluate the invasion ability of AGS cells. Briefly, transfected
cells with the presence or absence of 5-FU treatment in serum-free
DMEM were added to the upper chamber of the Transwell insert at a
density of 5x105 cells/ml. The lower chamber was
supplemented with 500 µl complete medium as chemoattractant.
Following incubation for 24 h at 37˚C, cells on the upper surface
were removed, while cells invaded onto the bottom of the membrane
were fixed and stained with 0.1% crystal violet for 10 min at room
temperature. The invaded cells were counted in five randomly
selected fields under a light microscope (Olympus Corporation).
Flow cytometric assay
Cell apoptosis was assessed using an Annexin V-FITC
apoptosis detection kit (Thermo Fisher Scientific, Inc.). Briefly,
2x105 cells were re-suspended in 300 µl binding buffer,
mixed gently with 5 µl Annexin V-FITC reagent at 4˚C for 15 min in
the dark prior to 10 µl PI staining fluid being added at 4˚C for 5
min. Finally, cell apoptosis was assessed using a flow cytometer
(FACSAria™; BD Biosciences) and FlowJo software (version 10.0.7;
Tree Star, Inc.). The total apoptosis rate is equal to the early
apoptotic rate plus the dead cell rate.
Chromatin immunoprecipitation
(ChIP)
The ChIP assay was performed using the Imprint ChIP
kit (Sigma-Aldrich; Merck KGaA) according to the manufacturer's
protocol. A total of 1x107 cells were cross-linked with
1% formaldehyde for 10 min at room temperature. The cell lysates
were sonicated using a 10 sec on and 10 sec off mode for 12 cycles,
on ice, to obtain chromatin fragments. DNA (8-40 µg) was diluted
with DNase-free water (Beyotime Institute of Biotechnology) and
incubated with the primary antibody at 4˚C overnight. The primary
antibody used was an anti-YY1 antibody (1:200, cat. no. ab109237;
Abcam). Next, the DNA that had bound to YY1 was collected using DNA
extraction buffer of the kit in DNase-free water and amplified
using qPCR to detect NLRC5. PCR products were separated by 1% gel
electrophoresis using agarose gels prestained with ethidium
bromide. Bands were analyzed using ImageJ software (version 1.8.0;
National Institutes of Health).
Luciferase reporter assay
Cell were plated in 6-well plates and after the
cells had adhered, 0.5 µg vectors containing the 3'-untranslated
region (UTR) of wild-type NLRC5 or mutant 3'-UTR NLRC5, with
control vector or YY1 overexpression vector and pMIR-Renilla
vector (Shanghai GeneChem Co., Ltd) were co-transfected with the
transfection kit (Polybrene; Shanghai GeneChem, Co., Ltd.) into the
cells (1x106 cells/well) and cells were incubated for 48
h at 37˚C. Cells were collected 24 h after transfection. Finally,
the luciferase activity was detected using a Renilla-Glo
Luciferase Assay System (cat. no. E2710; Promega Corporation) at
room temperature and a spectrophotometer at 490 nm (Thermo Fisher
Scientific, Inc.). Renilla luciferase activity was used to
normalize the firefly luciferase activity.
Statistical analysis
Data are expressed as the mean ± SD. All results
were analyzed using SPSS 18.0 software (SPSS, Inc.). The
differences in the present study except for the CCK8 results among
multiple groups were compared with one-way ANOVA followed by
Tukey's post hoc test. CCK8 results were analyzed by Two-way ANOVA
followed by Tukey's post hoc test. Each experiment was performed at
least three times. P<0.05 was considered to indicate a
statistically significant difference.
Results
NLRC5 knockdown attenuates the
proliferation, invasion and migration of GC cells
The expression levels of NLRC5 in GC cells were
detected by RT-qPCR and western blot analysis. The results showed
that NLRC5 was significantly upregulated in AGS, MKN-45, KATO III
and NCI-87 cells compared with GES-1 cells (Fig. 1A and B). The expression of NLRC5 was notably
higher in AGS cells compared with the other GC cell lines.
Therefore, AGS cells were selected for the follow-up experiments.
