Introduction
Recently, Japan's population has started aging and
the risk of contracting various age-related diseases has increased.
Therefore, analyzing component characteristics of aging and
examining their relationship with diseases to detect age-related
diseases at an early stage is necessary (1). Aging is often accompanied by an
increase in lifestyle-related diseases and many other diseases, and
early detection of disease is one of the most important issues,
especially in maintaining a healthy life expectancy.
At the cell level, senescence is a state in which
cells cease to divide and undergo distinctive phenotypic changes,
including altered gene expression and the secretion of
senescence-associated secretory phenotype (SASP) factors (2). SASP has profound effects, not only on
the senescence cells themselves, but also on the surrounding tissue
microenvironment to promote inflammation, tissue dysfunction, and
progression of aging- and senescence-related diseases (3,4).
Cellular senescence may be readily induced by repeated passage of
cultured cells or irradiation (5,6).
Recently, extracellular vesicles (EVs), such as
exosomes, have been reported to be involved in the onset and
progression of aging and aging-related diseases (7). EVs are small particles released from
cells that contain DNA, RNA, proteins, and lipids (8,9).
They may serve as useful biomarkers for various diseases because
they reflect the physiological state of the releasing cells
(10-12).
miRNAs (microRNAs) are known to be abundant among the various RNAs
contained in EVs (13). miRNAs are
approximately 21-24 nucleotides in length and are small noncoding
RNAs that bind to target mRNAs and either suppress translation or
promote mRNA degradation to post-transcriptionally regulate gene
expression (14). The role of
miRNAs in cellular senescence has been shown to influence many
biological processes associated with aging, including cell
proliferation, apoptosis, and the inflammatory response (15,16).
For example, miR-125b, miR-504, miR-25, and miR-30d directly act on
p53 and suppress its function (17). miR-21-5p is a small noncoding RNA
that was first reported as an miRNA in cancer-related studies
(18). It has been reported that
miR-21 increases with age in the heart of mice (19). It reportedly inhibits cell cycle
progression via CDC25A (20).
EVs are abundant in body fluids, such as blood, and
they are secreted by various cells throughout the body and are
present in body fluids in a mixed state (21). Blood is in constant contact with
vascular endothelial cells, and it is believed that EVs-derived
from vascular endothelial cells are abundant in the blood (22,23).
RNA-sequencing analysis of endothelial cells and fibroblasts
induced into senescence by irradiation revealed gene expression
changes when senescence is induced, including senescence-specific
gene expression patterns (24).
However, the relationship between changes in the expression of
blood miRNAs and vascular endothelial cell senescence has not been
examined.
In this study, we examined the characteristics of
serum miRNA expression in aging mice and the expression of miRNAs
inside and outside the cells of senescence-induced vascular
endothelial cells. Our results provide insight into
senescence-induced and secreted miRNAs in vascular endothelial
cells.
Materials and methods
Mice and blood collection
C57BL6NJcl male mice were purchased from CLEA Japan.
They were acclimated for at least one week before initiating the
experiments. The mice were provided a solid diet of CE2 (CLEA
Japan) and water ad libitum and were housed in a
conventional animal room with 12 h light/dark cycles at room
temperature and 40-50% humidity. The mice were housed up to 5 mice
per cage containing bedding, feed, and water, which were changed
weekly. The mice were observed 2-3 times/day for monitoring. No
abnormalities in mouse health or behavior were observed. Five
8-week-old mice were designated ‘Young’ mice, and five 82-week-old
and three 102-week-old mice were designated ‘Aging’ mice. Small
animal anesthesia machines (Muromachi Kikai) were used to
anesthetize the mice. Isoflurane vaporized to a concentration of
4-5% was administered to the mice and maintained at 2-3% throughout
the experiment. Following anesthesia, approximately 0.5-1.0 ml of
blood was drawn from the heart, and the mice were promptly
cervically dislocated to minimize distress. The time from the
beginning of anesthesia to the end of blood collection was less
than 10 min per animal. Death was confirmed by respiratory and
cardiac arrest. Blood was placed in a Microtainer (cat. no. 365967,
Becton Dickinson) for serum separation. After checking for
coagulation, the blood was centrifuged at 6,000 x g for 3 min. The
serum was separated and stored at -80˚C until use. The experiment
was approved by the Hirosaki University Animal Experiment Ethics
Committee and conducted based on the Hirosaki University Animal
Experiment Guidelines (Approval No. AE01-2023-097-1).
