Introduction
Liver cancer, a malignancy of the digestive system,
accounts for <2 million deaths in China (1). Due to the lack of symptoms, patients
with liver cancer are commonly diagnosed at an advanced stage of
the disease, making them contraindicated for curative surgical
therapy (2). Chemotherapy is the
primary treatment approach for patients with advanced liver cancer,
while 5-fluorouracil (5-FU) is one of the first-line chemotherapy
drugs (3). However, due to its
inherent toxicity to normal cells and multidrug resistance, 5-FU
has limited clinical application (4). Currently, several natural products,
such as curcumin (5) and quercetin
(6), have been identified to exert
synergistic effects with 5-FU, thus enhancing its efficacy.
Consequently, novel chemosensitizers are urgently needed to treat
patients with liver cancer.
Purpurogallin carboxylic acid (PCA; molecular
formula, C12H8O7; IUPAC name,
2,3,4,6-tetrahydroxy-5-oxobenzo[7]annulene-8-carboxylic acid) is a
natural phenol compound derived from Macleaya microcarpa
(Maxim.) Fedde, which is also the oxidation product of gallic acid
in fermented tea (7). Zeng et
al (8) demonstrated that
treatment with PCA could relieve corneal endothelial cell injury by
reducing oxidative stress. Furthermore, Rambabu et al
(9) revealed that PCA exhibited
significant suppressive effects on the proliferation ability of
MCF7 and A549 cancer cells by targeting claudin-4. However, its
effects and molecular mechanisms on liver cancer cells remain
elusive.
P-glycoproteins are promising drug efflux pumps,
that have been extensively studied for their association with
resistance to chemotherapeutic drugs (10,11).
ATP binding cassette subfamily G member 2 (ABCG2) is a member of
the P-glycoprotein family, which is upregulated in several types of
cancer, including liver cancer (12). A previous study indicated that
ABCG2 upregulation was associated with poor outcome in patients
with liver cancer (13).
Additionally, pharmacology- and genetic engineering
technology-mediated ABCG2 downregulation could increase the
sensitivity of liver cancer cells to 5-FU (14). Therefore, exploring novel molecules
to inhibit ABCG2 is a potential strategy for treating liver
cancer.
The present study aimed to uncover the effects and
underlying molecular mechanism of PCA on the behavior of liver
cancer cells, thus suggesting that PCA could exert synergistic
effects with 5-FU on liver cancer cells in vitro via
targeting ABCG2.
Materials and methods
Network pharmacological analysis
The secondary structure of PCA was downloaded from
PubChem Compound (https://www.ncbi.nlm.nih.gov/pccompound; accession no.
137628491). Subsequently, the secondary structure of PCA was
imported into TargetNet (http://targetnet.scbdd.com/calcnet/index/) and
Super-PERD (https://prediction.charite.de/) to predict the
potential targets of PCA, with a possibility of ≥0.8. The predicted
targets of PCA were visualized using Cytoscape (version 3.9.1; The
Cytoscape Consortium). Finally, the intersected targets were used
for further analysis.
Computer virtual docking
The crystal structure of ABCG2 (accession no. 6VXI),
G protein-coupled receptor 35 (GPR35; accession no. 8H8J) and
thyroid stimulating hormone receptor (TSHR; accession no. 4QT5)
were downloaded from Protein Data Bank (https://www.rcsb.org/). Subsequently, the crystal
structure of the aforementioned proteins was imported into SYBYL-X
software (version 2.0; Tripos, Inc.; Certara) to perform structural
optimization, including deletion of primary ligands, hydrogenation
and adhesive end repair. Finally, the aforementioned proteins and
the secondary structure of PCA were imported into AutoDock software
(v1.5.6; UCSF Computer Graphics Laboratory) to perform flexible
docking. Among them, ML230, a recognized ABCG2 inhibitor, was used
as a positive control to compare the score of the binding between
PCA and ABCG2. The binding sites between PCA and ABCG2 were
visualized using PYMOL software (version 2.5; Schrödinger).
Surface plasmon resonance (SPR)
Purified ABCG2 proteins were obtained from Sino
Biological Inc. and were then desalinated using the AKTA protein
purification system (Cytiva). Subsequently, proteins were dissolved
in buffer solution (200 mM HEPES, 2 mM NaCl and 0.5% DMSO) and were
conjugated with sodium acetate solution (pH 4.0) using the Biacore
T200 system (Cytiva). Following conjugation with human
metal-organic framework, an ethanolamine solution (1.0 M; Shanghai
Ruji Biological Technology, Co., Ltd.) was used to block the
uncoupled proteins at 28˚C for 6 h. The chip was then added to a
buffer solution for 10 h. PCA was dissolved in the buffer solution
(10 mmol/l HEPES, 150 mmol/l NaCl and 3 mmol/l EDTA) of the mobile
phase. The binding constants between ABCG2 proteins and PCA were
determined using a multi-cycle model. The flow rate of the mobile
phase, the binding time and the dissociation time were set at 30
µl/sec, 240 and 300 sec, respectively.
Enzymatic activity determination
Firstly, all molecular reagents were dissolved in 1X
Assay Buffer (5 mM MgCl2, 50 µM NADP+, NaCl
150 mM, pH 8.0 Tris-HCl 50 mM). Additionally, the purified ABCG2
proteins were diluted in a final concentration of 25 nM.
Subsequently, a total of 35 µl/well protein dilution was added into
a 384-well plate. Each well was then supplemented with 10 µl PCA
reagent (0.01-100 µM) and incubated at room temperature for 1 h in
the dark. Finally, the enzymatic reaction was initiated following
the addition of 5 µl ATP/well. Following incubation for 30 min, the
fluorescence intensity values were measured at a wavelength of 450
nm using the Varioskan LUX multifunctional microplate reader
(Thermo Fisher Scientific, Inc.).
Cell culture and cell transfection
with short interfering (si)RNA
The normal hepatocyte cell line, THLE-2, and the
HepG2, Huh7 and Huh1 liver cancer cell lines were obtained from
Procell Life Science & Technology Co., Ltd. All cell lines were
authenticated by short tandem repeat (STR) DNA profiling analysis.
THLE-2 cells were cultured in the corresponding special medium
(https://www.procell.com.cn/view/10030.html; Procell
Life Science & Technology Co., Ltd.) supplemented with 10% FBS,
while HepG2, Huh7 and Huh1 cells were cultured in DMEM with 10% FBS
(both from Hyclone; Cytiva). All cell lines were incubated at 37˚C
in an incubator with 5% CO2. The siRNAs targeting ABCG2
(si-ABCG2; sense, 5'-CUGGAGAUGUUCUGAUAAA-3' and antisense,
5'-UUUAUCAGAACAUCUCCAG-3') and the normal control siRNAs (si-NC;
sense, 5'-UUCUCCGAACGUGUCACGU-3' and antisense,
5'-ACGUGACACGUUCGGAGAA-3') were purchased from GenePharma Co., Ltd.
Cells were transfected with the aforementioned siRNAs (both 40
pmol) using Lipofectamine 2000® (Sangon Biotech Co.,
Ltd.), according to the manufacturer's instructions at 37˚C for 24
h. After 48 h, cells were used for performing biological
experiments.