Subsequently, GC cells were transfected with si-NLRC5-1 or
si-NLRC5-2 clones using the cell transfection technology. The
silencing activity of si-NLRC5-1 was more potent compared with that
of si-NLRC5-2 and it was therefore selected for subsequent
experiments (Fig. 1C and D). Furthermore, cells were divided into
the control, si-NC and si-NLRC5 groups. CCK-8 assays revealed that
compared with the si-NC group, the cell viability was significantly
decreased in the si-NLRC5 group in a time-dependent manner
(Fig. 1E). Additionally, the
colony formation assay showed that the proliferation ability of GC
cells was notably reduced in the si-NLRC5 group (Fig. 1F). Furthermore, wound healing and
Transwell assays demonstrated that the invasion and migration
abilities of GC cells were significantly attenuated in the si-NLRC5
group compared with the si-NC group (Fig. 2A and B). Finally, western blot analysis
revealed that the expression levels of the migration-related
proteins MMP2 and MMP9 were markedly reduced in NLRC5-depleted GC
cells (Fig. 2C).
NLRC5 silencing enhances the
sensitivity of GC cells to 5-FU
The aforementioned findings indicated that NLRC5
silencing could inhibit the proliferation, invasion and migration
of GC cells. However, whether NLRC5 is associated with the
prognosis of GC remains elusive. Therefore, 5-FU, a commonly used
postoperative chemotherapy drug, was selected to assess whether
NLRC5 knockdown could enhance the sensitivity of GC cells to
chemotherapy. It was found that the expression of NLRC5 in AGS
cells with 5-Fu resistance was decreased compared with that in AGS
cells (Fig. 3A). Cells were
divided into the control, 5-FU, 5-FU + si-NC and 5-FU + si-NLRC5
groups. Cell apoptosis was assessed by flow cytometry and the
results showed that 5-FU promoted AGS cell apoptosis. Accordingly,
cell treatment with 5-FU notably upregulated the expression of
cleaved caspase 3 and p53. However, cell apoptosis and the
expression levels of cleaved caspase 3 and p53 were further
increased in the 5-FU + si-NLRC5 group, compared with the 5-FU +
si-NC group (Fig. 3B and C). The above results suggested that NLRC5
silencing could increase the sensitivity of GC cells to 5-FU.
YY1 binds to NLRC5 promoter
Bioinformatics analysis using the JASPAR database
predicted that YY1 could bind to NLRC5 promoter (Fig. 4A). The above finding was verified
by overexpression and silencing experiments. Therefore, GC cells
were transfected with YY1 overexpression or silencing constructs
and the transfection efficiency was then evaluated. The mRNA and
protein expression levels of YY1 in YY1 overexpressing or depleted
GC cells were assessed by RT-qPCR and western blot analysis,
respectively (Fig. 4B and C). The results showed that YY1
overexpression in GC cells significantly upregulated NLRC5. By
contrast, YY1 knockdown notably inhibited NLRC5 expression
(Fig. 4D and E). These findings indicated that YY1
could regulate NLRC5 expression. Furthermore, luciferase reporter
assay was carried out to measure NLRC5 promoter activity in cells
overexpressing YY1. The results demonstrated that NLRC5 promoter
activity was significantly enhanced in GC cells transfected with
YY1 overexpression plasmid (Fig.
4F). Accordingly, the binding capacity of YY1 on NLRC5 promoter
was further verified by chromatin immunoprecipitation assay
(Fig. 4G).
NLRC5 knockdown inhibits the promotive
effect of YY1 on GC cell proliferation, invasion and migration
To further investigate the regulatory mechanism of
NLRC5 on GC cell invasion, migration and sensitivity to 5-FU
chemotherapy, GC cells were divided into the following five groups:
The control group; the ov-NC group; the ov-YY1 group; the ov-YY1 +
si-NC group; and the ov-YY1 + si-NLRC5 group. CCK-8 and colony
formation assays showed that the proliferation ability of GC cells
in the ov-YY1 group was significantly increased compared with that
in the ov-NC group. Additionally, compared with the ov-YY1 + si-NC
group, the cell proliferation ability was notably decreased in the
ov-YY1 + si-NLRC5 group (Fig. 5A
and B). Furthermore, wound healing
and Transwell assays revealed that compared with the ov-NC group,
the migration and invasion abilities of GC cells were markedly
enhanced in the ov-YY1 group. However, the cell migration and
invasion was significantly reduced in the ov-YY1 + si-NLRC5 group
compared with the ov-YY1 + si-NC group (Fig. 5C and D). Finally, western blotting results also
showed that MMP2 and MMP9 were notably upregulated in YY1
overexpressing cells, while the expression levels of both molecules
were restored in the ov-YY1 + si-NLRC5 group compared with the
ov-YY1 + si-NC group (Fig.
5E).