Serum RNA extraction
The serum was filtered through a 0.20 µm filter and
the RNAs derived from EVs were extracted from 200 µl of serum using
the exoRNeasy midi kit (cat. no. 77144, Qiagen) and cel-miR-39 was
added as a spike-in RNA. The concentration of the extracted
EV-derived RNAs was measured using a Qubit™ microRNA Assay Kit
(cat. no. Q32880, ThermoFisher Scientific) and a Qubit 4
Fluorometer (cat. no. Q33238, ThermoFisher Q33238, ThermoFisher
Scientific) based on the manufacturer's protocol. To confirm the
size of the RNAs in the serum EVs, an Agilent 2100 Bioanalyzer
(Agilent Technologies, Inc.) and the Agilent RNA 6000 Pico kit
(cat. no. 5067-1513, Agilent Technologies, Inc.) were used to
perform RNA electrophoresis based on the manufacturer's
protocol.
miRNA microarray
To examine miRNA expression in mouse serum, a miRNA
microarray analysis was performed using 1.8 ng of RNAs extracted by
the method described above. Microarray analysis was performed as
previously described according to the manufacturer's instructions
(25,26). The miRNA Complete Labeling Reagent
and Hyb kit (cat. no. 5190-0456, Agilent Technologies) was used to
label mouse serum EVs-derived RNAs with Cyanine-3 and hybridized to
SurePrint G3 Mouse 8x60 K miRNA microarray slides (version 21.0)
(cat. no. G4872A, design ID: 070155, Agilent Technologies) at 55˚C
for 20 h. The fluorescence signals were detected using a SureScan
Microarray Scanner (Agilent Technologies) and quantified using
Feature Extraction software (Agilent Technologies). The microarray
data were analyzed using R (version 4.3.1) and quantile
normalization was done using limma (27). Microarray data were analyzed for
genes with a fold-change cutoff of ≥1.5 (28,29),
as the commonly used cutoff in various studies detecting genes in
aging mice compared with young mice. A t-test was performed to
identify genes with P-values <0.05. The resulting microarray
data were registered to Gene Expression Omnibus (GSE274943).
Reverse transcription-quantitative PCR
(RT-qPCR) of serum EV miRNAs
RT-qPCR was used to validate the miRNAs in mouse
serum. Total RNA and reverse transcriptase were incubated at 37˚C
for 1 h for cDNA synthesis. Then, qPCR was performed using 10-fold
diluted cDNA, TB Green Advantage qPCR Premix (cat. no. 639676,
Takara Bio), and miRNA-specific primers (Table SI) based on the manufacturer's
protocol. The conditions for qPCR were as follows: cDNA was
denatured at 95˚C for 10 sec in a total volume of 25 µl, followed
by 40 cycles of 95˚C for 5 sec and 60˚C for 20 sec. The cDNA was
then incubated at 95˚C for 5 sec and 60˚C for 20 sec for 40 cycles.
Cel-miR-39 was used as an external control. The quantitation of
gene expression was done using the 2-ΔΔCq method
(30).
Reanalysis of RNA-sequence data
To evaluate gene expression in senescence-induced
HUVECs, RNA-sequence data of HUVECs exposed to 4 Gy of radiation
for 10 days were downloaded from the Sequence Read Archive (SRA)
(GSE130727, SRR9016151-SRR 9016156). FASTQ files downloaded from
SRA were quality-checked and trimmed using Fastp (31). Gene expression levels were
calculated from FASTQ files using salmon (32) and tximport (33). R (version 4.3.1) was used for data
analysis and quasi-likelihood F-tests were used with the expression
analysis packages, limma (27) and
edgeR (34). The p-values for
multiple correction by the Benjamini-Hochberg method were less than
0.05. In addition, genes with a fold-change ≥1.5 are selected.
Hierarchical clustering and principal component analysis (PCA) were
done to examine differences in gene expression between the 4
Gy-irradiated and unirradiated HUVECs. Gene Set Enrichment Analysis
(GSEA) and Gene Ontology (GO) analysis were performed using
ClusterProfiler (35), fgsea, and
the AnnotationDbi package. Kyoto Encyclopedia of Genes and Genomes
(KEGG) and pathway analysis using ReactomePA were also conducted
(36).