Rhodamine efflux assay
HepG2, Huh7 and Huh1 cells were seeded into a 6-well
plate at a density of 1x105 cells/well. Subsequently,
adhered cells were treated with various concentrations of PCA (0,
2.5, 5 and 10 µM) for 24 h. Cells were then digested and
resuspended in a 1-ml culture medium followed by incubation in the
presence of 10 mmol/l rhodamine 123 (Thermo Fisher Scientific,
Inc.) for 30 min at 37˚C. After washing three times with PBS, cells
were analyzed using a flow cytometer (NovoCyte Advanteon; Agilent
Inc.) at an excitation wavelength of 488 nm.
Cell Counting Kit-8 (CCK-8) assay and
synergy index assessment
THLE-2, HepG2, Huh7 and Huh1 cells were seeded into
a 6-well plate at a density of 1x103 cells/well. Cells
were then co-treated with various concentrations of PCA (0, 2.5, 5
and 10 µM) combined with 5-FU (0, 2.5, 5, 10 and 20 µM). Following
incubation for 48 h at 37˚C, each well was supplemented with 10 µl
CCK-8 reagent (Shanghai Yeasen Biotechnology Co., Ltd.) and cells
were cultured for an additional 2 h at 37˚C. The proliferation rate
of liver cells was determined by calculating the optical density of
each well at a wavelength of 450 nm using the Varioskan LUX
multifunctional microplate. The synergistic index of PCA and 5-FU
was measured using Compusyn software (version 2.0; ComboSyn, Inc.).
A synergistic index of >0.9, 0.9-0.6, 0.6-0.3 and <0.3
indicated no synergy, weak synergy, moderate synergy and strong
synergy, respectively.
Cell cycle analysis
HepG2, Huh7 and Huh1 cells were seeded into a 6-well
plate at a density of 1x105 cells/well. Subsequently,
cells were synchronized for 12 h with DMEM without FBS, followed by
treatment with DMSO, PCA (10 µM) or 5-FU (10 µM) and their
combination for 48 h. Following digestion, cells were resuspended
and fixed in cold 75% ethyl alcohol at 4˚C for 24 h. After washing
two times with PBS, cells were stained with propidium iodide
(Shanghai Univ Biotechnology Co., Ltd.) for 30 min in the dark at
28˚C. Cell cycle distribution was assessed using a flow cytometer
(NovoCyte Advanteon; Agilent Inc.) and analyzed with the FlowJo
software (version 7.6.2; FlowJo LLC).
Western blot analysis
Total proteins were extracted from HepG2, Huh7 and
Huh1 cells using RIPA reagent supplemented with 1% PMSF (both from
Shanghai Univ Biotechnology Co., Ltd.). Following protein
quantification using a BCA kit (cat no. abs9232-500T), proteins
were separated by 10% SDS-PAGE (Shanghai Univ Biotechnology Co.,
Ltd.), followed by transferring onto a PVDF membrane (Shanghai Univ
Biotechnology Co., Ltd.). Following blocking with 8% skim milk
powder in TBS-Tween-20 (0.1%) (TBST) for 2 h at 28˚C, the membranes
were incubated with primary antibodies against cyclin-dependent
kinase (CDK) 4 (1:2,000; cat no. 11026-1-AP), CDK6 (1:1,000; cat
no. 14052-1-AP), ABCG2 (1:1,000; cat no. 27286-1-AP) and GAPDH
(1:50,000; cat no. 60004-1-Ig) all from Proteintech Group, Inc. for
16 h at 4˚C. After washing free antibodies three times with TBST,
the membranes were incubated for 2 h at 28˚C with the corresponding
HRP-conjugated Affinipure Goat Anti-Mouse (1:3,000; cat no.
SA00001-1) or HRP-conjugated Affinipure Goat Anti-Rabbit (1:3,000;
cat no. SA00001-2) all from Proteintech Group, Inc. secondary
antibodies. Finally, the protein bands were visualized using an ECL
reagent (Proteintech Group, Inc.), while the expression levels of
CDK4, CDK6 and ABCG2 were normalized to those of GAPDH using Image
J (version 1.8.0; National Institutes of Health).
Reverse transcription-quantitative
(RT-q)PCR experiments
Total RNA was isolated from HepG2, Huh7 and Huh1
cells utilizing TRIzol® reagent (Shanghai Yeasen
Biotechnology Co., Ltd.), followed by reverse transcription into
cDNA with an mRNA First Strand cDNA Synthesis kit (Takara Bio,
Inc.) as per the manufacturer's instructions. Subsequently,
quantitative PCR was conducted using the SYBR® Green
Master Mix (Takara Bio, Inc.). Following primers were used in the
experiments: CDK4 forward, 5'-ATGGCTACCTCTCGATATGAGC-3' and
reverse, 5'-CATTGGGGACTCTCACACTCT-3'; CDK6 forward,
5'-GCTGACCAGCAGTACGAATG-3' and reverse,
5'-GCACACATCAAACAACCTGACC-3'; and GAPDH forward,
5'-GGAGCGAGATCCCTCCAAAAT-3' and reverse,
5'-GGCTGTTGTCATACTTCTCATGG-3'. Relative mRNA levels of CDK4 and
CDK6 were measured using the 2-ΔΔCq method (15), while GAPDH was set as loading
control. The qPCR thermocycling protocol consisted of an initial
denaturation step at 95˚C for 5 min, followed by 40 cycles of
denaturation at 95˚C for 25 sec, annealing at 60˚C for 40 sec and a
final elongation step at 72˚C for 30 sec.
Colony formation and 3D sphere
formation assays
HepG2, Huh7 and Huh1 cells were plated in a 6-well
plate and were then treated with DMSO, PCA (10 µM), 5-FU (10 µM) or
their combination for 48 h at 37˚C. For colony formation assays,
cells in each group were first digested and were then seeded into a
6-well plate at a density of 500 cells/well. After culturing for 14
days at 37˚C, cell colonies were fixed with 4% paraformaldehyde for
20 min at 28˚C and stained with 1% crystal violet for 30 min at
28˚C (both from Skillbio; Beijing Siqi Biotechnology Co., Ltd.).
Following washing with PBS, cell colonies were collected using a
vidicon (Sony Group Corporation) while colonies consisting of
>50 cells were counted using Image J (version 1.8.0).
Furthermore, for 3D sphere formation assay, cells in each group
were digested and seeded in a 24-well ultra-low adsorption culture
plate (U-section bottom; Absin Biotechnology Co., Ltd.) at a
density of 2,000 cells/well. After culturing for 21 days at 37˚C,
images of the formed spheres were captured under a light microscope
(magnification, x4; Olympus Corporation), while sphere volume was
calculated using the following formula: Volume=(length x
width2)/2.
Statistical analysis
All results are presented as the mean ± standard
deviation, and all experiments were performed three times. All data
were analyzed using SPSS 19.0 software (IBM Corp.). The differences
among multiple groups were analyzed by one-way ANOVA followed by
Bonferroni's post hoc test. P<0.05 was considered to indicate a
statistically significant difference.