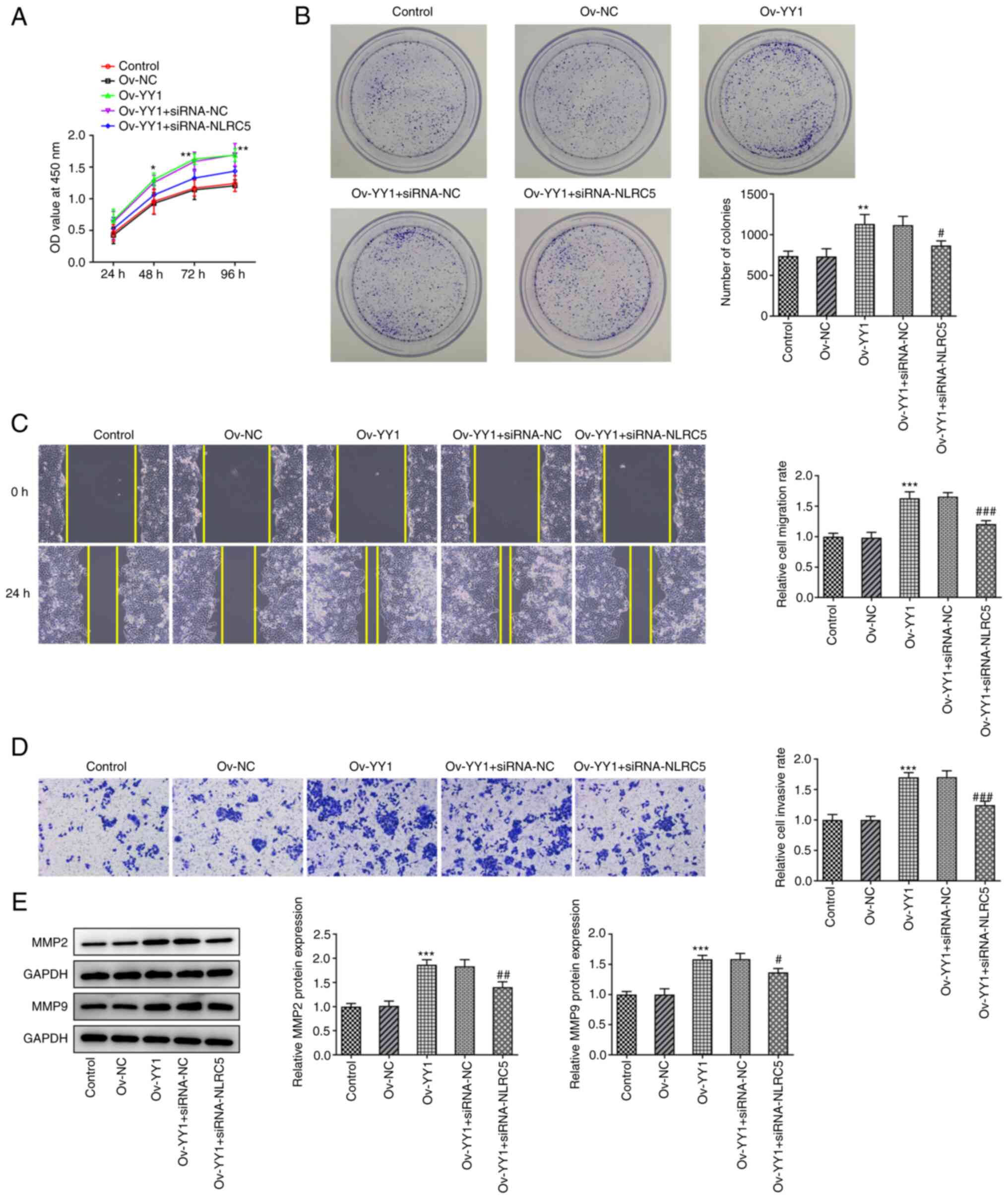 | Figure 5Inhibition of NLRC5 blocks the
promotion of YY1 on proliferation, invasion and migration. (A)
CCK-8 detected the cell viability. (B) Colony formation assay was
used to detected the cell proliferation. (C) Wound Healing detected
the ability of cell migration (magnification, x100). (D) Transwell
detected the ability of cell invasion (magnification, x100). (E)
Western blotting detected the expression of MMP2 and MMP9.
*P<0.05, **P<0.01,
***P<0.001 vs. Ov-NC. #P<0.05,
##P<0.01, ###P<0.001 vs.
