Introduction
As one of the most prevalent primary malignant bone
tumors, osteosarcoma (OS) commonly occurs in adolescents, children
and elderly individuals (>60 years old), and is characterized by
rapid lung metastasis (1,2). It was reported that the 5-year
survival rate of patients with OS without lung metastasis is
#x003C;70%, while the survival rate reduces significantly to
#x003C;30% with lung metastasis (3,4). At
present, the first-line treatment strategy for OS is surgical
excision combined with chemotherapy. However, it becomes
increasingly more challenging to manage OS due to the development
of chemotherapy resistance and tumor relapse (5-7).
Therefore, it is crucial to explore the molecular mechanism that
governs lung metastasis, relapse and drug resistance of OS. Recent
studies found that autophagy and ferroptosis are involved in the
progression and chemotherapy resistance of OS (8-11).
Substantial evidence demonstrated that epigenetics,
such as RNA methylation, play vital role in the pathogenesis of
various cancers (12-14).
Based on methylation sites, RNA methylation can be divided into
several types, such as N6-methyladenosine (m6A), 5-methylcytidine
(m5C), N1-methyladenosine, 2'-O-methylation,
3-methylcytidine, 1-methylguanosine, N2-methylguanosine,
7-methylguanosine, 2-methylthio-N6-isopentenyl-adenosine and
2-methylthio-N6-threonylcarbamoyl-adenosine (15,16).
Particularly, m5C widely exists in tRNA, mRNA and long
non-coding (lnc)RNA, and mainly impacts the progression of diseases
by regulating the export, stability and translation efficiency of
mRNA (17-20).
Like other RNA methylation types, m5C is dynamic and
catalyzed by a methyltransferase (writer), removed by demethylase
(eraser) and recognized by m5C-binding proteins
(reader). NOP2/Sun RNA methyltransferase family members 2 and 6
(NSUN2 and NSUN6, respectively) were suggested to be the most
important methyltransferases (writer) to catalyze m5C
(21,22). Previous studies reported that NSUN2
and NSUN6 could regulate the expression of mRNA, tRNA and lncRNA in
an m5C-dependent manner, thus affecting the progression
of nervous system diseases and different cancers, including
hepatocellular carcinoma, bladder cancer, gallbladder carcinoma,
and gastric cancer (23-32).
M6A, catalyzed by methyltransferase like 3 (METTL3)
and Wilms tumor 1 associated protein (WTAP), were shown to
contribute to the progression of OS (2,33).
Additionally, METTL3 could promote the progression of OS by
improving the stability of differentiation antagonizing non-protein
coding RNA (34) and histone
deacetylase 5 mRNAs (35), while
WTAP could trigger OS tumorigenesis by downregulating HMBOX1
expression (3). Nonetheless, the
role of m5C in the pathogenesis of OS remains unclear. A
recent study performed by the current authors demonstrated the
impact of epigenetics on OS progression (36). The current study, through
bioinformatics analysis, in vitro and in vivo
experiments, found that NSUN6 mainly regulates OS progression by
regulating the stability of eukaryotic elongation factor 1 α-2
(EEF1A2) mRNA in an m5C-dependent manner, which is known
to induce OS progression by activating Akt/mTOR signaling pathway
(37).
Materials and methods
Bioinformatics analysis
The present study analyzed the datasets for
GSE126209 downloaded from the Gene Expression Omnibus (GEO)
database (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE126209),
as well as the transcriptome data and the corresponding survival
information contained in TARGET-OS from UCSC-XENA (http://xena.ucsc.edu/). Gencode platform (https://www.gencodegenes.org/) was used and
transcripts per kilobase million (TPM) normalization was performed
in TARGET-OS. The data were analyzed by the software R 3.6.2
(https://www.r-project.org/).
Cell culture and cell
transfection
In this study, human OS cell lines 143b, MG63, HOS,
U2-OS, SAOS2 and bone mesenchymal stem cells (BMSCs) were cultured
at 37˚C in a 5% CO2 incubator (Thermo Fisher Scientific,
Inc.). All cell lines were obtained from the China Centre for Type
Culture Collection. The 143b cells were cultured in RPMI-1640
(Gibco; Thermo Fisher Scientific, Inc.; cat. no. 11875119), the
MG63 cells were cultured in DMEM (Gibco; Thermo Fisher Scientific,
Inc.; cat. no. 12430047), the U2-OS and SAOS2 cells were cultured
in McCoy's 5A (Gibco; Thermo Fisher Scientific, Inc.; cat. no.
12330031), the HOS cells were cultured in MEM (Gibco; Thermo Fisher
Scientific, Inc.; cat. no. 51200038), the BMSCs were cultured in
αMEM (Gibco; Thermo Fisher Scientific, Inc.; cat. no. 41061029),
containing 1% penicillin/streptomycin and 10% Fetal Bovine Serum
(FBS, Gibco; Thermo Fisher Scientific, Inc.; cat. no.
12484028).
In the section of functional rescue assays, Akt
signaling pathway activator SC79 (5 µM; cat. no. SF2730; Beyotime
Institute of Biotechnology) was used.
Lentiviruses with puromycin tags were purchased from
Hanbio Biotechnology Co., Ltd. The name of the lentiviral plasmid
containing shRNA was pHBLV-U6-MCS-EF1-ZsGreen-T2A-Luc. The
multiplicity of infection (MOI) used to infect 143b cells was 50,
and to infect U2OS cells, the MOI was 30. The cells were stably
transfected with NSUN6-knockdown lentiviruses
(short-hairpin(sh)-NSUN6#1 and sh-NSUN6#2) and the corresponding
negative control (sh-ctrl), as well as with the
EEF1A2-overexpression (OE) lentivirus (OE-EEF1A2) and empty vector
negative control (OE-ctrl). The duration of transduction into cells
was 24 h, and time interval between transduction and subsequent
experimentation was 72 h. Subsequently, puromycin (5 µg/ml) was
added to cells to select stably transfected OS cells for further
experiments. The following sequences were used for NSUN6-knockdown
transfections: sh-NSUN6#1, GAACAAAGGCGGTTAAACT; sh-NSUN6#2:
GCTGCAGCGATTTGATCCA. The sh-ctrl sequence was:
5'-TTCTCCGAACGTGTCACGTdTdT-3'.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from 1x106 cells
using TRIzol™ reagent (Invitrogen; Thermo Fisher Scientific, Inc.),
according to the manufacturer's instructions. Total RNA was
reverse-transcribed into cDNA using a reverse transcriptase kit
(HiScript II Q RT SuperMix for qPCR, +gDNA wiper; cat. no. R223;
Vazyme Biotech Co., Ltd.) according to the manufacturer's
instructions. qPCR was subsequently performed using the ChamQ
Universal SYBR qPCR Master Mix (Vazyme Biotech Co., Ltd.),
according to the manufacturer's instructions. The thermocycling
conditions used for qPCR were as follows: Stage 1, 95˚C for 30 sec;
stage 2, 95˚C for 10 sec and 60˚C for 30 sec, for 40 cycles; and
stage 3, 95˚C for 15 sec, 60˚C for 60 sec and 95˚C for 15 sec. The
mRNA levels were normalized to the reference gene GAPDH. The
2-∆∆Cq equation was applied to calculate the relative
expression level (38). The primer
pair sequences used for qPCR are listed in Table I.
 | Table IPrimer pair sequences used for
reverse transcription-quantitative PCR. |
Table I
Primer pair sequences used for
reverse transcription-quantitative PCR.
| Genes | Forward | Reverse |
|---|
| NOP2/Sun RNA
methyltransferase family member 6 |
5'-TCTCAGCCCTTCATTTGACAGT-3' |
5'-TCCAGTGCTATAACTTCTCCCTG-3' |
| Eukaryotic
elongation factor 1 α-2 |
5'-GAAGACCCACATCAACATCGT-3' |
5'-CTCCGCATTTGTAGATGAGGTG-3' |
| GAPDH |
5'-CAAATTCCATGGCACCGTCA-3' |
5'-GACTCCACGACGTACTCAGC-3' |
Cell Counting Kit-8 (CCK-8) and colony
formation assay
CCK-8 and colony formation assays were performed to
measure the proliferation of stably transfected OS cells. For the
CCK-8 assay, cells were seeded in 96-well plates at a density of
2,000 cells/well and the optical density value of each well was
detected at 450 nm using a microplate reader at 24, 48 and 72 h
after plating. The CCK-8 working fluid was purchased from Beyotime
Institute of Biotechnology.
For colony formation assay, OS cells were seeded in
6-well plates at a density of 1,000 cells/well and cultured for 2
weeks. Then, the cells were fixed in 4% paraformaldehyde (Beijing
Solarbio Science & Technology Co., Ltd.) at 25˚C for 15 min and
stained with 1% crystal violet (Beijing Solarbio Science &
Technology Co., Ltd.) at 25˚C for 15 min. Subsequently, the
colonies were photographed and counted. Colonies were defined as
>50 cells. The colonies were quantified using ImageJ V2.6
(National Institutes of Health).