Results
ABCG2 is a target of PCA
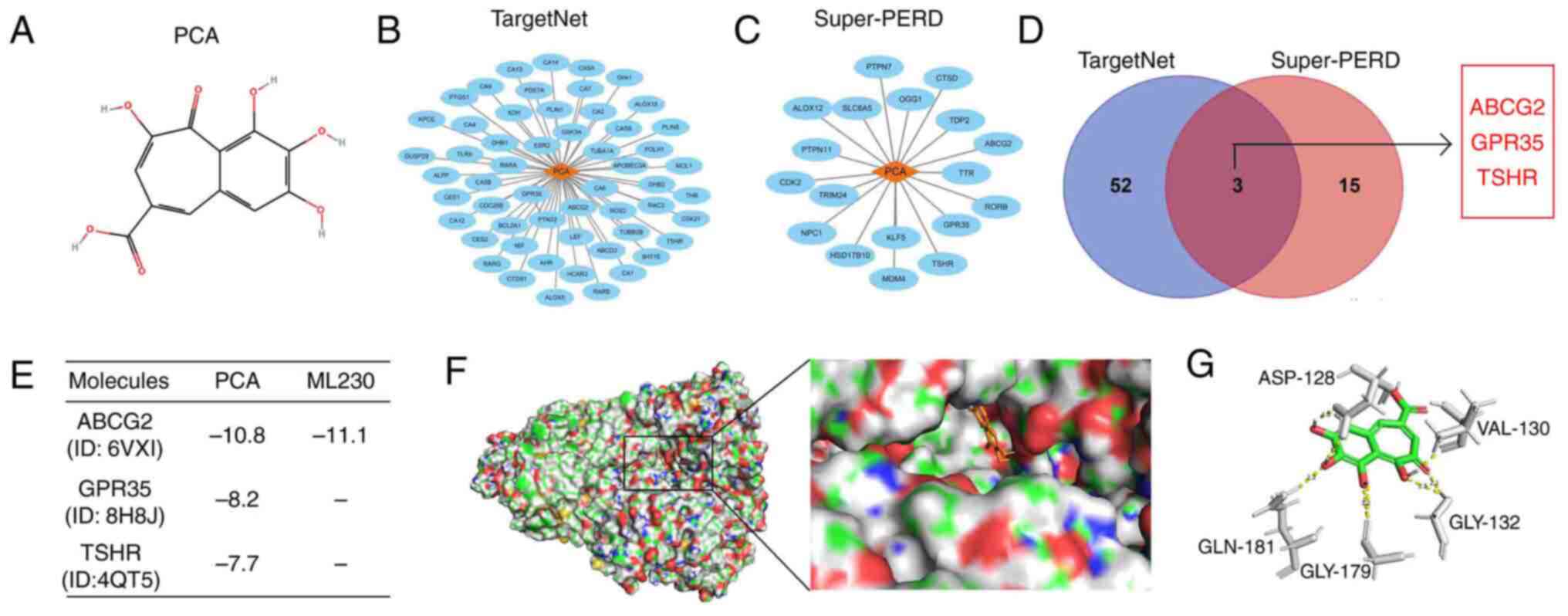
To identify the targets of PCA, the secondary
structure of PCA was downloaded from PubChem Compound (Fig. 1A). A total of 55 proteins with
binding possibility of ≥0.8 were predicted as targets of PCA using
the TargetNet online tool (Fig.
1B). Similarly, a total of 18 proteins with binding possibility
of ≥0.8 were predicted as targets of PCA using the Super-PERD
database (Fig. 1C). Finally, three
proteins, namely ABCG2, GPR35 and TSHR, were intersected in the
aforementioned two databases (Fig.
1D). To further analyze the binding capacity between the
aforementioned three target proteins and PCA, computer virtual
docking was performed using Autodock software. The analysis
revealed that ABCG2 displayed the best binding score to PCA
(Fig. 1E). The binding score of
ABCG2 to PCA was equivalent to that of its positive inhibitor,
ML230 (Fig. 1E). In detail, PCA
could form five hydrogen bonds with the ASP-128 (2.0 Å), VAL-130
(2.1 Å), GLY-132 (2.5 Å and 2.1 Å), GLY-179 (3.2 Å) and GLN-181
(2.2 Å) sites of ABCG2 (Fig. 1F
and G). Overall, the
aforementioned findings indicated that ABCG2 could be considered as
a key target of PCA.
PCA has high affinity with ABCG2 and
inhibits ABCG2 activity in liver cancer cells
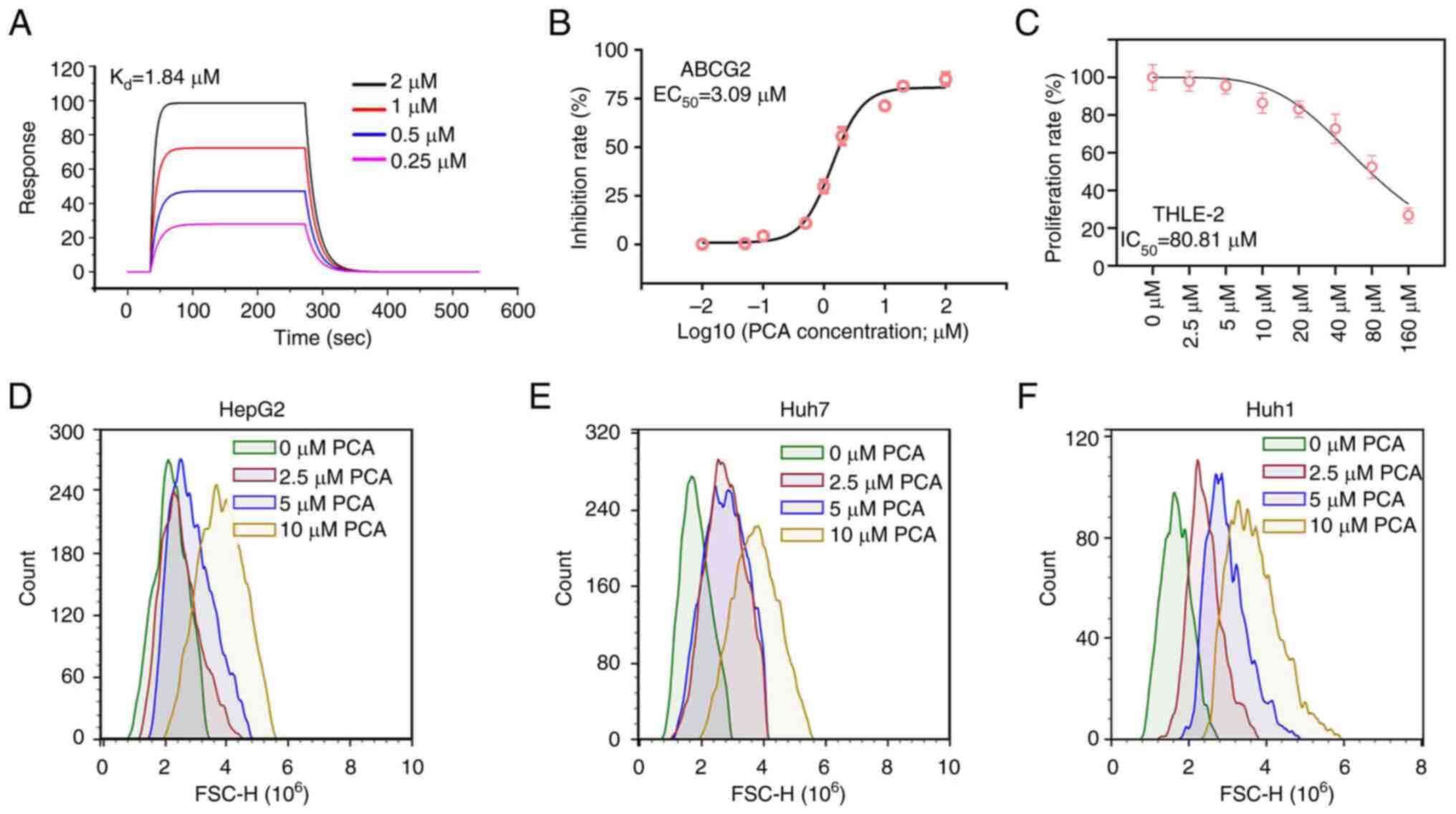
To further verify the binding potential between PCA
and ABCG2, SPR was carried out. The analysis revealed that the
equilibrium dissociation constant (Kd) of PCA to ABCG2
protein was 1.84 µM (Fig. 2A),
thus indicating that PCA had high binding affinity to ABCG2.