Cell culture
HUVECs, single donor P1 (cat. no. C-12200, Promo
Cell) were purchased from Takara Bio, and cultured in Basic Cell
Growth Medium 2 Kit (cat. no. C-22011, Promo Cell) with 5% Fetal
Calf Serum (FCS), 5.0 ng/ml Epidermal Growth Factor (recombinant
human), 10 ng/ml Basic Fibroblast Growth Factor (recombinant
human), 10 ng/ml Insulin-like Growth Factor (Long-type), 20 ng/ml
Insulin-like Growth Factor (Long R3 IGF, recombinant human), 1.0
µg/ml ascorbic acid, 22.5 µg/ml heparin, and 0.2 µg/ml
hydrocortisone. For the recovery of EVs, exosome-depleted fetal
bovine serum (cat. no. 558-39501, Fujifilm Wako) was used instead
of FCS in the above medium. 41220, Promo Cell) to pass the
HUVECs.
X-ray irradiation
HUVECs from P2 to P6 were seeded at 1x105
cells/60 mm dish and incubated overnight at 37˚C in a 5%
CO2 atmosphere. HUVECs were irradiated with 4 Gy X-rays
(MBR-1520R-3 X-ray machine, Hitachi Ltd.) at a dose rate of 1.0
Gy/min (150 kVp, 20 mA, 0.5 mm aluminum, and 0.3 mm copper
filters).
RNA extraction from HUVECs
Total RNA was extracted from HUVECs that were
cultured for 10 days after 4 Gy irradiation. The control group
consisted of HUVECs that were passaged on day 5 and cultured for 10
days to avoid becoming 100% confluent. Intracellular RNA was
extracted from cell pellets after centrifuging at 300 g for 3 min
using the miRNeasy mini kit (cat. no. 217004, Qiagen) based on the
manufacturer's protocol. The concentration of the extracted RNA was
measured using a Nanodrop instrument (ThermoFisher Scientific)
based on the manufacturer's protocol.
The HUVEC culture supernatant was filtered through a
0.20 µm filter and RNAs from the EVs were extracted from 2 ml of
culture supernatant using the exoRNeasy midi kit (cat. no. 77144,
Qiagen). The concentration of RNAs in the extracted EVs was
determined using the Qubit™ microRNA Assay Kit (cat. no. Q32880,
ThermoFisher Scientific) and Qubit 4 Fluorometer (cat. no. Q33238,
ThermoFisher Scientific) based on the manufacturer's protocol.
RT-qPCR from HUVECs
RT-qPCR was performed to confirm the expression
change of miR-21-5p in HUVECs cultured for 10 days after 4 Gy
irradiation. Total RNA was reverse-transcribed into cDNA at 37˚C
for 1 h. qPCR was performed with 10-fold diluted cDNA using the TB
Green Advantage qPCR Premix (cat. no. 639676, Takara Bio) and
miRNA-specific primers (Table SI)
based on the manufacturer's protocol. U6 small nuclear RNA was used
as an internal control. In the Mir-X miRNA First-Strand Synthesis
kit (cat. no. 638313, Takara Bio) containing the U6 primer, the U6
primer sequence is a trade secret and not disclosed.
RT-qPCR was used to validate the messenger RNAs
(mRNAs) of the candidate miR-21-5p target genes and genes that were
downregulated by RNA-sequencing reanalysis in HUVECs cultured for
10 days following 4 Gy irradiation. Candidate target genes were
downloaded from TargetScan (https://www.targetscan.org/vert_72/). cDNA synthesis
was done using 200 ng of total RNA extracted from HUVECs cultured
for 10 days after 4 Gy irradiation with the High Capacity cDNA
Reverse Transcriptase Kit (cat. no. 4368814, ThermoFisher
Scientific) based on the manufacturer's protocol. Total RNA (200
ng) and 2x Reverse Transcriptase Master Mix solution were mixed in
a total volume of 20 µl. The reverse transcription reaction was
performed at 25˚C for 10 min, 37˚C for 120 min, and 85˚C for 5 min.
Next, qPCR was done using 5-fold diluted cDNA, Power SYBR Green PCR
Master Mix (2x) (cat. no. 4367659, ThermoFisher Scientific), and
gene-specific primer pairs (Table
I) based on the manufacturer's protocol. Of the primers used,
primer 3 (version 4.1.0) was designed for RT-qPCR of mRNA. Actin
beta was used as an internal control.
 | Table IPrimer pairs for reverse
transcription-quantitative PCR. |
Table I
Primer pairs for reverse
transcription-quantitative PCR.