Transwell and migration assays
Transwell assays were used to assess cell invasion
of stably transfected OS cells, while wound-healing assays were
used to evaluate their cell migration. For the Transwell assay, the
upper chambers were precoated with Matrigel (Corning, Inc.; pore
size 8.0 µm) at room temperature until dry. Cells were seeded in
the upper chamber at a density of 2x104 cells/well with
culture medium supplemented with 1% FBS, while the bottom chambers
contained culture medium supplemented with 20% FBS. After 24 h of
incubation at 37˚C, the cells in the upper chamber were removed
with a cotton swab, while cells invading the lower surfaces of the
chambers were fixed in 4% paraformaldehyde (Beijing Solarbio
Science & Technology Co., Ltd.) at 25˚C for 15 min, and then
stained with 1% crystal violet (Beijing Solarbio Science &
Technology Co., Ltd.) at 25˚C for 15 min. Stained cells were
counted in three randomly selected fields using an optical
microscope (ThermoFisher).
For the wound-healing assay, stably transfected
1x105 OS cells were seeded in 6-well plates and an
artificial wound was created using a 1,000-µl pipette tip when the
cell confluence reached 90%. The cells were then cultured with
culture medium containing 1% FBS at 37˚C for 24 h. Images were
taken to record the width of the wound at 0 and 24 h. Cell
migration was evaluated using the following formula:
(Width0h-width24h)/width0h.
RNA immunoprecipitation (RIP)
After mixing with 5 µg of antibody against NSUN6
(Invitrogen; Thermo Fisher Scientific, Inc.; cat. no. PA5-96608) or
IgG (Proteintech Group, Inc.; cat. no. 30000-0-AP), magnetic beads
(MedChemExpress) were added to OS cell lysates and the mixtures
were incubated at 4˚C for 12 h. The lysis buffer was purchased from
Thermo Fisher Scientific, Inc. Subsequently, proteinase K digestion
buffer was added to digest the complexes for 45 min at 55˚C. After
that, extraction buffer (phenol:chloroform:isoamyl
alcohol=125:24:1) was used to isolate RNA from the mixture. The
extracted RNA was subsequently analyzed using RT-qPCR. IgG was used
as a negative control to preclude nonspecific binding.
Methylated-RIP
The protocol used for methylated RIP assay was very
similar to that used for RIP. Briefly, ~150 µg of total RNA
isolated from stably transfected OS cells was immunoprecipitated
with magnetic beads (MedChemExpress) precoated with 10 µg of
anti-m5C antibody (Abcam; cat. no. ab214727).
Subsequently, the complexes were digested using proteinase K
digestion buffer so that the RNA combined with the antibody could
be eluted from the complex. Similarly, buffer
(phenol:chloroform:isoamyl alcohol=125:24:1) was used to extract
the RNA from the mixture. Then, the extracted RNA was further
analyzed using RT-qPCR. The relative m5C enrichment of
mRNA was normalized to the Input, a positive control, which was RNA
solution without antibodies added.
Dot blot assay
mRNA was isolated from the total RNA using an mRNA
Purification Kit (Beyotime Institute of Biotechnology) and
denatured at 65˚C for 5 min. Subsequently, mRNAs were loaded onto
an Amersham™ Hybond™ N+ membrane (GE Healthcare). After being
crosslinked with ultraviolet light for 5 min, the membrane was
stained with 0.02% methylene blue at 25˚C for 5 min (Sangon Biotech
Co. Ltd.). Subsequently, the membrane was blocked with 5% non-fat
dried milk in PBS with 0.02% Tween-20 (at 25˚C for 1 h) and then
incubated with an anti-m5C antibody (1:1,000, cat. no.
ab214727; Abcam) overnight at 4˚C. Following primary incubation,
the membrane was incubated with the secondary antibody (1:5,000;
cat. no. PR30011; Proteintech Group, Inc.), the membrane was
visualized with an Odyssey Infrared Imaging System (LI-COR
Biosciences).
mRNA stability assay
To analyze mRNA stability, stably transfected OS
cells were treated with actinomycin D (ActD, 5 µg/ml).
Subsequently, the cells were collected by scraping them from the
plates and total RNA was extracted using TRIzol reagent at
different time points (0, 1, 2 and 4 h after ActD treatment).
Thereafter, mRNA content was measured using RT-qPCR. The mRNA
remaining was normalized to the expression at 0 h.
Western blotting
Total proteins were extracted from OS cells with IP
cell lysate (Thermo Fisher Scientific, Inc.), quantified by BCA
assay (Beyotime Institute of Biotechnology) and then separated by
SDS-PAGE on a 10% gel. The separated proteins were subsequently
transferred onto PVDF membranes (Beyotime Institute of
Biotechnology). Next, the membranes were blocked with 5% fat-free
milk powder (Beyotime Institute of Biotechnology) at 25˚C for 2 h.
The membranes were incubated with primary antibodies at 4˚C for 12
h, followed by incubation with secondary antibodies (goat
anti-rabbit; 1:5,000; cat. no. PR30011; Proteintech Group, Inc.)
for 1 h at room temperature after washes. Subsequently, protein
band visualization was performed using an Odyssey Infrared Imaging
System (LI-COR Biosciences) and quantified using ImageJ software
V2.6 (National Institutes of Health). Detailed information about
the primary antibodies is provided in Table II.
 | Table IIDetailed information about primary
antibodies. |
Table II
Detailed information about primary
antibodies.
| Antibodies | Manufacturer (cat.
no.) | Dilution |
|---|
| NOP2/Sun RNA
methyltransferase family member 6 | Proteintech Group,
Inc. (17240-1-AP) | 1:1,000 |
| Eukaryotic
elongation factor 1 α-2 | Proteintech Group,
Inc. (16091-1-AP) | 1:1,000 |
| P-AKT | Proteintech Group,
Inc. (66444-1-Ig) | 1:1,000 |
| AKT | Proteintech Group,
Inc. (60203-2-Ig) | 1:1,000 |
| P-mTOR | Proteintech Group,
Inc. (67778-1-Ig) | 1:1,000 |
| mTOR | Proteintech Group,
Inc. (66888-1-Ig) | 1:1,000 |
| c-Myc | Proteintech Group,
Inc. (10828-1-AP) | 1:1,000 |
| P21 | Proteintech Group,
Inc. (10355-1-AP) | 1:1,000 |
| GAPDH | Proteintech Group,
Inc. (60004-1-Ig) | 1:1,000 |
| 5-methylcytidine
methyltransferase | Abcam
(ab214727) | 1:1,000 |
Tumor xenograft model
The animal study was approved by the Ethics
Committee of Zhongnan Hospital of Wuhan University (approval no.
WQ20210015). A total of 18 4-week-old male BALB/c nude mice were
purchased from GemPharmatech Co., Ltd. All the mice were evenly
divided into three groups: Sh-ctrl; sh-NSUN6#1; and sh-NSUN6#2.
Stably transfected 143b cells were collected and then resuspended
in PBS (approximately 107/ml). Next, 100 µl of cells
(containing ~106 cells) was injected into the upper
right side of each mouse's back. Then the health status of each
mouse and measured the size of each tumor were monitored every two
days. Mice with tumor tissue diameter >20 mm were euthanized.
Mice were observed for a total of 3 weeks and no mice were
euthanized in advance. After 3 weeks, all 18 mice were euthanized
and all the tumor tissues were harvested. To minimize the suffering
and distress of the mice, the mice were euthanized with an
intraperitoneal injection of pentobarbital sodium at a dose of 150
mg/kg. Death was confirmed when the mouse lost breathing and lost
its response after being stimulated. Subsequently, all the tumor
tissues were weighed and measured, and the volume of each tumor was
calculated using the formula: Tumor volume (mm3)=length
x width2 x 0.5. Immunohistochemical analysis of the
tumor tissues was performed by Wuhan Servicebio Technology Co.,
Ltd.
Statistical analysis
In this study, all data are presented as mean ±
standard deviation from three independent experiments. Statistical
significance was determined using one-way ANOVA or unpaired
two-tailed Student's t-test. To compare >2 groups one-way ANOVA
was used followed by Bonferroni multiple comparisons post hoc test.
Statistical analysis was performed with GraphPad Prism 8.0
(GraphPad Software; Dotmatics). P#x003C;0.05 was considered to
indicate a statistically significant difference.