Subsequently, enzyme activity assay was performed and the results
revealed that the median effective concentration (EC50)
of PCA for ABCG2 was 3.09 µM (Fig.
2B). Notably, the half-maximal inhibitory concentration
(IC50) of PCA in the THLE-2 normal hepatocyte cell line
was 80.81 µM (Fig. 2C), thus
suggesting that the non-specific toxic effects of PCA on normal
cells were low. It has been reported that ABCG2 can efflux drugs
from the inside of the cell to the outside. Therefore, rhodamine
efflux assays were performed to assess whether PCA could inhibit
the efflux capacity of ABCG2. The results demonstrated that PCA
could markedly increase the accumulation of rhodamine inside HepG2
(Fig. 2D), Huh7 (Fig. 2E) and Huh1 (Fig. 2F) cells.
PCA exhibits synergistic effects with
5-FU for the inhibition of the proliferation of liver cancer
cells
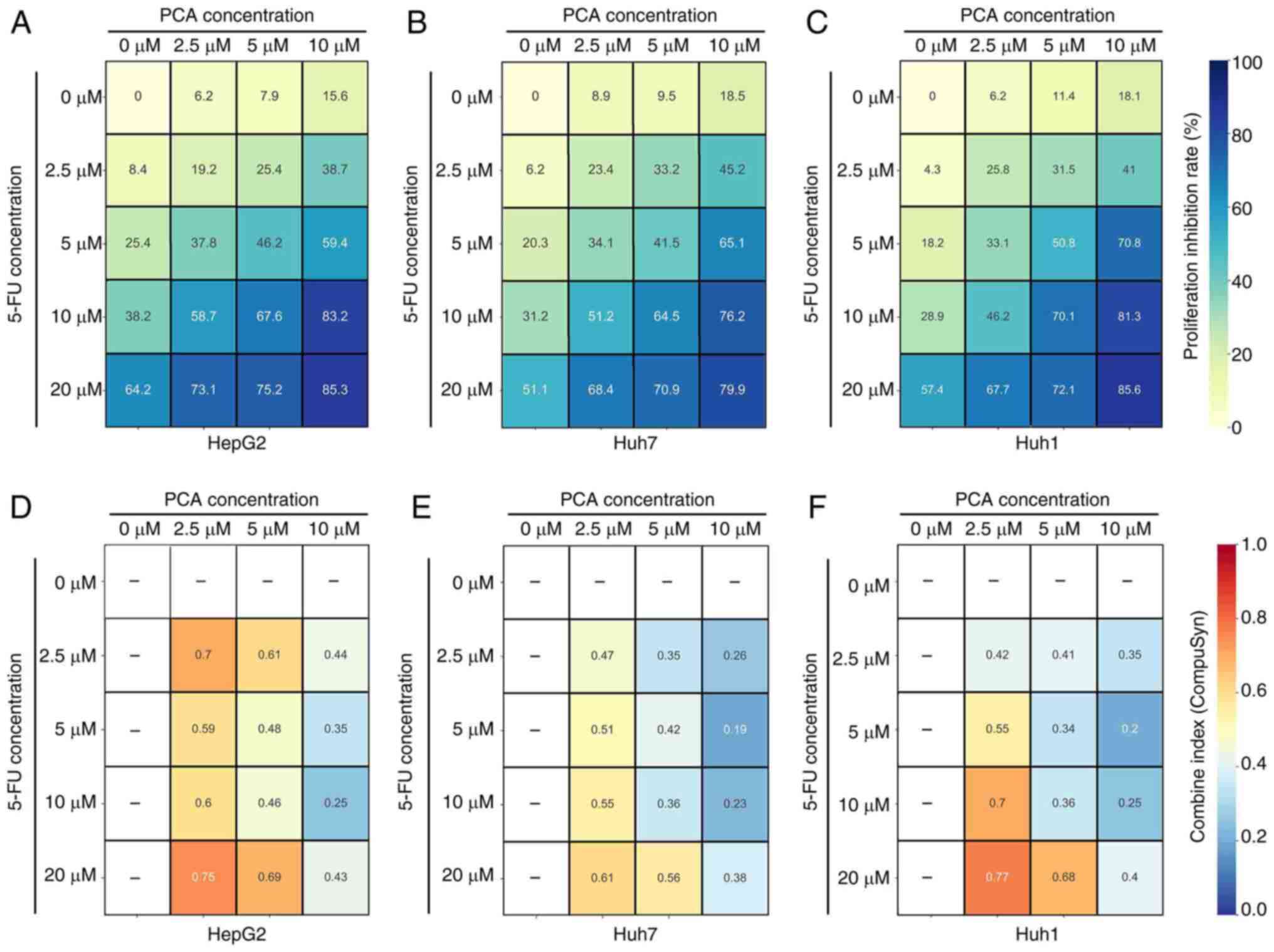
A previous study suggested that targeting ABCG2
could increase the sensitivity of liver cancer cells to
chemotherapeutic compounds, including 5-FU (14). Therefore, to assess whether PCA
could display synergistic effects with 5-FU, liver cancer cells
were treated with various concentrations of PCA (0, 2.5, 5 and 10
µM) combined with 5-FU (0, 2.5, 5 and 10 µM) and then a CCK-8 assay
was performed. Detailed proliferation inhibition rates of
concentration of each combination (PCA + 5-FU) in HepG2 (Fig. 3A), Huh7 (Fig. 3B) and Huh1 (Fig. 3C) are shown. Based on the
inhibition rates, Compusyn software was used to calculate the
synergistic index (also named combined index, CI) of each
combination. The analysis indicated that 10 µM PCA combined with 10
µM 5-FU had a strong synergistic effect in HepG2 (CI=0.25; Fig. 3D, Huh7 (CI=0.23; Fig. 3E) and Huh1 (CI=0.25; Fig. 3F) cells. Therefore, 10 µM PCA
combined with 10 µM 5-FU were considered to present the optimal
synergistic profile and these concentrations were therefore used
for the subsequent experiments.