| Primer name | Sequence
(5'-3') | Amplicon size,
bp |
|---|
| CDC25A
forward |
CTACTGATGGCAAGCGTGTC | 88 |
| CDC25A
reverse |
TCTCTCTCACATACCGGCAC | |
| MSH2
forward |
CATGTCACAGCACTCACCAC | 99 |
| MSH2
reverse |
GCTCTGCAACATGAATCCCA | |
| MTAP
forward |
TTCTTGTGCCAGAGGAGTGT | 102 |
| MTAP
reverse |
CACCGGAGTCCTAGCTTCTT | |
| MTHFD1
forward |
AAAGAGAGGGCGAGCTTCAT | 97 |
| MTHFD1
reverse |
AACGCTTGGCACTCTCTACT | |
| ZNF367
forward |
CACATCAGCGTCTTCACACC | 83 |
| ZNF367
reverse |
CGGTTTGCATGGGTGAATCT | |
| ZNF704
forward |
CTCGCTCCATCTGTCTCCTT | 116 |
| ZNF704
reverse |
CATTGCTGCTGTCACCTTGT | |
| ACTB
forward |
CCAACCGCGAGAAGATGA | 97 |
| ACTB
reverse |
CCAGAGGCGTACAGGGATAG | |
RT-qPCR was performed for changes in the expression
of miR-21-5p in EVs in culture supernatants secreted from HUVECs
cultured for 10 days after 4 Gy irradiation. Total RNA (100 µg) was
extracted from EVs in the culture supernatants secreted from HUVECs
10 days after 4 Gy irradiation. The Mir-X miRNA First-Strand
Synthesis kit (cat. no. 638313, Takara Bio) was used for cDNA
synthesis based on the manufacturer's protocol. Total RNA and
reverse transcriptase were incubated at 37˚C for 1 h. Then, qPCR
was performed using 10-fold diluted cDNA using TB Green Advantage
qPCR Premix (cat. no. 639676, Takara Bio) and miRNA-specific
primers (Table SI) based on the
manufacturer's protocol. The respective miRNA sequences published
on miRbase were used as primer sequences. Cel-miR-39 was used as an
external control. The quantitation of gene expression was done
using the 2-ΔΔCq method (30).
Senescence-associated β-galactosidase
(SA-β-GAL) staining
To detect cell senescence in HUVECs cultured for 10
days following 4 Gy irradiation, the senescence β-galactosidase
staining kit (cat. no. 9860, Cell Signaling Technology) was used.
HUVECs (1x105 cells) were treated with 1x Fixative
Solution. After washing three times with phosphate-buffered saline
(PBS), 1.0 ml of β-galactosidase stain was added and incubated at
37˚C overnight. HUVECs were observed under an optical microscope
with the β-galactosidase staining solution remaining.
To quantify cellular senescence in HUVECs, the
cellular senescence detection kit SPiDER-βGal (cat. no. SG02,
Dojindo Molecular Technologies, Inc.) was used. HUVECs
(1x105 cells) were collected and incubated with Hank's
Balanced Salt Solution (HBSS). After washing twice with HBSS, 1.0
ml of the prepared SPiDER-βGal working solution was added and
incubated in a 5% CO2 incubator at 37˚C for 1 h. After
washing twice with HBSS, 1.0 ml of the prepared SPiDER-βGal working
solution was added and incubated at 37˚C in a 5% CO2
incubator for 30 min. The supernatant was removed by aspiration,
the cells were washed twice with HBSS, and detached with the
DetachKit (cat. No. C-41220, Promo Cell). The fluorescence of the
detached HUVECs was analyzed using CytoFLEX flow cytometer and
CytExpert software version 2.4 (both Beckman Coulter) and kaluza2.2
software (Beckman Coulter).
Cell cycle analysis
To confirm the cell cycle of 4 Gy-irradiated and
nonirradiated HUVECs, each exfoliated cell was fixed by adding 70%
ethanol and incubated at -20˚C overnight or longer. Then, each
exfoliated cell was washed three times with PBS, and 200 µl of 250
µg/ml RNase A (cat. no. 318-06391, Fujifilm Wako) was added and
incubated at room temperature for 30 min. Propidium iodide was then
added at 50 µg/ml and fluorescence was detected with a CytoFLEX
flow cytometer and CytExpert software version 2.4 (both Beckman
Coulter). All events were measured up to 10,000 events per sample.
For data analysis, histograms were generated using kaluza2.2
software (Beckman Coulter).