Results
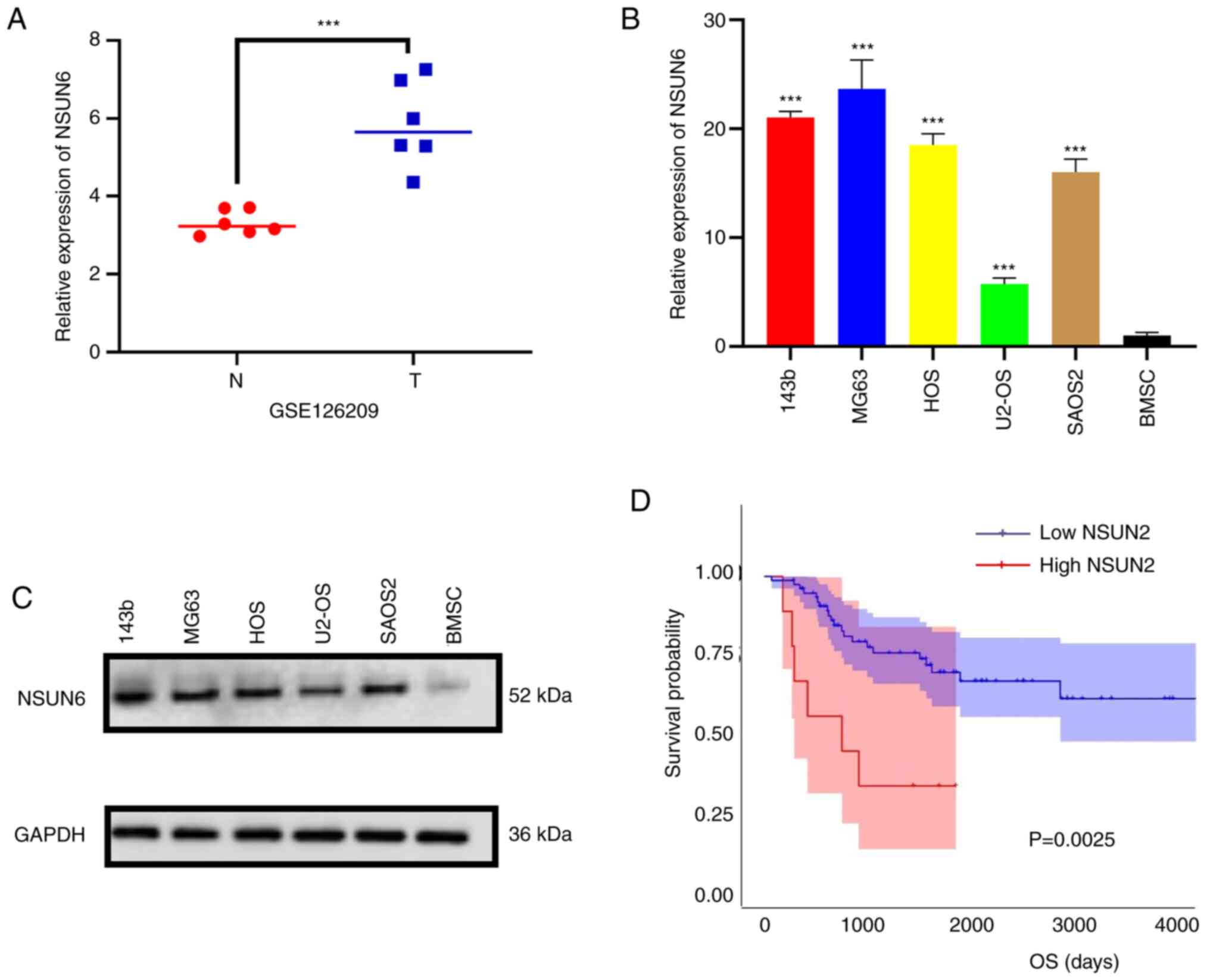
NSUN6 is highly expressed in
osteosarcoma
To characterize the expression profile of NSUN6 in
human OS, the data of GSE126209 from the GEO database were first
analyzed and the results showed that NSUN6 expression was higher in
osteosarcomas tumors than that in the adjacent normal tissues
(Fig. 1A). Subsequently, RT-qPCR
and Western Blot assays were performed to validate NSUN6 expression
in OS cell lines and bone mesenchymal stem cells (BMSCs). Results
from both assays showed that the NSUN6 expression was increased in
OS cell lines compared with that in BMSCs (Fig. 1B and C). In addition, after analyzing the
transcriptome data and corresponding survival information in
TARGET-OS from UCSC-XENA (http://xena.ucsc.edu/), it was observed that higher
NSUN6 expression was correlated with poorer prognosis in patients
with OS (Fig. 1D). Collectively,
these results indicate that NSUN6 is highly expressed in human OS
and may contribute to poor prognosis.
NSUN6 deficiency significantly
inhibits OS progression in vitro
To explore the functional roles of NSUN6 in OS
progression, stable NSUN6-knockdown OS cell lines were generated by
transfecting 143b and U2-OS cells with lentivirus encoding
sh-NSUN6#1 and sh-NSUN6#2; thereafter proliferation, invasion and
migration of these cells were examined. NSUN6 expression was
successfully downregulated in these cells when compared with the
control group (sh-ctrl) (Fig. 2A).
As shown through the CCK-8 and colony formation assays, the
knockdown of NSUN6 inhibited the proliferation of OS cells
(Fig. 2B and C). Interestingly, overexpressing NSUN6 in
BMSC (Fig. S1A) could enhance
proliferation and colony formation, mimicking the cancerous
phenotype of OS cells (Fig. S1B
and C). Furthermore, knocking
down NSUN6 significantly attenuated the invasion and migration of
OS cells, as evidenced by results from Transwell and wound-healing
assays, respectively (Fig. 2D and
E). Overall, the aforementioned
findings indicated that NSUN6 deficiency could suppress OS
progression in vitro.
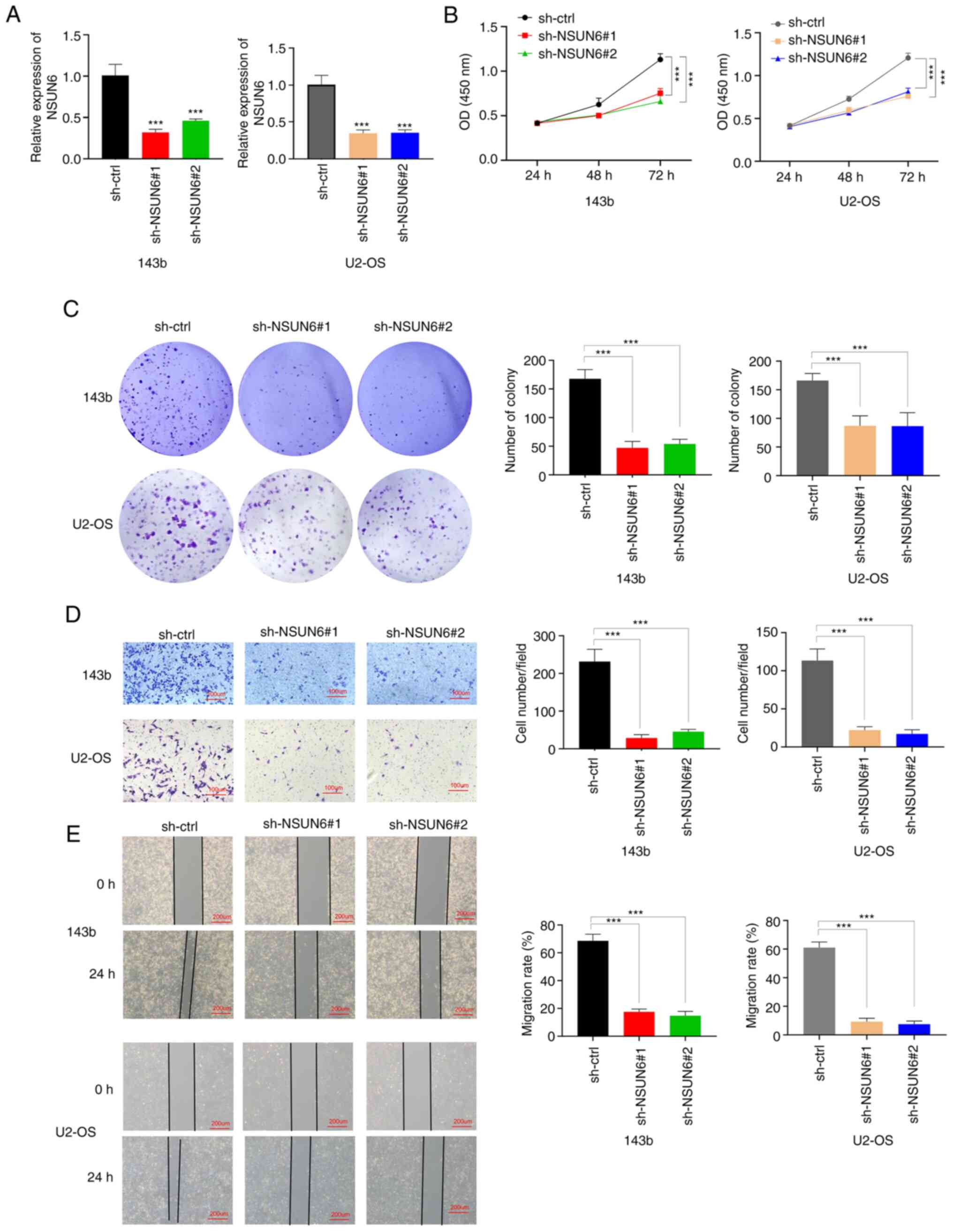 | Figure 2NSUN6 deficiency significantly
inhibits OS progression in vitro. (A) The expression of
NSUN6 mRNA in sh-ctrl, sh-NSUN6#1 and sh-NSUN6#2 groups of 143b and
U2-OS cells. Results of (B) Cell Counting Kit-8, (C) Colony
formation, (D) Transwell (scale bar, 100 µM) and (E) Wound-healing
(scale bar, 200 µM) assays. ***P#x003C;0.001. NSUN6,
NOP2/Sun RNA methyltransferase family member 6; OS, osteosarcoma;
sh, short-hairpin; ctrl, control; OD, optical density. |
EEF1A2 is the potential target of
NSUN6 in human OS
To dissect the underlying molecular mechanism of
NSUN6-mediated OS progression, the data in TARGET-OS from UCSC-XENA
(http://xena.ucsc.edu/) were analyzed to screen
for downstream targets of NSUN6. First, all the patients in
TARGET-OS were divided into the ‘high NSUN6’ group and ‘low NSUN6’
group using the median value of NSUN6 expression. Subsequently, all
genes were screened using two criteria: P#x003C;0.05; and the
absolute value of Log2(Fold Change) >1. The following
four target genes with the largest fold changes were identified:
EEF1A2; melanoma antigen preferentially expressed in tumors
(PRAME); fibroblast growth factor binding protein 2 (FGFBP2); and
TP53 (Fig. 3A). Further analysis
revealed that EEF1A2 expression had the strongest correlation with
the expression of NSUN6 (P=0.0017) (Fig. 3B-E). This finding was further
validated experimentally; the expression of EEF1A2 mRNA was
substantially reduced in cells transfected with sh-NSUN6#1 and
sh-NSUN6#2 lentivirus (Fig. 3F and
G). Lastly, a RIP assay was
performed to determine whether NSUN6 regulated EEF1A2 expression
through direct or indirect action. The result of the RIP assay
showed that EEF1A2 mRNA could be pulled down with NUSN6 antibody,
indicating that NSUN6 protein could directly bind to EEF1A2 mRNA
(Fig. 3H and I). Collectively, these data suggested
that EEF1A2 expression is positively correlated with the expression
of NSUN6, which could directly bind to EEF1A2 mRNA.