PCA exerts synergistic effects with
5-FU on inducing the G1 phase arrest of liver cancer
cells
Cell cycle analysis indicated that 5-FU (10 µM)
slightly increased the number of HepG2, Huh7 and Huh1 cells in the
G1 phase of the cell cycle and slightly decreased those
in the S phase (Fig. 4A).
Additionally, PCA significantly amplified the effects of 5-FU on
regulating the cell cycle in liver cancer cells (Fig. 4A). Subsequently, the expression
levels of two biomarkers associated with the G1 phase of
the cell cycle, namely CDK4 and CDK6, were detected in liver cancer
cells treated with PCA (10 µM), 5-FU (10 µM) or their combination.
Therefore, cell treatment with PCA and 5-FU significantly
downregulated the protein and mRNA levels of CDK4 and CDK6 in HepG2
(Fig. 4B and E) and Huh7 (Fig. 4C and F) cells. However, the downregulation rate
was relatively low. Cell co-treatment with PCA and 5-FU could
significantly and acutely reduce the expression levels of both CDK4
and CDK6 in the aforementioned cell lines. In Huh1 cells, cell
treatment with PCA or 5-FU alone significantly downregulated the
protein and mRNA levels of CDK4, but not those of CDK6 (Fig. 4D and G). Cell co-treatment with both compounds
significantly and acutely reduced the expression levels of both
CDK4 and CDK6 (Fig. 4D and
G). The aforementioned results
suggested that PCA could exert a synergistic effect with 5-FU on
inducing G1 phase arrest in liver cancer cells.
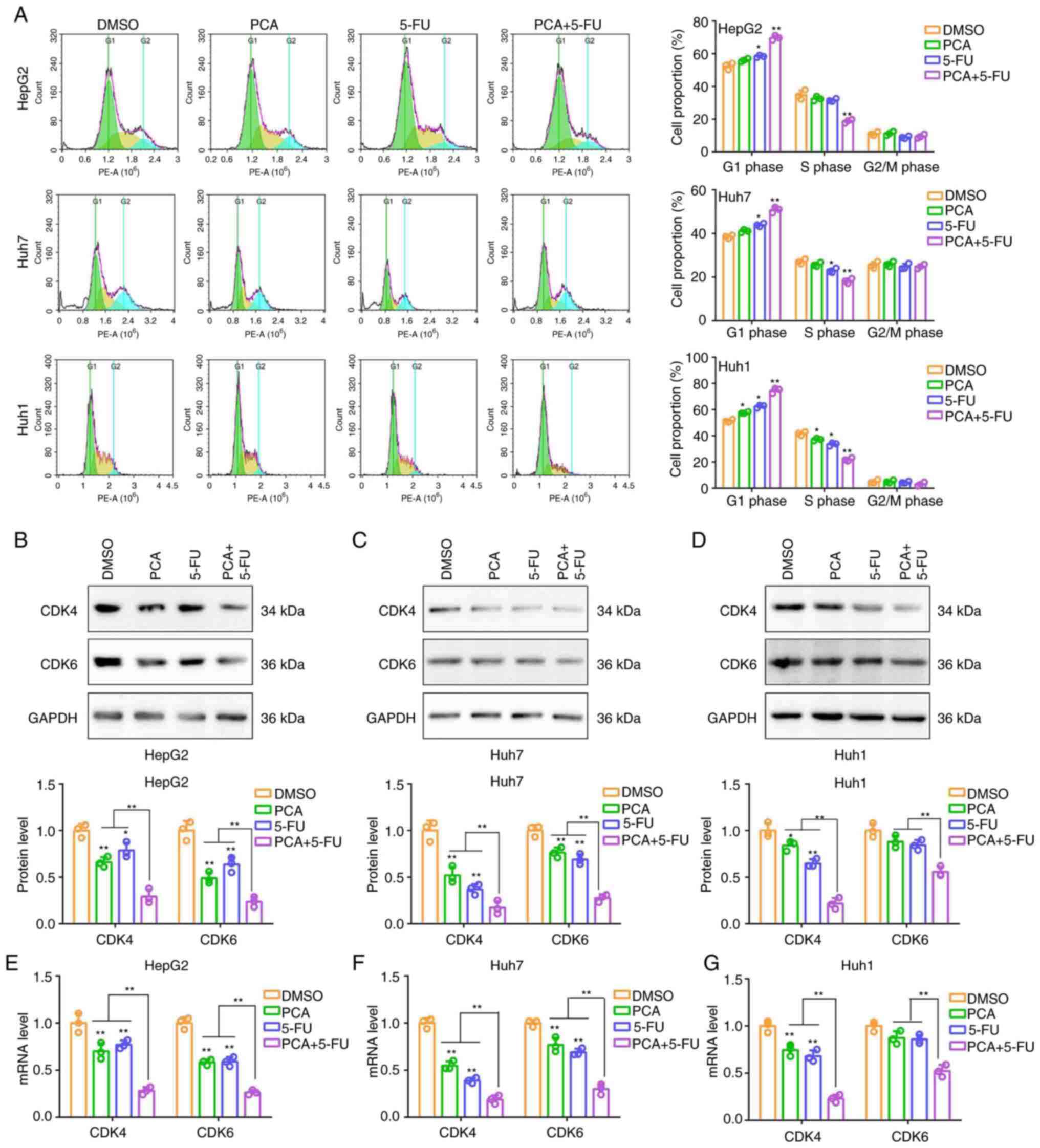 | Figure 4PCA combined with 5-FU induces the G1
phase cell cycle arrest in liver cancer cells. (A) Flow cytometry
was carried out to evaluate the cell cycle distribution in HepG2,
Huh7 and Huh1 cells treated with DMSO, 10 µM PCA, 10 µM 5-FU or