Detection of EV markers
HUVEC culture supernatant (2 ml) was centrifuged at
300 g for 3 min. The collected supernatant was passed through a
0.20 µm filter and enriched for EVs by ultrafiltration using an
Amicon Ultra 0.5 ml centrifugal filter (cat. No. UFC510096, Merke
Millipore). EVs were then collected using the PS capture™ Exosome
Flow Cytometry Kit (cat. no. 297-79701, Fujifilm Wako) based on the
manufacturer's recommended protocol. Fluorescein isothiocyanate
(FITC) anti-human CD63 Antibody (cat. no. 353005 BioLegend), FITC
anti-human CD9 Antibody (cat. no. 312103, BioLegend), FITC
anti-human CD81 Antibody (cat. no. 349503, BioLegend), and FITC
Mouse IgG1, κ isotype Ctrl antibody (cat. no. 400110, BioLegend)
were used. The data were analyzed using a CytoFLEX flow cytometer
and CytExpert software version 2.4 (both Beckman Coulter). For data
analysis, histograms were generated using kaluza2.2 software
(Beckman Coulter).
Statistical analysis
All statistical analyses were performed using R
(version 4.3.1). Two-group tests were subject to the Shapiro-Wilk
test to confirm normality, followed by the F test, and Welch's
T-test for data without equal variance. Mann-Whitney's U test was
performed if normality was rejected. The significance level was set
at P<0.05. For all experiments using HUVECs, the number of
samples was n=4.
Results
Expression of blood EV mmu-miR-21a-5p
is increased in aging mice
In our previous study, we found that cognitive
function was impaired in 58-week-old mice in the Morris water maze
(28). To further confirm changes