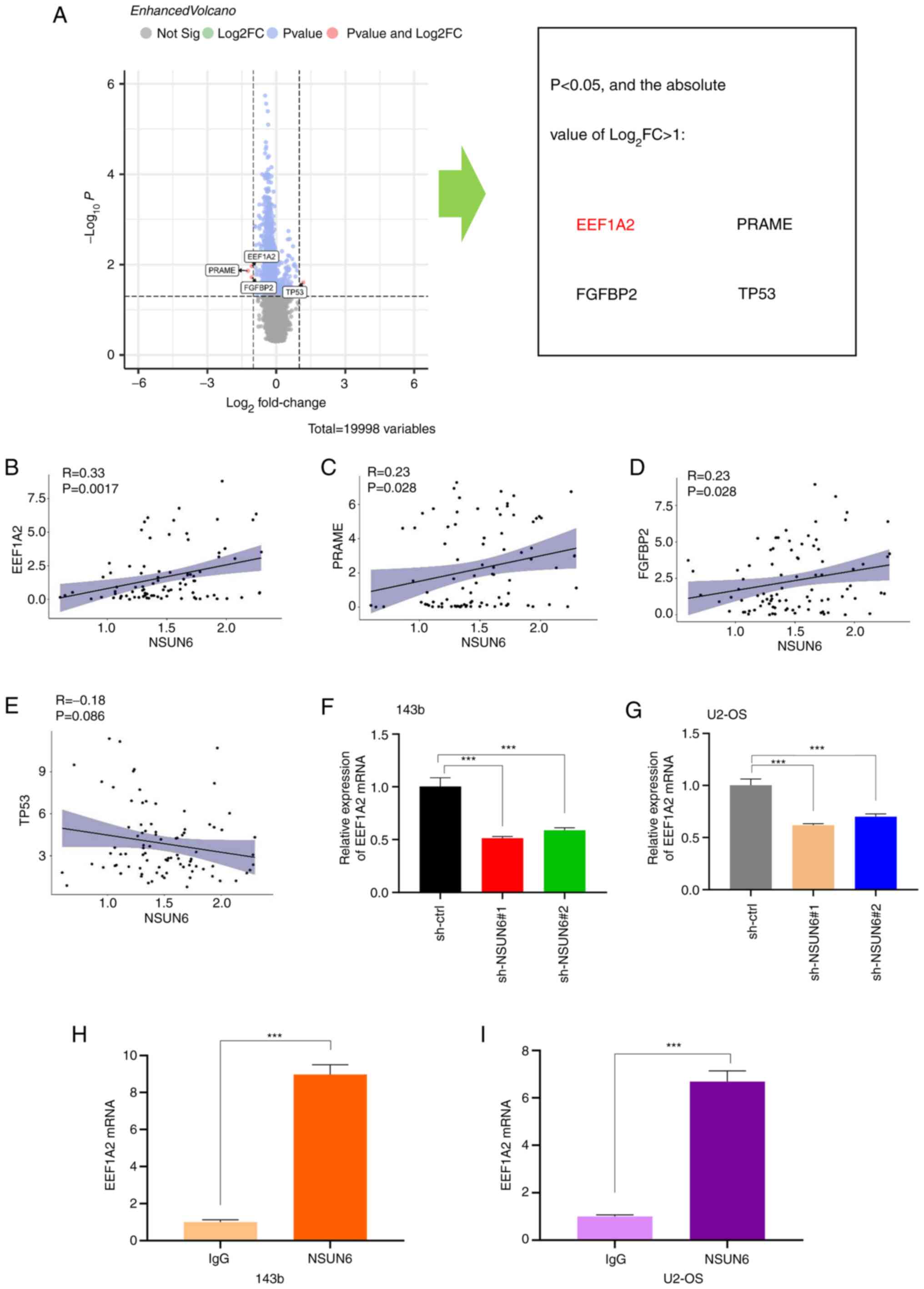 | Figure 3EEF1A2 is the potential target of
NSUN6 in human OS. (A) Volcano plot showing that expression
difference of EEF1A2, PRAME, FGFBP2 and TP53 was the largest
between high- and low-NSUN6 groups. Correlation of NSUN6 with (B)
EEF1A2 (P=0.0017), (C) PRAME (P=0.028), (D) FGFBP2 (P=0.028) and
(E) TP53 (P=0.086) in human OS. Expression of EEF1A2 mRNA in
sh-ctrl, sh-NSUN6#1 group and sh-NSUN6#2 groups of (F) 143b and (G)
U2-OS cells. RIP assay results in (H) 143b and (I) U2-OS cells. IgG
was used as a negative control. ***P#x003C;0.001. NSUN6,
NOP2/Sun RNA methyltransferase family member 6; OS, osteosarcoma;
EEF1A2, eukaryotic elongation factor 1 α-2; PRAME, melanoma antigen
preferentially expressed in tumors; FGFBP2, fibroblast growth
factor binding protein 2; sh, short-hairpin; ctrl, control. |
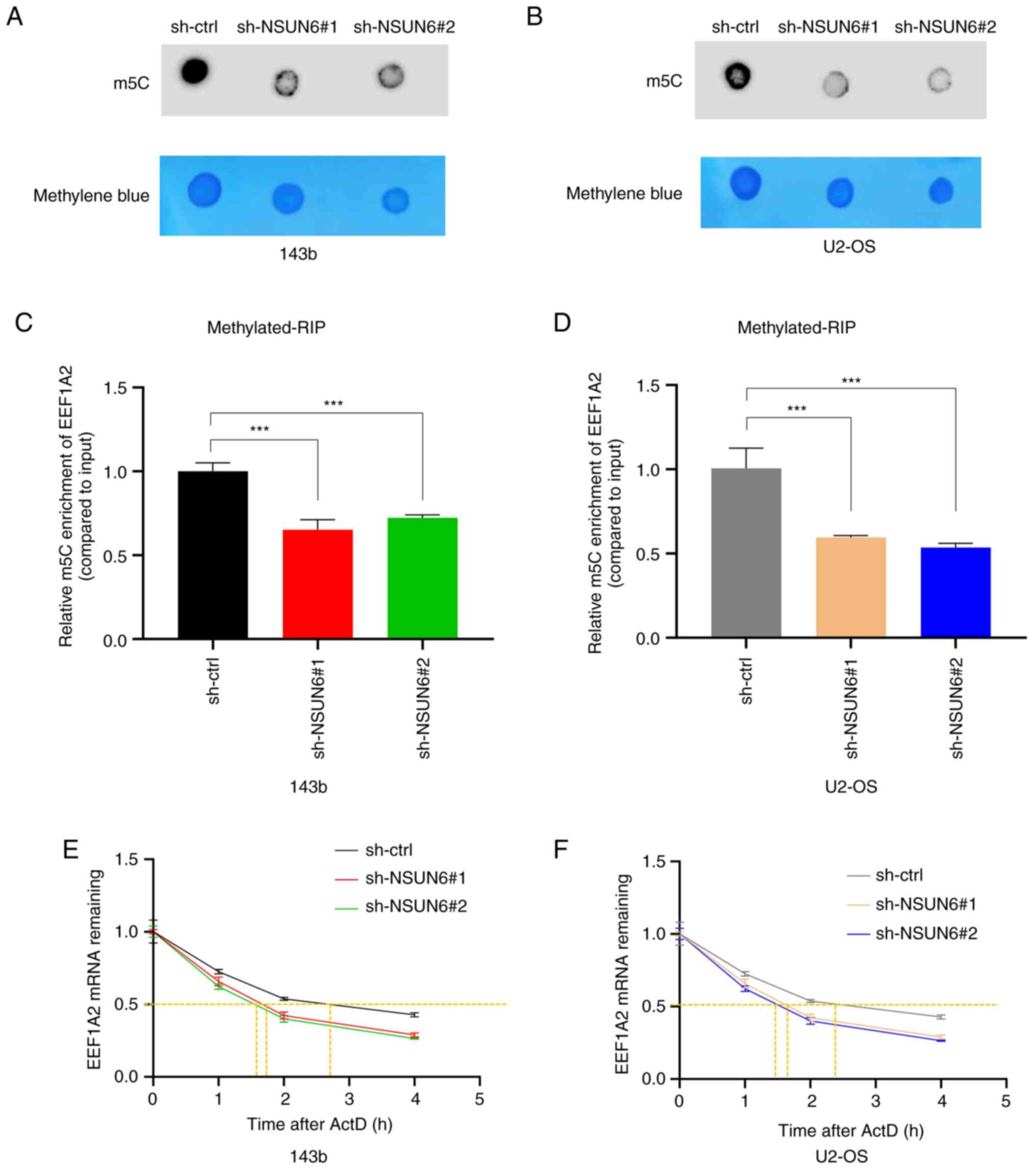
NSUN6 regulates the stability of
EEF1A2 mRNA via m5C
Given that NSUN6 is a well-known methylase with a
function to induce m5C modification (22), a Dot Plot assay was performed to
evaluate whether NSUN6 could regulate the m5C level in
OS cells. As shown in Fig. 4A and
B, the knockdown of NSUN6 led to a
decreased level of m5C in 143b and U2-OS cells. Since
EEF1A2 mRNA was confirmed as a direct target of NSUN6, a
methylated-RIP assay was utilized to further explore whether NSUN6
could regulate EEF1A2 expression by modulating the m5C
level of EEF1A2 mRNA. The results confirmed the presence of
m5C modification on EEF1A2 mRNA; more importantly, loss
of NSUN6 resulted in less m5C modification of EEF1A2
mRNA (Fig. 4C and D). A previous study demonstrated that
NSUN6 primarily targets the consensus sequence motif CTCCA on the
3'-untranslated region of mRNA, which is mainly related to mRNA
stability (39); therefore, the
present study evaluated whether NSUN6 affects EEF1A2 mRNA
stability. The current data showed a faster degradation rate of
EEF1A2 mRNA in sh-NSUN6#1 and sh-NSUN6#2 groups, when compared with
the sh-ctrl group (Fig. 4E and
F), suggesting that loss of NSUN6
reduced the stability of EEF1A2 mRNA in 143b and U2-OS cells.