their combination. Western blot analysis was performed to detect
the protein expression levels of CDK4 and CDK6 in (B) HepG2, (C)
Huh7 and (D) Huh1 cells treated with DMSO, 10 µM PCA, 10 µM 5-FU or
their combination. Reverse transcription- quantitative PCR was
performed to detect the mRNA levels of CDK4 and CDK6 in (E) HepG2,
(F) Huh7 and (G) Huh1 cells treated with DMSO, 10 µM PCA, 10 µM
5-FU or their combination. *P<0.05 and
**P<0.01. PCA, purpurogallin carboxylic acid; 5-FU,
5-fluorouracil; CDK, cyclin-dependent kinase. |
PCA has synergistic effects with 5-FU
on suppressing the colony and spheroid formation abilities of liver
cancer cells
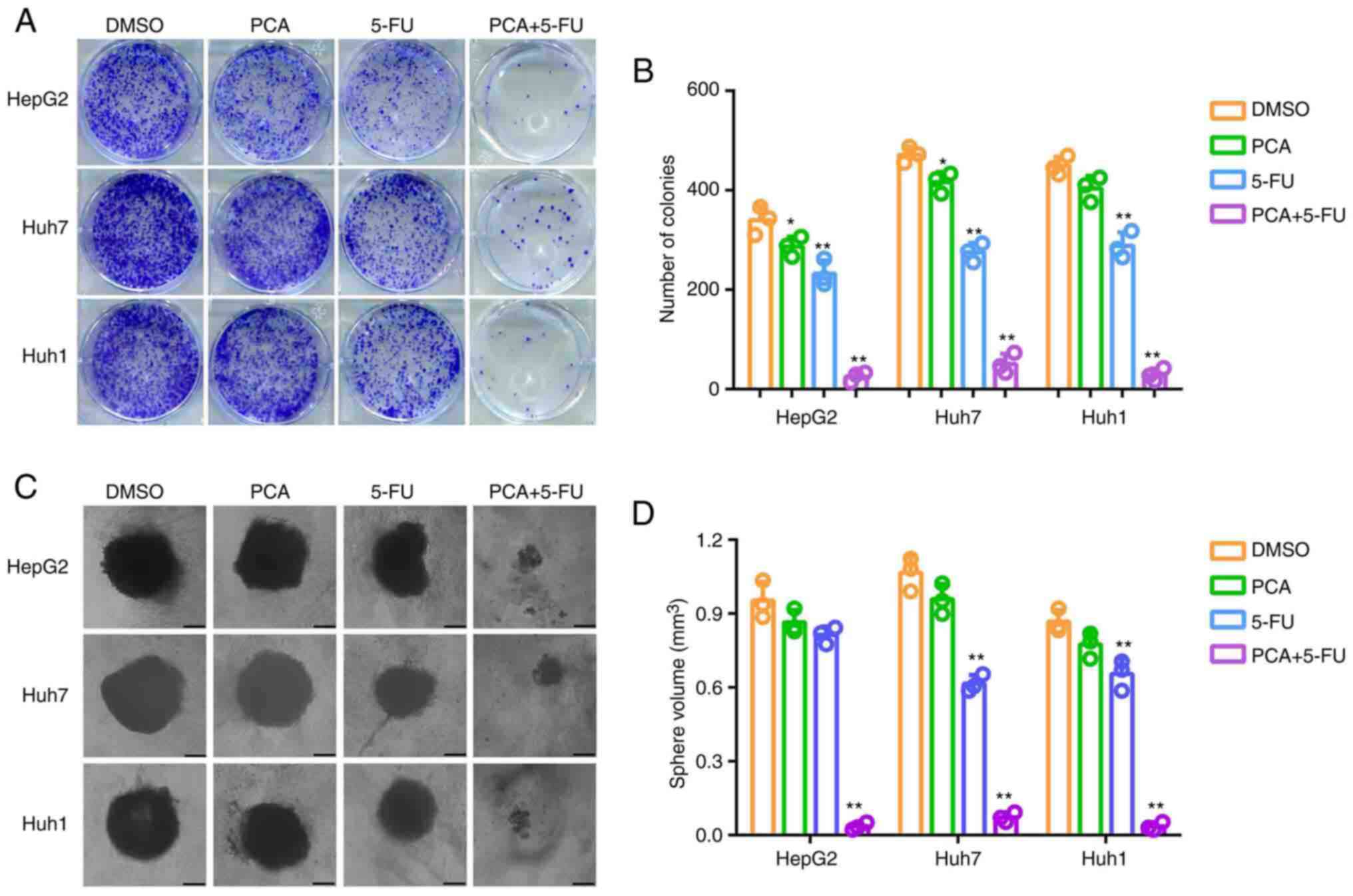
Furthermore, colony formation assays revealed that
5-FU significantly reduced the colony formation ability of liver
cancer cells, and PCA significantly amplified the effects of 5-FU
(Fig. 5A and B). Additionally, 3D sphere formation
assays demonstrated that compared with the number of spheres
derived from DMSO-treated cells, that derived for 5-FU-treated
cells was slightly lower. However, the number of spheres derived
from cells co-treated with 5-FU and PCA was significantly lower
(Fig. 5C and D). Taken together, the aforementioned
findings indicated that PCA exhibited synergistic effects with 5-FU
on attenuating the colony and spheroid formation abilities of liver
cancer cells.
PCA has no synergistic effects with
5-FU on inhibiting the proliferation and colony formation abilities
of ABCG2-depleted liver cancer cells
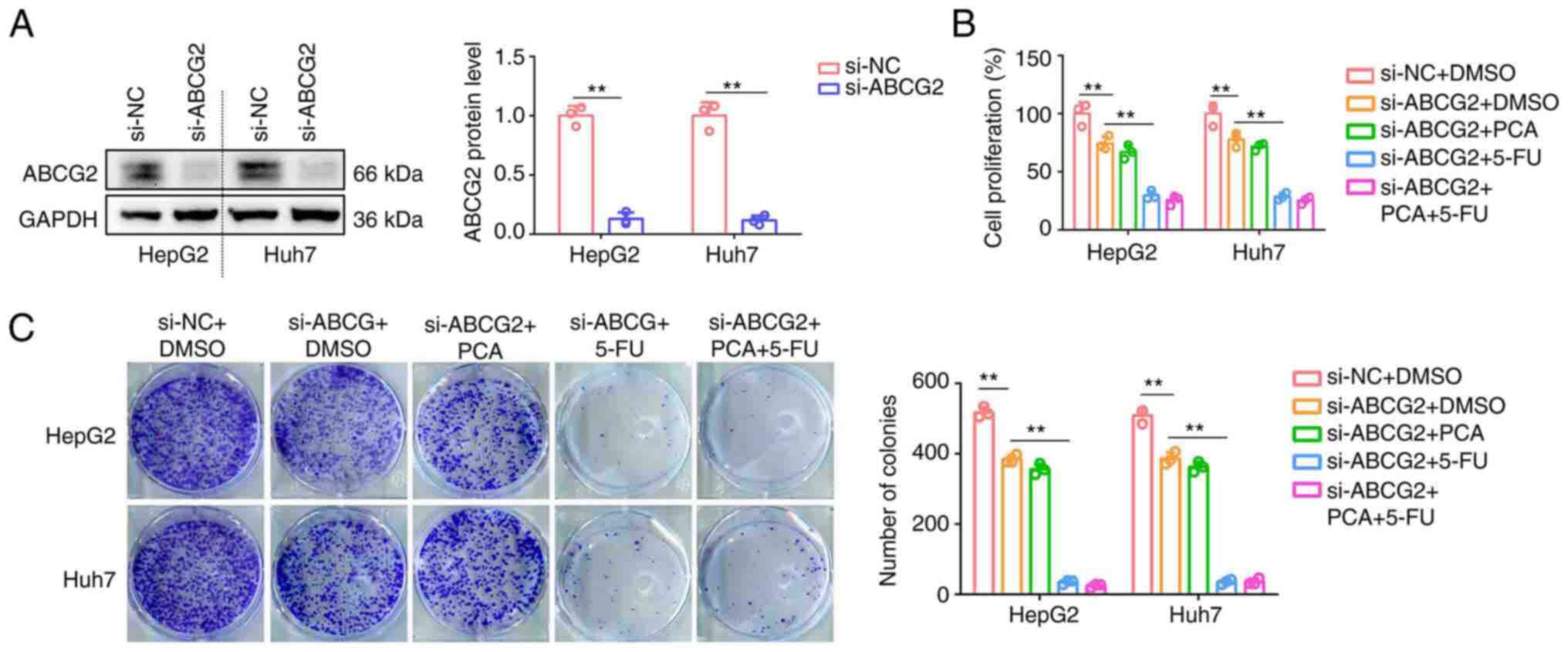
To reveal whether the synergistic effects of PCA and
5-FU were dependent on ABCG2, the expression of ABCG2 was silenced
in HepG2 and Huh7 cells (Fig. 6A).