Ov-YY1+siRNA-NC. NLRC5, NOD like receptor family, CARD domain
containing 5; YY1, Yin Yang 1; Ov, overexpression; NC, negative
control; si, short interfering. |
NLRC5 silencing attenuates the
enhancing effect of YY1 on the sensitivity of GC cells to 5-FU
It was then found that the expression of YY1 in AGS
cells with 5-Fu resistance was decreased compared with that in AGS
cells (Fig. 6A). Subsequently,
cell apoptosis was assessed by flow cytometry and western blot
analysis. The results showed that AGS cell apoptosis and the
protein expression levels of cleaved caspase 3 and p53 were notably
increased following cell exposure to 5-FU. However, following YY1
overexpression, cell apoptosis and the expression levels of cleaved
caspase 3 and p53 were markedly reduced compared with the 5-FU +
ov-NC group. Additionally, compared with the 5-FU + ov-YY1 + si-NC
group, cell apoptosis and the expression of both apoptosis-related
proteins were significantly enhanced in the 5-FU + ov-YY1 +
si-NLRC5 group (Fig. 6B and
C). These findings suggested that
NLRC5 knockdown abrogated the enhancing effect of YY1 on the
sensitivity of GC cells to 5-FU.
Discussion
GC is a common tumor of the gastrointestinal system.
Although the pathogenesis of GC has not been fully elucidated, its
occurrence and development is associated with a series of molecular
changes, including the activation of several major signal pathways,
mutations, and abnormal expression and regulation of related genes
(20,21). The present study revealed that
NLRC5 expression was increased in several GC cell lines. A previous
study showed that NLRC5 is significantly upregulated in cells
infected with Helicobacter pylori (22). In addition, another study
demonstrated that NLRC5 is upregulated in mucosal organs such as
the stomach (23). These findings
were consistent with the results of the current study demonstrating
that the expression levels of NLRC5 were significantly elevated in
the GC cell lines AGS, MKN-45, KATO III and NCL-87.
It has been previously reported that NLRC5 regulates
the proliferation, invasion and migration of tumor cells (5,6).
Therefore, NLRC5 knockdown can significantly inhibit the malignant
biological behavior of glioma cells by attenuating the activation
of the Wnt/β-catenin signaling pathway (24). In endometrial cancer, NLRC5 can
promote the migration and invasion of endometrial cancer cell by
activating the PI3K/AKT signaling pathway (25). Another study revealed that the
increased expression levels of NLRC5 in GC tissues are associated
with worse prognosis (7). However,
the effect of NLRC5 on regulating the proliferation, invasion and
migration of GC cells has not been previously investigated.
Therefore, the results of the present study demonstrated that NLRC5
knockdown in the GC cell line AGS markedly attenuated the
proliferation, migration and invasion of these cells.
Gene heterogeneity, differences in protein
expression levels and the clinicopathological characteristics of
patients with GC may result in huge differences in the response of
patients to chemotherapeutic and targeted therapy drugs. Therefore,
a previous study revealed that NLRC5 was a poor prognostic
indicator in patients with non-small cell lung cancer (26). Additionally, increased NLRC5
expression was associated with advanced stage and poor prognosis in
patients with renal clear cell carcinoma (5). However, the effect of NLRC5 on the
prognosis of GC has not been previously reported. 5-FU combined
with platinum is the recommended first-line drug regimen for
postoperative systemic chemotherapy, with an effective rate of
30-50% (27). Therefore, 5-FU was
selected to explore whether NLRC5 silencing could enhance the
sensitivity of AGS cells to chemotherapy. The results showed that
NLRC5 knockdown enhanced the sensitivity of GC cells to 5-FU and
upregulated the expression of apoptosis-related proteins.
To further investigate the regulatory mechanism of
NLRC5 in GC cell proliferation, invasion, migration and sensitivity
to chemotherapeutic drugs, the binding capacity of YY1 on NLRC5
promoter was evaluated. Bioinformatics analysis using the
ALGGEN-PROMO and JASPAR databases predicted that YY1 could activate
NLRC5 by binding to its promoter region. This finding was
consistent with the results obtained by Guo et al (28). YY1 serves a significant role in the
occurrence and development of GC. A previous study demonstrated
that YY1 can regulate the coiled-coil domain containing 43/adhesion
regulating molecule 1 axis to promote the proliferation and
metastasis of GC cells (15).
Zhang et al (16) also
found that YY1 is significantly upregulated in tumor tissues and
serum from patients with GC. Therefore, it was hypothesized that
NLRC5 could be activated by the transcription factor YY1 to
regulate the proliferation, invasion and migration of GC cells. The
role of YY1 in the above processes and sensitivity of GC cells to
5-FU chemotherapy was therefore explored. The results showed that
NLRC5 knockdown could inhibit the effects of YY1 on promoting GC
cell proliferation, invasion and migration. Additionally, NLRC5
silencing attenuated the effect of YY1 on enhancing the sensitivity
of GC cells to 5-FU.