Altogether, the aforementioned results revealed that NSUN6 could
bind to EEF1A2 mRNA and regulate its m5C modification,
thus regulating the stability of EEF1A2 mRNA in OS.
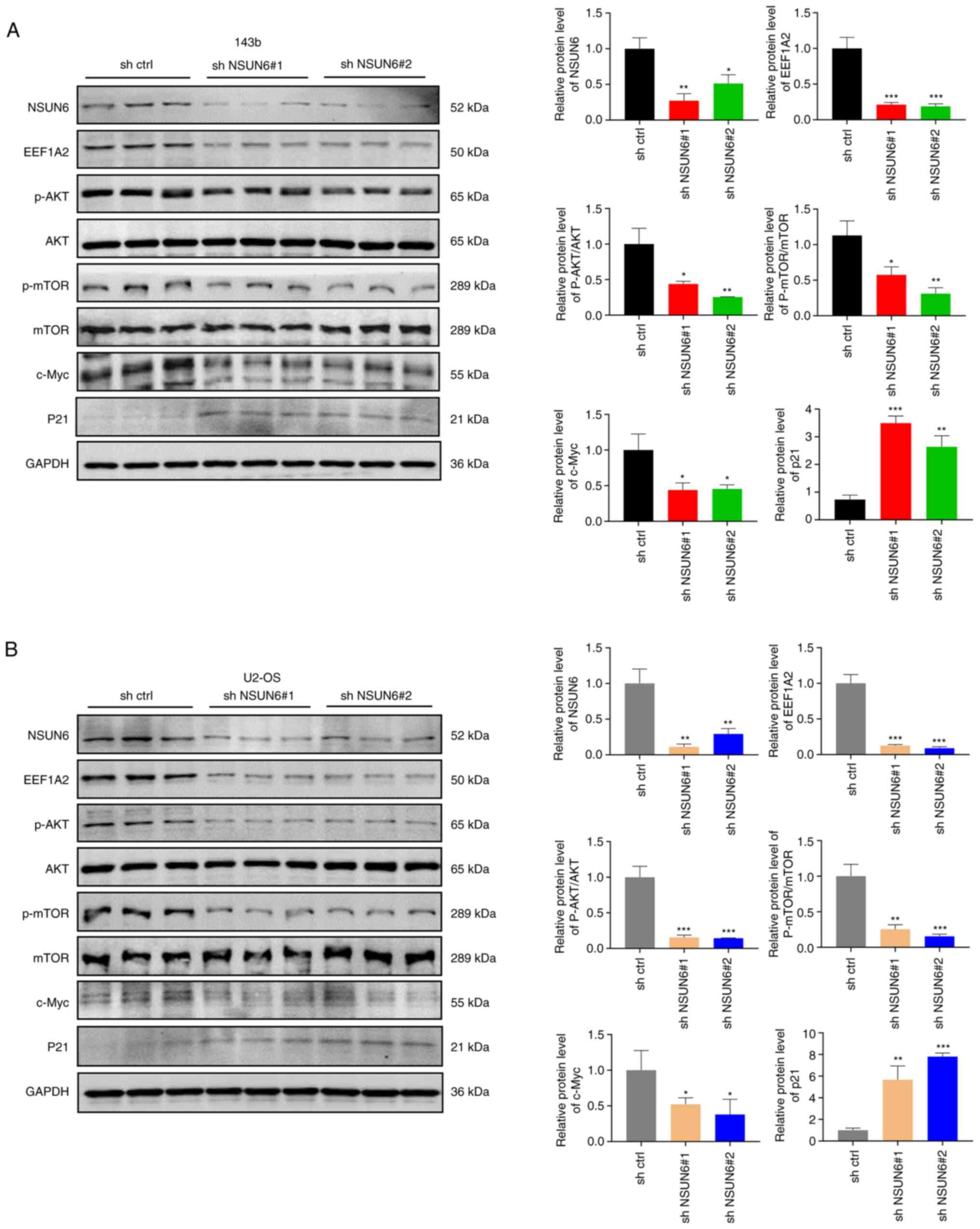
Loss of NSUN6 reduced the activation
of the Akt/mTOR signaling pathway in OS
A previous report showed that EEF1A2 could promote
the progression of OS by activating Akt/mTOR signaling pathway
(37). Thus, the present study
aimed to evaluate whether the Akt/mTOR signaling pathway is a
downstream effector of the NSUN6/EEF1A2 signaling in OS. The level
of NSUN6 protein was significantly decreased following sh-NSUN6#1
and sh-NSUN6#2 transfections in 143b and U2-OS cells (Fig. 5A and B). This was accompanied by a significant
decrease in protein levels of EEF1A2 and c-Myc, and phosphorylation
of Akt and mTOR; whereas p21 protein levels were increased in
NSUN6-deficient OS cells (Fig. 5A
and B). Taken together, NSUN6
knockdown decreased the expression of EEF1A2 protein and
consequently suppressed Akt/mTOR signaling pathway.