in the expression of miRNAs in blood EVs in aged mice, we compared
EVs RNAs from ‘Aging’ (82 and 102 weeks old) and ‘Young’ (8 weeks
old) mice. The size of the EV RNA in the serum of the Aging and
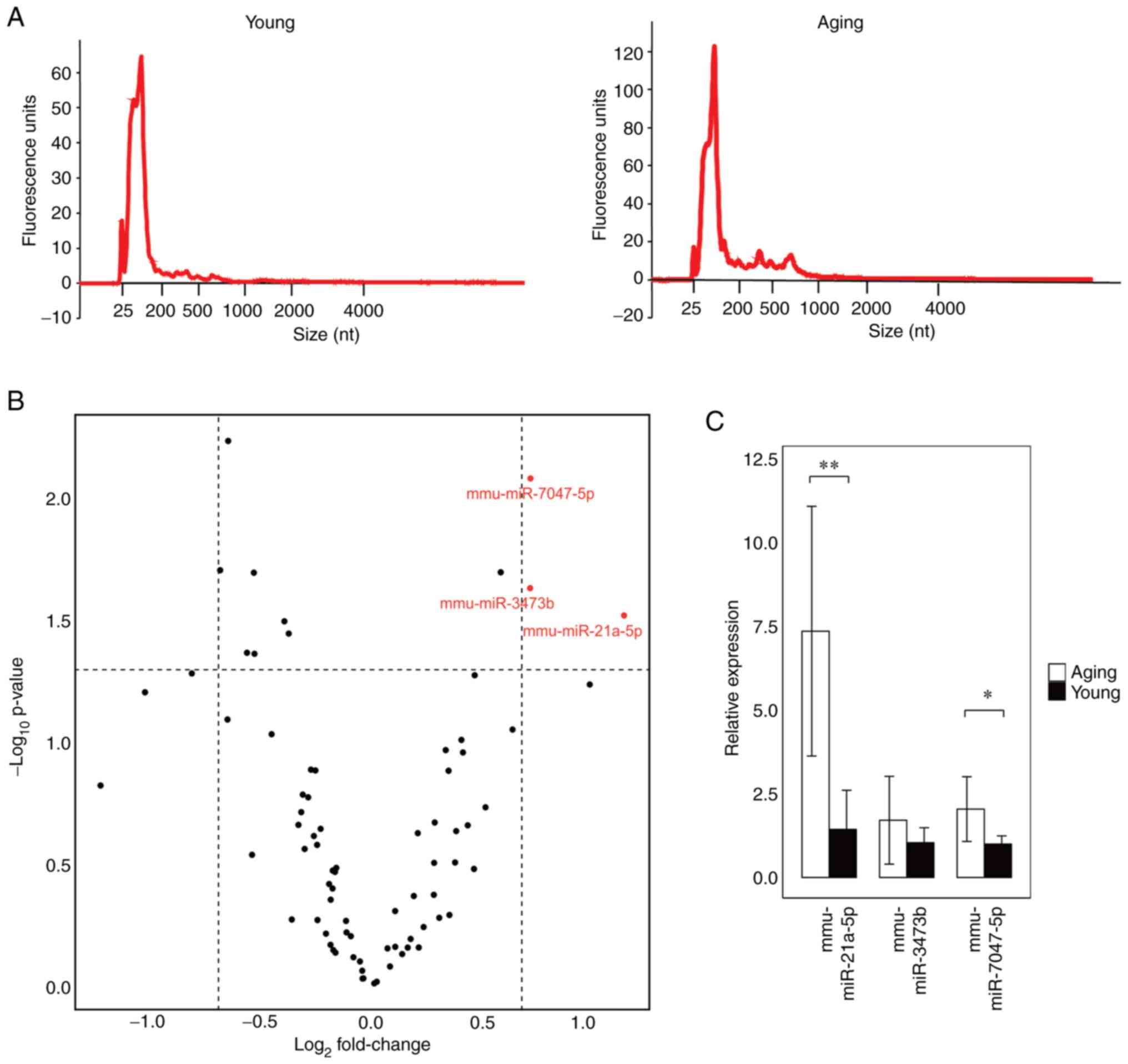
Young mice was found to be in the range of 25-200 nucleotides (nt)
(Fig. 1A). We performed a mouse
miRNA microarray using the extracted serum RNAs to identify miRNAs
whose expression was significantly altered in Aging mice compared
with Young mice. We found that mmu-miR-21a-5p, mmu-miR-3473b, and
mmu-miR-7047-5p were significantly increased in the serum of the
Aging mice (Fig. 1B). RT-qPCR was
used to confirm the expression of the three miRNAs that were
significantly increased. We found that mmu-miR-21a-5p and
mmu-miR-7047-5p were significantly increased in Aging mice
(Fig. 1C). This suggests that
these two blood miRNAs are molecules associated with aging. One
possible reason for the discrepancy between PCR results and
microarray results is that microarrays and RT-qPCR have different
sensitivities, so a sample that is statistically significant by
microarray may not be statistically significant by RT-qPCR.
Differentially expressed genes in
HUVECs irradiated with 4 Gy
The altered miRNAs described above may be derived
from the senescence of vascular endothelial cells (22,23).
Therefore, we considered the possibility that the senescence of
vascular endothelial cells affects the expression of miRNAs in
blood EVs. To confirm the gene expression changes in
senescence-induced HUVECs, we downloaded and reanalyzed the