The present study has some limitations. First it
only discussed cell experiments, which were not verified in animal
experiments or other cell lines and the expression of NLRC5 in GC
patients cannot be detected. The experimental results of the
present study will be further verified in animal experiments and
clinical experiments in the future. Moreover, the present study
discussed only one of the mechanisms by which NLRC5 regulates the
malignant progression of gastric cancer cells. NLRC5 can also serve
a role in regulating the malignant progression of gastric cancer by
regulating other signaling pathways or cytokines. Future
experiments will also explore the regulatory mechanism of NLRC5 in
gastric cancer from more perspectives.
In brief, the present study indicated that NLRC5
knockdown could reduce the malignant growth and enhance the
sensitivity of GC cells to 5-FU chemotherapy by inhibiting the
carcinogenic effect of YY1. These findings could provide a
theoretical basis for the treatment of GC via enhancing the
sensitivity of GC cells to chemotherapeutic drugs.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by the Chongqing
Natural Science Foundation of China (grant no.
cstc2019jcyj-msxmX0543).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
SLia performed the experiments. TX and SLiu analyzed
the data. WX designed the experiments, interpreted the data and
wrote the manuscript. SLiu and WX confirm the authenticity of all
the raw data. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Correa P: Gastric cancer: Overview.
Gastroenterol Clin North Am. 42:211–217. 2013.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Liu LP, Sheng XP, Shuai TK, Zhao YX, Li B
and Li YM: Helicobacter pylori promotes invasion and metastasis of
gastric cancer by enhancing heparanase expression. World J
Gastroenterol. 27:3138–3141. 2021.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Martinez-Jimenez F, Muinos F, Sentis I,
Deu-Pons J, Reyes-Salazar I, Arnedo-Pac C, Mularoni L, Pich O,
Bonet J, Kranas H, et al: A compendium of mutational cancer driver
genes. Nat Rev Cancer. 20:555–572. 2020.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Tang F, Xu Y and Zhao B: NLRC5: New cancer
buster? Mol Biol Rep. 47:2265–2277. 2020.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Wang Q, Ding H, He Y, Li X, Cheng Y, Xu Q,
Yang Y, Liao G, Meng X, Huang C and Li J: NLRC5 mediates cell
proliferation, migration, and invasion by regulating the
Wnt/β-catenin signalling pathway in clear cell renal cell
carcinoma. Cancer Lett. 444:9–19. 2019.PubMed/NCBI View Article : Google Scholar
|
|
6
|
He YH, Li MF, Zhang XY, Meng XM, Huang C
and Li J: NLRC5 promotes cell proliferation via regulating the
AKT/VEGF-A signaling pathway in hepatocellular carcinoma.
Toxicology. 359-360:47–57. 2016.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Li Y, Zhang M and Zheng X: High expression
of NLRC5 is associated with prognosis of gastric cancer. Open Med
(Wars). 13:443–449. 2018.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Oh SC, Sohn BH, Cheong JH, Kim SB, Lee JE,
Park KC, Lee SH, Park JL, Park YY, Lee HS, et al: Clinical and
genomic landscape of gastric cancer with a mesenchymal phenotype.
Nat Commun. 9(1777)2018.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Digklia A and Wagner AD: Advanced gastric
cancer: Current treatment landscape and future perspectives. World
J Gastroenterol. 22:2403–2414. 2016.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Catalano C, da Silva Filho MI, Jiraskova
K, Vymetalkova V, Levy M, Liska V, Vycital O, Naccarati A,
Vodickova L, Hemminki K, et al: Short article: Influence of
regulatory NLRC5 variants on colorectal cancer survival and
5-fluorouracil-based chemotherapy. Eur J Gastroenterol Hepatol.
30:838–842. 2018.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Naidoo K, Clay V, Hoyland JA, Swindell R,
Linton K, Illidge T, Radford JA and Byers RJ: YY1 expression
predicts favourable outcome in follicular lymphoma. J Clin Pathol.
64:125–129. 2011.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Kim H, Bang S, Jee S, Park S, Kim Y, Park
H, Jang K and Paik SS: Loss of YY1 expression predicts unfavorable
prognosis in stage III colorectal cancer. Indian J Pathol
Microbiol. 64 (Supplement):S78–S84. 2021.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Xia W, Li Y, Wu Z, Wang Y, Xing N, Yang W
and Wu S: Transcription factor YY1 mediates epithelial-mesenchymal
transition through the TGFβ signaling pathway in bladder cancer.