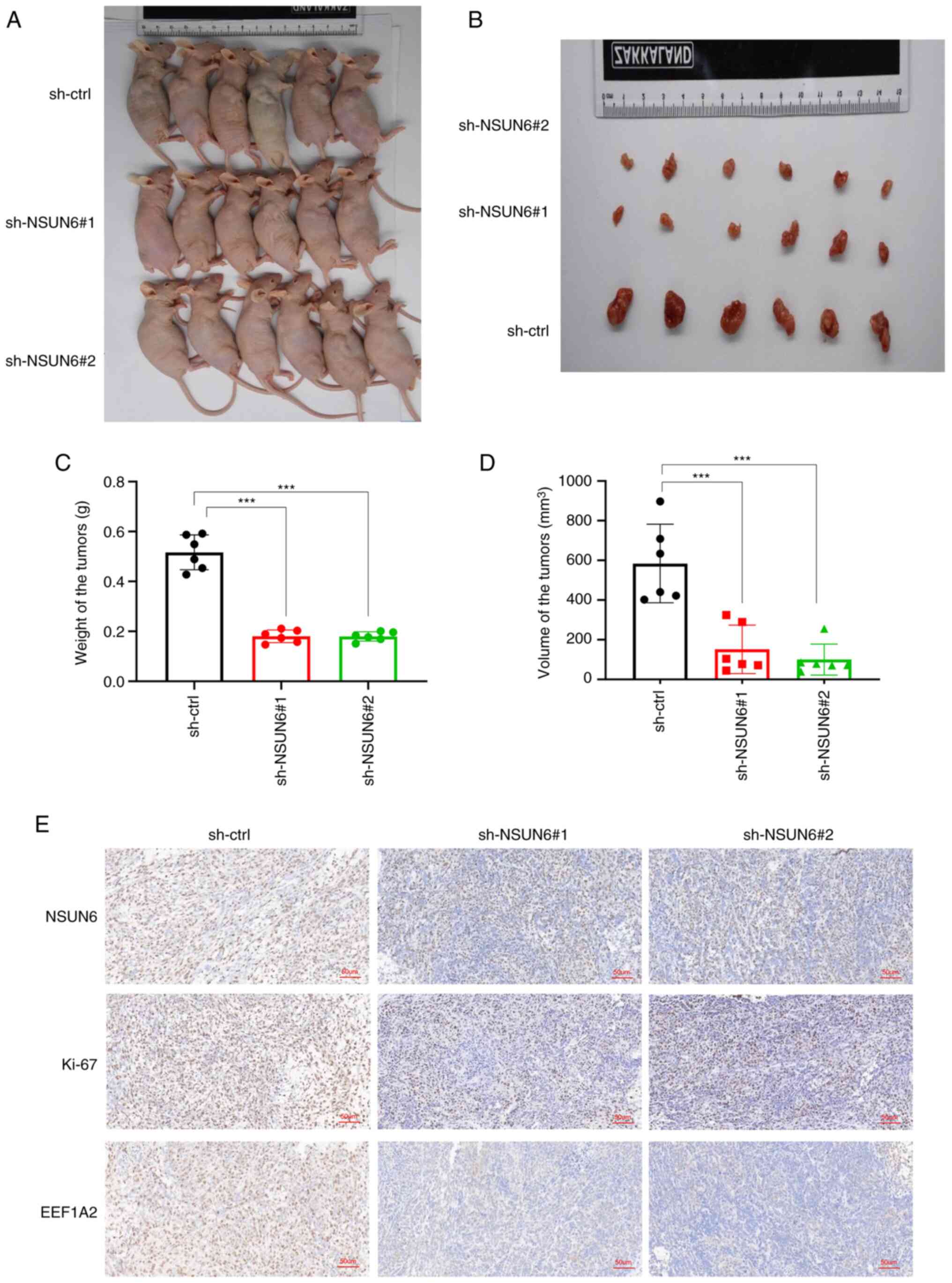
Loss of NSUN6 inhibits the
proliferation of OS in vivo
To evaluate whether NSUN6 could regulate the
proliferation of OS in vivo, tumor xenografts in 4-week-old
nude mice were generated using stably transfected 143b cells. The
mice were euthanized 3 weeks after the 143b cells were inoculated
and tumor tissue was harvested from each mouse for further
examination. As shown in Fig.
6A-D, the tumor size, weight and volume were smaller in mice in
the sh-NSUN6#1 and sh-NSUN6#2 groups compared with those in the
sh-ctrl group. In addition, the immunostaining results showed that
protein levels of NSUN6, EEF1A2 and Ki-67 were all significantly
lower in the tumors from mice injected with NSUN6-knockdown OS
cells compared with those in the control group (Fig. 6E). Overall, the aforementioned
results suggested a blunted tumor progression in vivo when
NSUN6 was knocked down.
EEF1A2 overexpression or Akt signaling
pathway activator SC79 can counterbalance the inhibitory effects of
NSUN6 deficiency on OS progression
To further validate the functional roles of EEF1A2
and Akt/mTOR signaling pathway action on OS suppression triggered
by NSUN6 deficiency, functional rescue assays were performed in
143b cells by overexpressing EEF1A2 or stimulating cells with Akt
signaling pathway activator SC79. As is shown in Fig. S2, lentiviral transfection
successfully increased EEF1A2 mRNA expression in 143b cells, while
an empty vector was used as OE-ctrl. Meanwhile, SC79 treatment
showed no effect on EEF1A2 gene expression (Fig. 7A). In Fig. 7, the cells transfected with sh-ctrl
lentiviral were used as control. Importantly, both EEF1A2
overexpression and SC79 stimulation improved the proliferation of
143b cells, as shown using CCK-8 and colony formation assays
(Fig. 7B and C). In addition, the results of the
Transwell and wound-healing assays demonstrated that EEF1A2
overexpression and SC79 administration significantly increased 143b
cell invasion and migration (Fig.
7D and E). Statistical chart
of colony formation, Transwell and wound-healing assays are shown
in Fig. 7 F-H. Collectively, the
present results indicated that EEF1A2 overexpression or Akt/mTOR
signaling pathway action could blunt the beneficial effect of NSUN6
knockdown on suppressing OS progression.
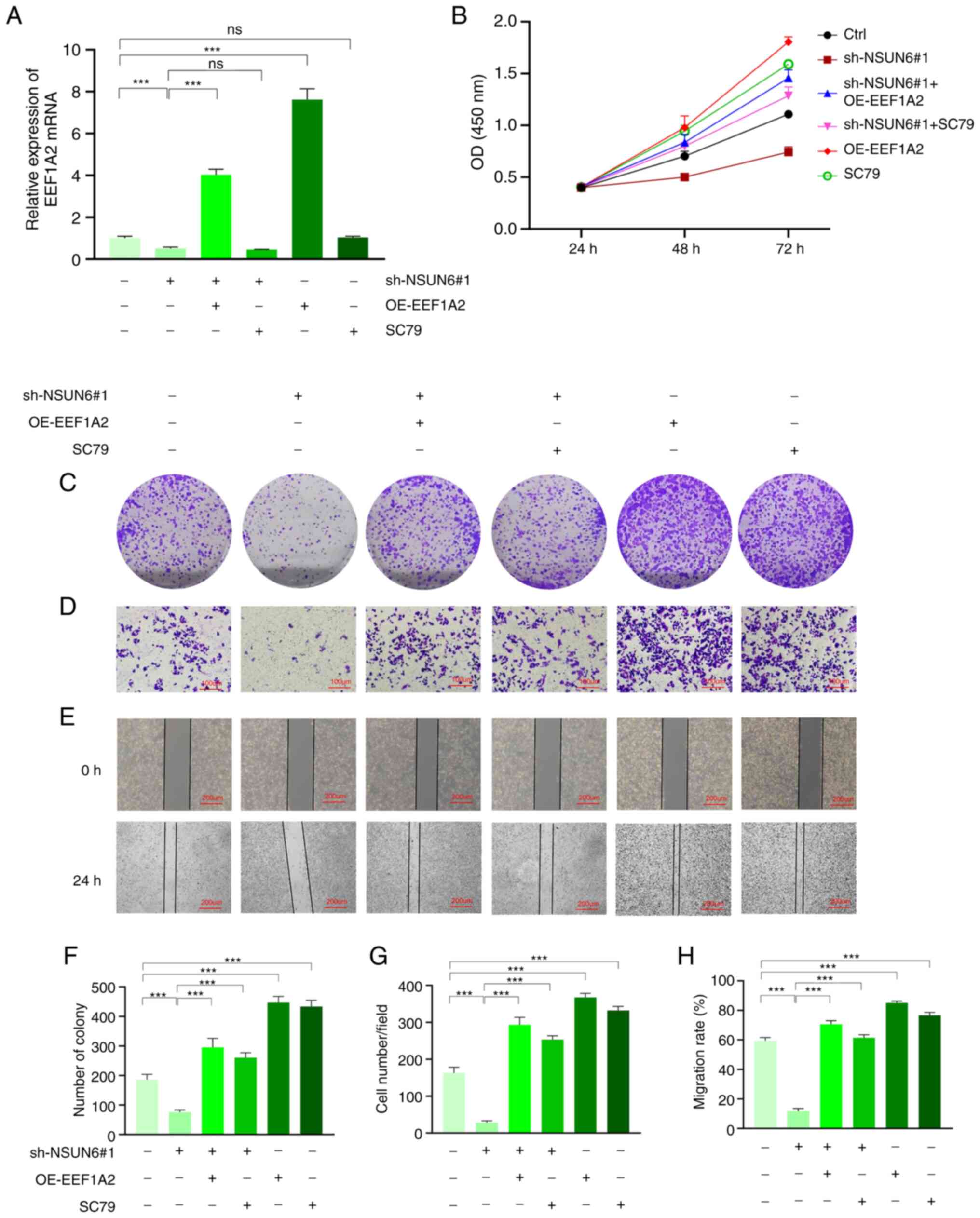 | Figure 7EEF1A2 overexpression and Akt
signaling pathway activator SC79 can counterbalance the inhibition
of NSUN6 deficiency on OS progression. (A) Results of reverse
transcription-quantitative PCR showing the expression level of
EEF1A2 mRNA in 143b cells of different groups. Results of (B) Cell
Counting Kit-8, (C) Colony formation, (D) Transwell (scale bar, 100
µM) and (E) Wound-healing assays (scale bar, 200 µM). Statistical
charts of (F) Colony formation, (G) Transwell, and (H)
Wound-healing assay. ***P#x003C;0.001. NSUN6, NOP2/Sun
RNA methyltransferase family member 6; EEF1A2, eukaryotic
elongation factor 1 α-2; sh, short-hairpin; OE, overexpression;
ctrl, control. |
Discussion
Currently, the primary treatment approach for OS
involves a combination of chemotherapy and surgery. However, OS has
a high propensity to metastasize to the lungs in a relatively short
time, leading to a significant number of cases where patients with
OS are diagnosed with pre-existing lung metastases, thereby
eliminating the possibility of surgical intervention (1,2).