Therefore, ABCG2 knockdown inhibited the proliferation and colony
formation abilities of HepG2 and Huh7 cells (Fig. 6B and C). Cell treatment with 5-FU could further
attenuate the proliferation and colony formation abilities of
ABCG2-depleted HepG2 and Huh7 cells (Fig. 6B and C). However, PCA had no effect on
inhibiting the aforementioned processes or enhancing the inhibitory
effects of 5-FU on ABCG2-depleted HepG2 and Huh7 cells (Fig. 6B and C). The aforementioned findings suggested
that the synergistic effects of PCA with 5-FU on liver cancer cells
were dependent on ABCG2 expression.
Discussion
5-FU, a nucleoside antimetabolite/analog of uracil,
is an effective antitumor drug, which is used to treat liver cancer
(16). However, patients with
liver cancer are at risk of developing acquired resistance to 5-FU,
thus limiting its clinical use (17). It has been reported that
p-glycoproteins can promote drug efflux, which is one of the key
factors associated with acquired drug resistance (18). Therefore, the development of drugs
that target p-glycoproteins could enhance the sensitivity of cancer
cells to 5-FU, thus promoting liver cancer therapy.
It has been reported that ABCG2, a member of the
p-glycoprotein family, plays a role in the resistance of cancer
cells to several therapeutic agents (19). As part of its normal function,
ABCG2 transports toxic metabolites from the fetal to the maternal
blood vessels of the placenta (20). ABCG2 upregulation in liver cancer
tissues was revealed to be associated with poor prognosis and
multi-drug resistance (13).
Targeting ABCG2 could be a significant strategy to enhance the
sensitivity of liver cancer cells to several drugs. A previous
study revealed that inhibition of ABCG2 could increase the effects
of doxorubicin on eliminating liver cancer stem cells (21). Similarly, ABCG2 knockdown could
enhance the sensitivity of liver cancer cells to sorafenib
(22). Therefore, the development
of drugs targeting ABCG2 could help liver cancer therapy. Actually,
several drugs targeting ABCG2 have been reported. For example,
isocorydine was demonstrated to suppress ABCG2 activity and induce
liver cancer cell apoptosis (23).
In addition, chrysin was found to inhibit the expression of ABCG2
and enhance the sensitivity of liver cancer cells to sorafenib
(24). However, the majority of
the aforementioned natural products exhibited indirect effects on
ABCG2. These drugs may mediate some toxic side effects through
other targets, while inhibiting the effects of ABCG2.
In the present study, network pharmacology, computer
virtual docking and enzymatic activity detection experiments
revealed that PCA had high binding affinity to ABCG2 and
discriminative activity against ABCG2. Consistently, PCA could
significantly inhibit the drug efflux capacity of liver cancer
cells. This evidence is consistent with previous theories (25,26)
that targeting ABCG2 reduces drug efflux. Actually, in comparison
with numerous previously discovered drugs, it is posited that PCA
exhibits enhanced specificity for ABCG2 binding, suggesting a
potential advantage of PCA.
Natural products are important molecular libraries
for screening the synergistic action of various drugs (27). In fact, the synergistic action
between 5-FU and several natural products has been widely reported.
Therefore, Zeng et al (28)
demonstrated that puerarin, a major isoflavone of the kudzu root,
displayed synergistic effects with 5-FU in liver cancer cells.
Additionally, Li et al (29) revealed that tetrandrine derivative
could target signal transducer and activator of transcription 3 to
enhance the efficacy of 5-FU on targeting liver cancer cells.
Furthermore, Cao et al (30) suggested that sinomenine combined
with 5-FU could synergistically suppress the proliferation of liver
cancer cells. However, the aforementioned drug combinations
revealed high non-specific toxicity, thus also inhibiting the
proliferation of normal cells. Furthermore, low synergy is another
issue.
In the present study, the results revealed that PCA,
within the pharmacological dose range, exhibited less non-specific
toxicity for normal hepatocytes. Further experiments demonstrated
that liver cancer cell (HepG2, Huh7 and Huh1) co-treatment with 10
µM PCA and 10 µM 5-FU exhibited a strong synergistic effect on
suppressing cell proliferation, and cell colony and spheroid
formation, as well as, on inducing G1 phase arrest.
However, the synergistic effect of PCA with 5-FU in liver cancer
cells was abrogated following ABCG2 knockdown, thus indicating that
the effects of PCA were dependent on ABCG2 expression. This
evidence aligned with prior research indicating that targeting
ABCG2 can increase the efficacy of chemotherapy drugs. Notably, in
comparison with numerous previously discovered drugs, low toxicity
is also an advantage of PCA. The present study may provide an
effective therapeutic strategy with low toxicity for liver cancer
treatment via increasing the sensitivity of cells to 5-FU.
In conclusion, the present study demonstrated that
PCA displayed synergistic effects with 5-FU on liver cancer cells
in vitro via targeting ABCG2. PCA combined with 5-FU could
be a potential strategy for liver cancer therapy. However, the
present study has some limitations. More importantly, the
synergistic effects of PCA and 5-FU on liver cancer was not
verified in vivo. Therefore, further studies should be
performed in the future to confirm the synergistic effect of these
two drugs in vivo.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by Task Book of TCM
Science and Technology Project in Shandong province (grant no.
2021M089).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
JL designed the experiments. PZ and WL performed the
experiments. SW analyzed the results and wrote the manuscript. All
authors read and approved the final version of the manuscript. JL
and PZ confirm the authenticity of all the raw data.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Cao W, Chen HD, Yu YW, Li N and Chen WQ:
Changing profiles of cancer burden worldwide and in China: A
secondary analysis of the global cancer statistics 2020. Chin Med J
(Engl). 134:783–791. 2021.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Chakraborty E and Sarkar D: Emerging
therapies for hepatocellular carcinoma (HCC). Cancers (Basel).
14(2798)2022.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Li QJ, He MK, Chen HW, Fang WQ, Zhou YM,
Xu L, Wei W, Zhang YJ, Guo Y, Guo RP, et al: Hepatic arterial
infusion of oxaliplatin, fluorouracil, and leucovorin versus
transarterial chemoembolization for large hepatocellular carcinoma:
A randomized phase III trial. J Clin Oncol. 40:150–160.