Med Oncol. 37(93)2020.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Kang W, Tong JH, Chan AW, Zhao J, Dong Y,
Wang S, Yang W, Sin FM, Ng SS, Yu J, et al: Yin Yang 1 contributes
to gastric carcinogenesis and its nuclear expression correlates
with shorter survival in patients with early stage gastric
adenocarcinoma. J Transl Med. 12(80)2014.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Wang J, Wu X, Dai W, Li J, Xiang L, Tang
W, Lin J, Zhang W, Liu G, Yang Q, et al: The CCDC43-ADRM1 axis
regulated by YY1, promotes proliferation and metastasis of gastric
cancer. Cancer Lett. 482:90–101. 2020.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Zhang L, Zou L and Sun P: Relationship
between miR-378c and YY1 expression in patients with gastric cancer
and the clinicopathological features. Cell Mol Biol Lett.
26(12)2021.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Messeguer X, Escudero R, Farre D, Nunez O,
Martinez J and Alba MM: PROMO: Detection of known transcription
regulatory elements using species-tailored searches.
Bioinformatics. 18:333–334. 2002.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Fornes O, Castro-Mondragon JA, Khan A, van
der Lee R, Zhang X, Richmond PA, Modi BP, Correard S, Gheorghe M,
Baranašić D, et al: JASPAR 2020: Update of the open-access database
of transcription factor binding profiles. Nucleic Acids Res.
48:D87–D92. 2020.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
20
|
El-Rifai W and Powell SM: Molecular
biology of gastric cancer. Semin Radiat Oncol. 12:128–140.
2002.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Frycz BA, Murawa D, Borejsza-Wysocki M,
Wichtowski M, Spychala A, Marciniak R, Murawa P, Drews M and
Jagodziński PP: mRNA expression of steroidogenic enzymes, steroid
hormone receptors and their coregulators in gastric cancer. Oncol
Lett. 13:3369–3378. 2017.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Yang T, Wang R, Zhang J, Bao C, Zhang J,
Li R, Chen X, Wu S, Wen J, Wei S, et al: Mechanism of berberine in
treating Helicobacter pylori induced chronic atrophic gastritis
through IRF8-IFN-ү signaling axis suppressing. Life Sci.
248(117456)2020.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Yang QY, Chen T, Chen YB and Lan DL:
Molecular characterization and expression analysis of the NLR
family CARD containing five transcripts in the pig. Pol J Vet Sci.
19:753–761. 2016.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Zong Z, Song Y, Xue Y, Ruan X, Liu X, Yang
C, Zheng J, Cao S, Li Z and Liu Y: Knockdown of LncRNA SCAMP1
suppressed malignant biological behaviours of glioma cells via
modulating miR-499a-5p/LMX1A/NLRC5 pathway. J Cell Mol Med.
23:5048–5062. 2019.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Fan Y, Dong Z, Shi Y, Sun S, Wei B and
Zhan L: NLRC5 promotes cell migration and invasion by activating
the PI3K/AKT signaling pathway in endometrial cancer. J Int Med
Res. 48(300060520925352)2020.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Li X, Guo F, Liu Y, Chen HJ, Wen F, Zou B,
Li D, Qin Q, Liu X, Shen Y and Wang Y: NLRC5 expression in tumors
and its role as a negative prognostic indicator in stage III
non-small-cell lung cancer patients. Oncol Lett. 10:1533–1540.
2015.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Ito S, Sano T, Mizusawa J, Takahari D,
Katayama H, Katai H, Kawashima Y, Kinoshita T, Terashima M,
Nashimoto A, et al: A phase II study of preoperative chemotherapy
with docetaxel, cisplatin, and S-1 followed by gastrectomy with D2
plus para-aortic lymph node dissection for gastric cancer with
extensive lymph node metastasis: JCOG1002. Gastric Cancer.
20:322–331. 2017.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Guo XM, Liu XP, Chang GB, Xu L, Bi YL,
Wang HZ, Zhang Y, Zhu PF, Wu Y and Chen GH: Characterization of the
NLRC5 promoter in chicken: SNPs, regulatory elements and CpG
islands. Anim Genet. 47:579–587. 2016.PubMed/NCBI View Article : Google Scholar
|




