This underscores the need for more effective treatment strategies
that can prevent or mitigate OS progression to distant sites,
particularly the lungs (1). The
efficacy of the current therapeutic approaches is limited due to
the onset of adverse effects, development of chemotherapy
resistance and high rates of relapse (5). To address this challenge, there is a
pressing need for in-depth studies that can elucidate the molecular
mechanisms involved in OS pathogenesis because these findings may
reveal novel therapeutic targets that can be exploited to develop
more effective treatments for OS.
Accumulating evidence demonstrated that RNA
methylation modification could contribute to the pathogenesis of
various diseases, especially in cancer (16,40).
Among different RNA modifications, m6A has been extensively studied
in cancer progression and molecular mechanism, whereas the roles of
m5C have been relatively neglected (41,42).
Preliminary studies suggested that m5C modification
solely occurred on tRNA (18,19);
however, more mature studies showed that m5C
modification could also occur on mRNA, significantly impacting mRNA
metabolism, including export, stability and translation efficiency
(17,18,20,43-45).
A previous study reported that m5C modification, induced
by NSUN2 and recognized by YBX1, could promote the pathogenesis of
bladder cancer by stabilizing different mRNA targets (24). Furthermore, NSUN2 was also shown to
promote cancer progression by destabilizing p57Kip2 mRNA or
stabilizing growth factor receptor-bound protein 2 mRNA via
m5C modification, and enhancing Autotaxin mRNA
translation (27,46,47).
As another important methyltransferase that induces
m5C modification, NSUN6 was found to suppress pancreatic
cancer development by regulating cell proliferation (28). Despite numerous studies (19,21,22)
highlighting the critical role of m5C modification
induced by NSUN2 or NSUN6 in various cancer types, its function in
OS progression remains unclear. A previous study from the present
authors indicated a high expression level of NSUN2 in OS cells
(36). In addition, the current
study found that NSUN6 was highly expressed in OS, which was
associated with poorer prognosis in patients with OS. It is
plausible that NSUN6 DNA undergoes various chemical modifications
during OS progression (i.e. acetylation or lactate), which loosens
its binding to histone resulting in higher transcription
efficiency. Further research is required to dissect the underlying
mechanisms involved in the upregulation of USUN6 expression in OS
progression.
The present findings further revealed that the
downregulation of NSUN6 in OS cells could inhibit cancer cell
proliferation, invasion and migration. Bioinformatics analysis
showed that EEF1A2 has the strongest correlation with NSUN6.
Mechanistically, the present study demonstrated that NSUN6 protein
could directly bind to EEF1A2 mRNA and the knockdown of NSUN6 led
to reduced EEF1A2 mRNA stability via m5C modification.
Importantly, EEF1A2 was previously reported to be involved in the
pathogenesis of various cancers by enhancing TGF-β/SMAD signaling,
binding to protein kinase R and modulating its activity or
modulating apoptosis (48-51).
Specifically concerning OS progression, Yang et al (37) has reported that EEF1A2 promotes OS
progression via activating Akt/mTOR pathway. In line with this
finding, the present data showed that NSUN6 deficiency decreased
the phosphorylation level of Akt/mTOR in OS cells. Furthermore,
EEF1A2 overexpression or Akt signaling pathway activation through
SC79 stimulation could counterbalance the inhibitory effects of
NSUN6 deficiency on OS progression.
Overall, the aforementioned results indicated that
NSUN6 could contribute to OS progression by upregulating EEF1A2 and
activating Akt/mTOR pathway. Lastly, further research is needed to
identify the ‘reader’ that recognizes the target mRNA methylated by
NSUN6.
The current study provided important insights into
the molecular mechanisms of OS progression and highlights NSUN6 as
a potential therapeutic target for OS. By identifying the role of
NSUN6 in regulating the Akt/mTOR signaling pathway, the present
findings could potentially inform the development of new treatment
strategies for patients with OS.
Supplementary Material
NSUN6 overexpression can enhance the
proliferation and colony formation of BMSC. (A) Expression of NSUN6
in the OE-NSUN6 and OE-ctrl group of BMSC. Results of (B) Cell
Counting Kit-8 and (C) Colony formation assays of BMSC.
**P<0.01 and ***P<0.001. NSUN6,
NOP2/Sun RNA methyltransferase family member 6; OE, overexpression;
ctrl, control; BMSC, bone mesenchymal stem cells.
The transfection efficiency of
eukaryotic elongation factor 1 α-2 overexpression in 143b cells.
***P<0.001. EEF1A2, eukaryotic elongation factor 1
α-2; OE, overexpression; ctrl, control.
Acknowledgements
Not applicable.
Funding
Funding: This work was supported by the National Natural Science
Foundation of China (grant no. 82103285) and the Fundamental
Research Funds for the Central Universities (grant no.
2042020kf0138).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
SH and MY performed most of the experiments and
wrote the manuscript. KX performed the bioinformatics analysis,
designed the cell experiments, and helped SH and MY perform western
blot assay and write the manuscript. ZY performed the CCK-8,
Transwell and wound-healing assays. LC helped RW design the study
and checked the manuscript. YX provided the funding, designed
animal experiments, and helped SH and MY perform the animal
experiments. LW helped RW design the study, and helped SH and MY
write and revise the manuscript. RW designed and supervised the
study. SH and MY confirm the authenticity of all the raw data. All
authors have read and approved the final manuscript.
Ethics approval and consent to
participate
The animal study was approved by the Ethics
Committee of Zhongnan Hospital of Wuhan University (approval no.
WQ20210015; Wuhan, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Durfee RA, Mohammed M and Luu HH: Review
of osteosarcoma and current management. Rheumatol Ther. 3:221–243.
2016.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Li S, Zhang H, Liu J and Shang G: Targeted
therapy for osteosarcoma: A review. J Cancer Res Clin Oncol: Feb
18, 2023 (Epub ahead of print).
|
|
3
|
Chen S, Li Y, Zhi S, Ding Z, Wang W, Peng
Y, Huang Y, Zheng R, Yu H, Wang J, et al: WTAP promotes
osteosarcoma tumorigenesis by repressing HMBOX1 expression in an
m6A-dependent manner. Cell Death Dis.
11(659)2020.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Xie L, Yao Z, Zhang Y, Li D, Hu F, Liao Y,
Zhou L, Zhou Y, Huang Z, He Z, et al: Deep RNA sequencing reveals
the dynamic regulation of miRNA, lncRNAs, and mRNAs in osteosarcoma
tumorigenesis and pulmonary metastasis. Cell Death Dis.
9(772)2018.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Jafari F, Javdansirat S, Sanaie S, Naseri
A, Shamekh A, Rostamzadeh D and Dolati S: Osteosarcoma: A
comprehensive review of management and treatment strategies. Ann
Diagn Pathol. 49(151654)2020.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Wang B, Yao L, Dong Y, Liu J and Wu J:
LncRNA PCED1B-AS1 knockdown inhibits osteosarcoma via
methylation-mediated miR-10a downregulation. J Orthop Surg Res.
17(464)2022.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Tsukamoto S, Righi A, Kido A, Honoki K,
Tanaka Y, Fujii H, Mavrogenis AF, Tanaka Y and Errani C: Effect of
adjuvant chemotherapy on periosteal osteosarcoma: A systematic
review. Jpn J Clin Oncol. 52:896–904. 2022.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Liu W, Zhao Y, Wang G, Feng S, Ge X, Ye W,
Wang Z, Zhu Y, Cai W, Bai J and Zhou X: TRIM22 inhibits
osteosarcoma progression through destabilizing NRF2 and thus
activation of ROS/AMPK/mTOR/autophagy signaling. Redox Biol.
53(102344)2022.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Lin H, Chen X, Zhang C, Yang T, Deng Z,
Song Y, Huang L, Li F, Li Q, Lin S and Jin D: EF24 induces
ferroptosis in osteosarcoma cells through HMOX1. Biomed
Pharmacother. 136(111202)2021.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Xiao X, Wang W, Li Y, Yang D, Li X, Shen
C, Liu Y, Ke X, Guo S and Guo Z: HSP90AA1-mediated autophagy
promotes drug resistance in osteosarcoma. J Exp Clin Cancer Res.
37(201)2018.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Liu Q and Wang K: The induction of
ferroptosis by impairing STAT3/Nrf2/GPx4 signaling enhances the
sensitivity of osteosarcoma cells to cisplatin. Cell Biol Int.
43:1245–1256. 2019.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Dong S, Wu Y, Liu Y, Weng H and Huang H:
N6 -methyladenosine steers RNA metabolism and regulation
in cancer. Cancer Commun (Lond). 41:538–559. 2021.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Kumar VE, Nambiar R, De Souza C, Nguyen A,
Chien J and Lam KS: Targeting epigenetic modifiers of tumor
plasticity and cancer stem cell behavior. Cells.
11(1403)2022.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Nacev BA, Jones KB, Intlekofer AM, Yu JSE,
Allis CD, Tap WD, Ladanyi M and Nielsen TO: The epigenomics of
sarcoma. Nat Rev Cancer. 20:608–623. 2020.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Yang B, Wang JQ, Tan Y, Yuan R, Chen ZS
and Zou C: RNA methylation and cancer treatment. Pharmacol Res.