RNA-sequence data (GSE130727, SRR9016151-SRR9016156) of 4
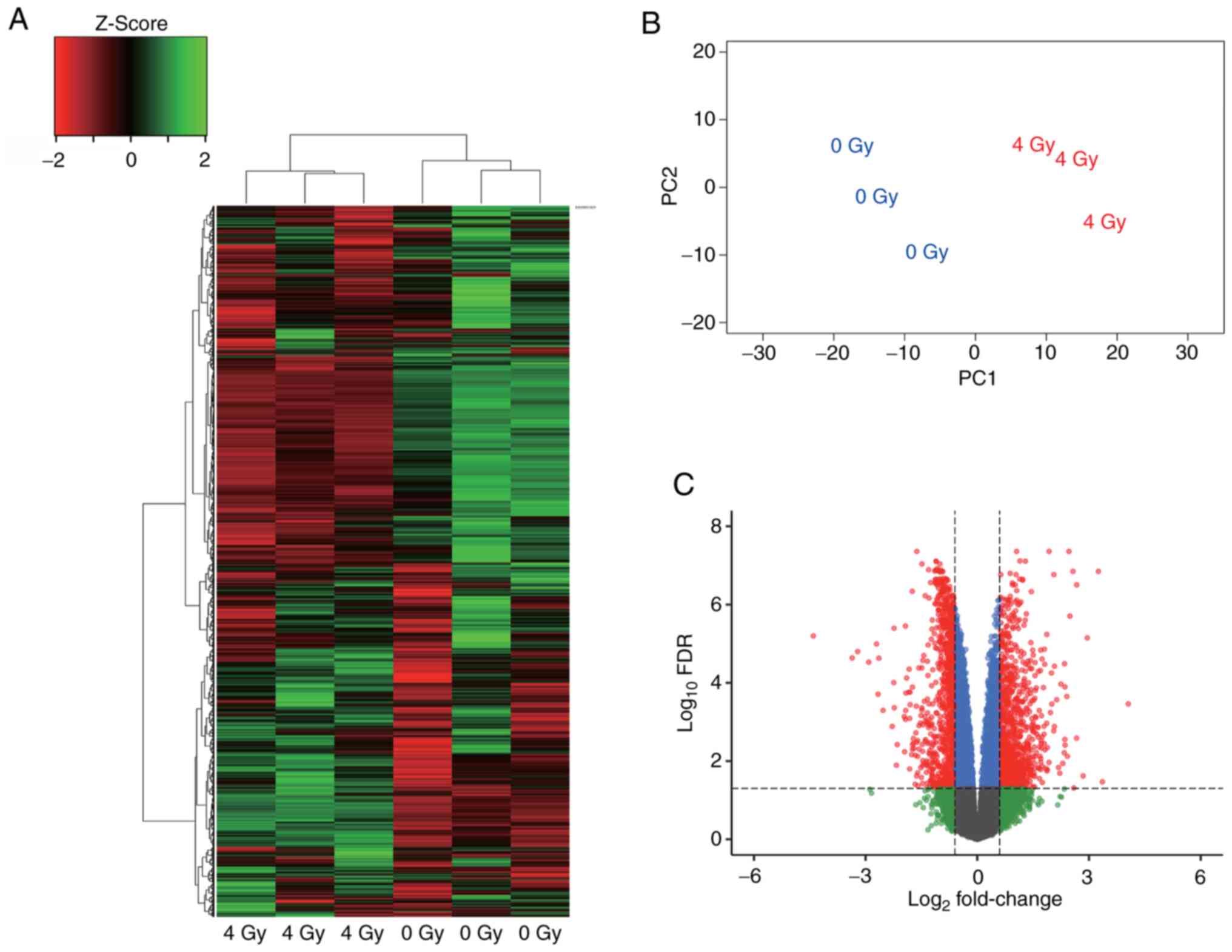
Gy-exposed HUVECs. Cluster analysis revealed that gene expression
was different between 4 Gy-irradiated and nonirradiated HUVECs
(Fig. 2A). Similarly, PCA showed a
significant change in gene expression between the 4 Gy-irradiated
and nonirradiated HUVECs (Fig.
2B). Furthermore, differentially expressed gene (DEG) analysis
using edgeR identified 1881 genes that were altered by ≥1.5-fold
(Fig. 2C). This reanalysis
indicated that 4 Gy irradiation alters the expression of many HUVEC
genes.
4 Gy irradiation induces changes in
the expression of genes involved in cell division and cell cycle in
HUVECs
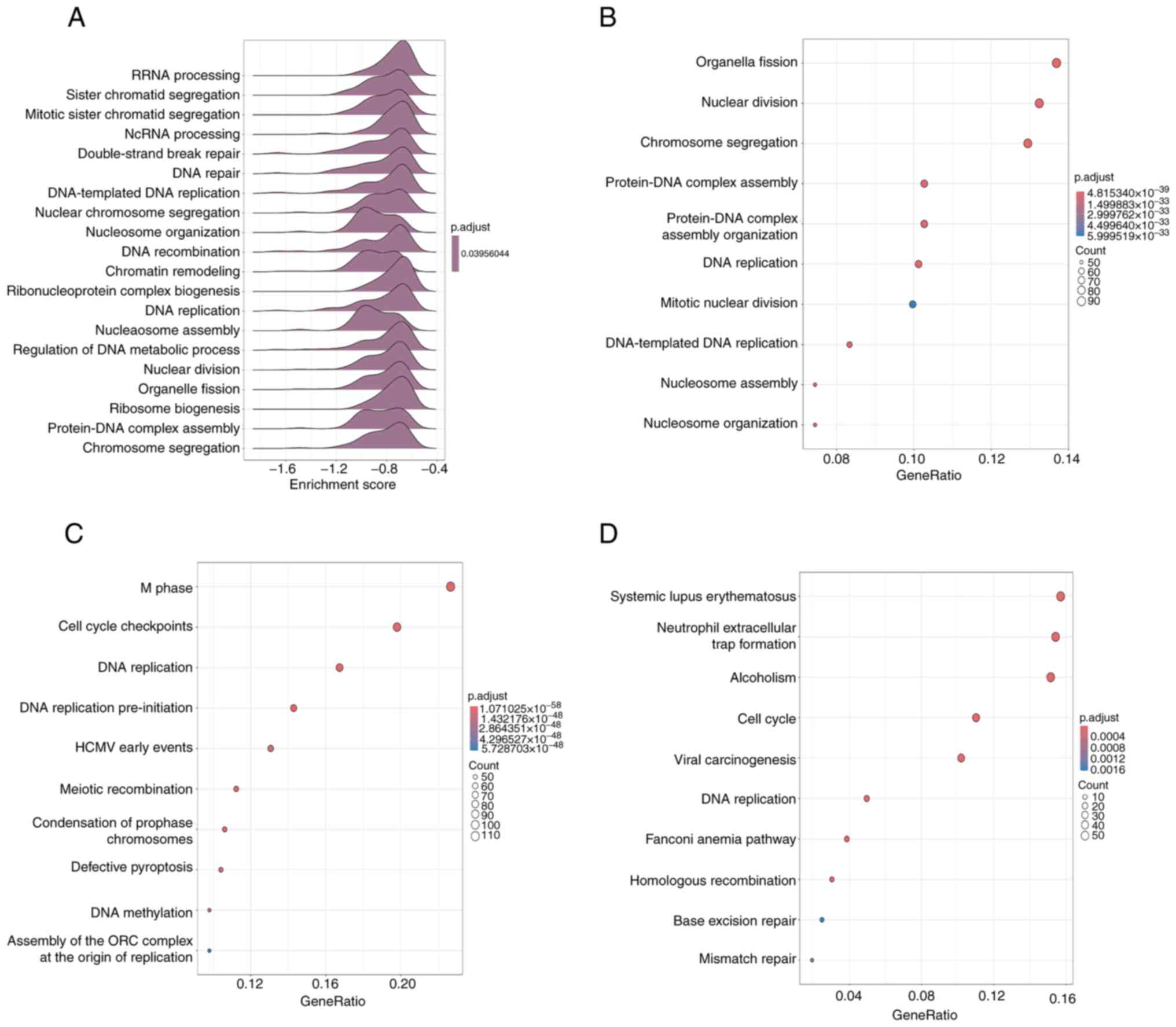
Enrichment analysis was performed on 1881 genes that
were altered by DEG analysis of the RNA sequences. A GSEA analysis
was performed on the GO terms to determine whether senescence
induction of HUVECs by 4 Gy irradiation was affected by a decrease
or increase of the altered genes. The results indicated that most
of the variable genes were enriched in the downregulated gene group
because the enrichment score was less than zero (Fig. 3A). Based on this result, GO
enrichment analysis was performed on the downregulated genes, and
terms associated with DNA replication and cell division were
identified (Fig. 3B). In addition,
KEGG pathway and Reactome pathway enrichment analyses were also
performed and pathways related to the cell cycle and DNA repair
were annotated (Fig. 3C and
D). These results suggest that