2022.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Ray EM and Sanoff HK: Optimal therapy for
patients with hepatocellular carcinoma and resistance or
intolerance to sorafenib: Challenges and solutions. J Hepatocell
Carcinoma. 4:131–138. 2017.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Xu T, Guo P, Pi C, He Y, Yang H, Hou Y,
Feng X, Jiang Q, Wei Y and Zhao L: Synergistic Effects of curcumin
and 5-fluorouracil on the hepatocellular carcinoma in vivo and
vitro through regulating the expression of COX-2 and NF-κB. J
Cancer. 11:3955–3964. 2020.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Dai W, Gao Q, Qiu J, Yuan J, Wu G and Shen
G: Quercetin induces apoptosis and enhances 5-FU therapeutic
efficacy in hepatocellular carcinoma. Tumour Biol. 37:6307–6313.
2016.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Zeng LH and Wu TW: Purpurogallin is a more
powerful protector of kidney cells than trolox and allopurinol.
Biochem Cell Biol. 70:684–690. 1992.PubMed/NCBI View
Article : Google Scholar
|
|
8
|
Zeng LH, Rootman DS, Burnstein A, Wu J and
Wu TW: Morin hydrate: A better protector than purpurogallin of
corneal endothelial cell damage induced by xanthine oxidase and
SIN-1. Curr Eye Res. 17:149–152. 1998.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Rambabu M and Jayanthi S: Virtual
screening of national cancer institute database for claudin-4
inhibitors: Synthesis, biological evaluation, and molecular
dynamics studies. J Cell Biochem. 120:8588–8600. 2019.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Mollazadeh S, Sahebkar A, Hadizadeh F,
Behravan J and Arabzadeh S: Structural and functional aspects of
P-glycoprotein and its inhibitors. Life Sci. 214:118–123.
2018.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Kodan A, Futamata R, Kimura Y, Kioka N,
Nakatsu T, Kato H and Ueda K: ABCB1/MDR1/P-gp employs an
ATP-dependent twist-and-squeeze mechanism to export hydrophobic
drugs. FEBS Lett. 595:707–716. 2021.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Zattoni IF, Delabio LC, Dutra JP, Kita DH,
Scheiffer G, Hembecker M, Pereira GDS, Moure VR and Valdameri G:
Targeting breast cancer resistance protein (BCRP/ABCG2): Functional
inhibitors and expression modulators. Eur J Med Chem.
237(114346)2022.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Chen YL, Chen PM, Lin PY, Hsiau YT and Chu
PY: ABCG2 overexpression confers poor outcomes in hepatocellular
carcinoma of elderly patients. Anticancer Res. 36:2983–2988.
2016.PubMed/NCBI
|
|
14
|
Kobayashi K, Higai K, Mukozu T, Matsui D,
Amanuma M, Yoshimine N, Ogino Y, Matsui T, Wakui N, Shinohara M, et
al: Tivantinib decreases hepatocyte growth factor-induced BCRP
expression in hepatocellular carcinoma HepG2 cells. Biol Pharm
Bull. 43:1421–1425. 2020.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Hou Z, Liu J, Jin Z, Qiu G, Xie Q, Mi S
and Huang J: Use of chemotherapy to treat hepatocellular carcinoma.
Biosci Trends. 16:31–45. 2022.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Li S, Gao M, Li Z, Song L, Gao X, Han J,
Wang F, Chen Y, Li W and Yang J: p53 and P-glycoprotein influence
chemoresistance in hepatocellular carcinoma. Front Biosci (Elite
Ed). 10:461–468. 2018.PubMed/NCBI View
Article : Google Scholar
|
|
18
|
Liu X: ABC family transporters. Adv Exp
Med Biol. 1141:13–100. 2019.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Mao Q and Unadkat JD: Role of the breast
cancer resistance protein (BCRP/ABCG2) in drug transport-an update.
AAPS J. 17:65–82. 2015.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Han LW, Gao C and Mao Q: An update on
expression and function of P-gp/ABCB1 and BCRP/ABCG2 in the
placenta and fetus. Expert Opin Drug Metab Toxicol. 14:817–829.
2018.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Yin W, Xiang D, Wang T, Zhang Y, Pham CV,
Zhou S, Jiang G, Hou Y, Zhu Y, Han Y, et al: The inhibition of
ABCB1/MDR1 or ABCG2/BCRP enables doxorubicin to eliminate liver
cancer stem cells. Sci Rep. 11(10791)2021.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Wang M, Wang Z, Zhi X, Ding W, Xiong J,
Tao T, Yang Y, Zhang H, Zi X, Zhou W and Huang G: SOX9 enhances
sorafenib resistance through upregulating ABCG2 expression in
hepatocellular carcinoma. Biomed Pharmacother.
129(110315)2020.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Lu P, Sun H, Zhang L, Hou H, Zhang L, Zhao
F, Ge C, Yao M, Wang T and Li J: Isocorydine targets the
drug-resistant cellular side population through PDCD4-related
apoptosis in hepatocellular carcinoma. Mol Med. 18:1136–1146.
2012.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Wei CT, Chen LC, Hsiang YP, Hung YJ, Chien
PH, Pan HL and Chen YJ: Chrysin-induced ERK1/2 phosphorylation
enhances the sensitivity of human hepatocellular carcinoma cells to
sorafenib. Anticancer Res. 39:695–701. 2019.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Guo X, To KKW, Chen Z, Wang X, Zhang J,
Luo M, Wang F, Yan S and Fu L: Dacomitinib potentiates the efficacy
of conventional chemotherapeutic agents via inhibiting the drug
efflux function of ABCG2 in vitro and in vivo. J Exp Clin Cancer
Res. 37(31)2018.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Bharathiraja P, Yadav P, Sajid A, Ambudkar
SV and Prasad NR: Natural medicinal compounds target signal
transduction pathways to overcome ABC drug efflux
transporter-mediated multidrug resistance in cancer. Drug Resist
Updat. 71(101004)2023.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Aung TN, Qu Z, Kortschak RD and Adelson
DL: Understanding the effectiveness of natural compound mixtures in
cancer through their molecular mode of action. Int J Mol Sci.
18(656)2017.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Zeng YP, Yang ZR, Guo XF, Jun W and Dong
WG: Synergistic effect of puerarin and 5-fluorouracil on
hepatocellular carcinoma. Oncol Lett. 8:2436–2442. 2014.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Li F, Wang J, Wu N, Zhang H, Li Z and Wei
N: H1, a derivative of tetrandrine, enhances the efficacy of 5-FU
in Bel7402/5-FU cells via suppressing STAT3/MCL-1 and inducing
PUMA. Biochem Biophys Res Commun. 520:93–98. 2019.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Cao J, Huang J, Gui S and Chu X:
Preparation, synergism, and biocompatibility of in situ liquid
crystals loaded with sinomenine and 5-fluorouracil for treatment of
liver cancer. Int J Nanomedicine. 16:3725–3739. 2021.PubMed/NCBI View Article : Google Scholar
|