174(105937)2021.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Han X, Wang M, Zhao YL, Yang Y and Yang
YG: RNA methylations in human cancers. Semin Cancer Biol.
75:97–115. 2021.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Dominissini D and Rechavi G:
5-methylcytosine mediates nuclear export of mRNA. Cell Res.
27:717–719. 2017.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Garcia-Vilchez R, Sevilla A and Blanco S:
Post-transcriptional regulation by cytosine-5 methylation of RNA.
Biochim Biophys Acta Gene Regul Mech. 1862:240–252. 2019.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Xue C, Zhao Y and Li L: Advances in RNA
cytosine-5 methylation: Detection, regulatory mechanisms,
biological functions and links to cancer. Biomark Res.
8(43)2020.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Yang X, Yang Y, Sun BF, Chen YS, Xu JW,
Lai WY, Li A, Wang X, Bhattarai DP, Xiao W, et al: 5-methylcytosine
promotes mRNA export-NSUN2 as the methyltransferase and ALYREF as
an m5C reader. Cell Res. 27:606–625. 2017.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Chellamuthu A and Gray SG: The RNA
methyltransferase NSUN2 and its potential roles in cancer. Cells.
9(1758)2020.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Zhang Q, Liu F, Chen W, Miao H, Liang H,
Liao Z, Zhang Z and Zhang B: The role of RNA m5C
modification in cancer metastasis. Int J Biol Sci. 17:3369–3380.
2021.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Sun Z, Xue S, Zhang M, Xu H, Hu X, Chen S,
Liu Y, Guo M and Cui H: Aberrant NSUN2-mediated m5C
modification of H19 lncRNA is associated with poor differentiation
of hepatocellular carcinoma. Oncogene. 39:6906–6919.
2020.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Chen X, Li A, Sun BF, Yang Y, Han YN, Yuan
X, Chen RX, Wei WS, Liu Y, Gao CC, et al: 5-methylcytosine promotes
pathogenesis of bladder cancer through stabilizing mRNAs. Nat Cell
Biol. 21:978–990. 2019.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Gao Y, Wang Z, Zhu Y, Zhu Q, Yang Y, Jin
Y, Zhang F, Jiang L, Ye Y, Li H, et al: NOP2/Sun RNA
methyltransferase 2 promotes tumor progression via its interacting
partner RPL6 in gallbladder carcinoma. Cancer Sci. 110:3510–3519.
2019.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Hu Y, Chen C, Tong X, Chen S, Hu X, Pan B,
Sun X, Chen Z, Shi X, Hu Y, et al: NSUN2 modified by SUMO-2/3
promotes gastric cancer progression and regulates mRNA m5C
methylation. Cell Death Dis. 12(842)2021.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Mei L, Shen C, Miao R, Wang JZ, Cao MD,
Zhang YS, Shi LH, Zhao GH, Wang MH, Wu LS and Wei JF: RNA
methyltransferase NSUN2 promotes gastric cancer cell proliferation
by repressing p57Kip2 by an m5C-dependent
manner. Cell Death Dis. 11(270)2020.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Yang R, Liang X, Wang H, Guo M, Shen H,
Shi Y, Liu Q, Sun Y, Yang L and Zhan M: The RNA methyltransferase
NSUN6 suppresses pancreatic cancer development by regulating cell
proliferation. Ebiomedicine. 63(103195)2021.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Awah CU, Winter J, Mazdoom CM and Ogunwobi
OO: NSUN6, an RNA methyltransferase of 5-mC controls glioblastoma
response to temozolomide (TMZ) via NELFB and RPS6KB2 interaction.
Cancer Biol Ther. 22:587–597. 2021.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Blaze J, Navickas A, Phillips HL, Heissel
S, Plaza-Jennings A, Miglani S, Asgharian H, Foo M, Katanski CD,
Watkins CP, et al: Neuronal Nsun2 deficiency produces tRNA
epitranscriptomic alterations and proteomic shifts impacting
synaptic signaling and behavior. Nat Commun.
12(4913)2021.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Lu L, Zhu G, Zeng H, Xu Q and Holzmann K:
High tRNA Transferase NSUN2 Gene expression is associated with poor
prognosis in head and neck squamous carcinoma. Cancer Invest.
36:246–253. 2018.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Haag S, Warda AS, Kretschmer J, Gunnigmann
MA, Hobartner C and Bohnsack MT: NSUN6 is a human RNA
methyltransferase that catalyzes formation of m5C72 in specific
tRNAs. RNA. 21:1532–1543. 2015.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Zhou L, Yang C, Zhang N, Zhang X, Zhao T
and Yu J: Silencing METTL3 inhibits the proliferation and invasion
of osteosarcoma by regulating ATAD2. Biomed Pharmacother.
125(109964)2020.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Zhou X, Yang Y, Li Y, Liang G, Kang D,
Zhou B and Li Q: METTL3 contributes to osteosarcoma progression by
increasing DANCR mRNA stability via m6A modification. Front Cell
Dev Biol. 9(784719)2021.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Jiang R, Dai Z, Wu J, Ji S, Sun Y and Yang
W: METTL3 stabilizes HDAC5 mRNA in an m6A-dependent
manner to facilitate malignant proliferation of osteosarcoma cells.
Cell Death Discov. 8(179)2022.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Yang M, Wei R, Zhang S, Hu S, Liang X,
Yang Z, Zhang C, Zhang Y, Cai L and Xie Y: NSUN2 promotes
osteosarcoma progression by enhancing the stability of FABP5 mRNA
via m5C methylation. Cell Death Dis.
14(125)2023.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Yang J, Tang J, Li J, Cen Y, Chen J and
Dai G: Effect of activation of the Akt/mTOR signaling pathway by
EEF1A2 on the biological behavior of osteosarcoma. Ann Transl Med.
9(158)2021.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Selmi T, Hussain S, Dietmann S, Heiβ M,
Borland K, Flad S, Carter JM, Dennison R, Huang YL, Kellner S, et
al: Sequence- and structure-specific cytosine-5 mRNA methylation by
NSUN6. Nucleic Acids Res. 49:1006–1022. 2021.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Wu S, Zhang S, Wu X and Zhou X:
m6A RNA methylation in cardiovascular diseases. Mol
Ther. 28:2111–2119. 2020.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Shen H, Lan Y, Zhao Y, Shi Y, Jin J and
Xie W: The emerging roles of N6-methyladenosine RNA methylation in
human cancers. Biomark Res. 8(24)2020.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Wang S, Sun C, Li J, Zhang E, Ma Z, Xu W,
Li H, Qiu M, Xu Y, Xia W, et al: Roles of RNA methylation by means
of N6-methyladenosine (m6A) in human cancers.
Cancer Lett. 408:112–120. 2017.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Shinoda S, Kitagawa S, Nakagawa S, Wei FY,
Tomizawa K, Araki K, Araki M and Suzuki T and Suzuki T: Mammalian
NSUN2 introduces 5-methylcytidines into mitochondrial tRNAs.
Nucleic Acids Res. 47:8734–8745. 2019.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Auxilien S, Guerineau V,
Szweykowska-Kulinska Z and Golinelli-Pimpaneau B: The human tRNA m
(5) C methyltransferase Misu is multisite-specific. RNA Biol.
9:1331–1338. 2012.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Hussain S: The emerging roles of
cytosine-5 methylation in mRNAs. Trends Genet. 37:498–500.
2021.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Xu X, Zhang Y, Zhang J and Zhang X: NSUN2
promotes cell migration through methylating autotaxin mRNA. J Biol
Chem. 295:18134–18147. 2020.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Su J, Wu G, Ye Y, Zhang J, Zeng L, Huang
X, Zheng Y, Bai R, Zhuang L, Li M, et al: NSUN2-mediated RNA
5-methylcytosine promotes esophageal squamous cell carcinoma
progression via LIN28B-dependent GRB2 mRNA stabilization. Oncogene.
40:5814–5828. 2021.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Jia L, Ge X, Du C, Chen L, Zhou Y, Xiong
W, Xiang J, Li G, Xiao G, Fang L and Li Z: EEF1A2 interacts with
HSP90AB1 to promote lung adenocarcinoma metastasis via enhancing
TGF-β/SMAD signalling. Br J Cancer. 124:1301–1311. 2021.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Losada A, Munoz-Alonso MJ, Martinez-Diez
M, Gago F, Dominguez JM, Martinez-Leal JF and Galmarini CM: Binding
of eEF1A2 to the RNA-dependent protein kinase PKR modulates its
activity and promotes tumour cell survival. Br J Cancer.
119:1410–1420. 2018.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Sun Y, Du C, Wang B, Zhang Y, Liu X and
Ren G: Up-regulation of eEF1A2 promotes proliferation and inhibits
apoptosis in prostate cancer. Biochem Biophys Res Commun. 450:1–6.
2014.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Lee MH, Choi BY, Cho YY, Lee SY, Huang Z,
Kundu JK, Kim MO, Kim DJ, Bode AM, Surh YJ, et al: Tumor suppressor
p16 (INK4a) inhibits cancer cell growth by downregulating eEF1A2
through a direct interaction. J Cell Sci. 126:1744–1752.
2013.PubMed/NCBI View Article : Google Scholar
|