there are changes in DNA replication and cell cycle-related genes
in 4 Gy-irradiated senescence-induced HUVECs.
Senescence-induced HUVECs cause
decreased expression of 6 genes, a miR-21-5p target gene, and
change the rate of cell cycle
We identified mmu-miR-21a-5p and mmu-miR-7047-5p as
miRNAs that are significantly up-regulated in Aging mice (Fig. 1); however, because no miRNA
corresponding to mmu-miR-7047-5p was found in humans, we focused
only on miR-21-5p in this study. The results of an RNA-sequence
enrichment analysis showed that the expression of 435 genes was
decreased by 4 Gy irradiation. The common genes among the 378
candidate target genes of miR-21-5p were examined by target scan,
and cell division cycle 25A (CDC25A), mutS homolog 2
(MSH2), methylthioadenosine phosphorylase (MTAP),
methylenetetrahydrofolate dehydrogenase, cyclohydrolase and
formyltetrahydrofolate synthetase 1 (MTHFD1), zinc finger
protein 367 (ZNF367), and zinc finger protein 704
(ZNF704) were identified (Fig.
4A). Next, to induce senescence in HUVECs, the cells were
cultured for 10 days after 4 Gy irradiation. Total RNA was
extracted from the HUVECs for RT-qPCR of intracellular miR-21-5p
and the candidate target genes. The results indicated that 4 Gy
irradiation increased miR-21-5p expression in HUVECs and decreased
the expression of five of the six target candidate mRNAs (Fig. 4B and C). Next, SA-β-GAL staining of 4
Gy-irradiated HUVECs revealed that the cytoplasm of 4 Gy-irradiated
HUVECs was stained darker and bluer compared with that of
nonirradiated HUVECs (Fig. 4D). To
quantitate the staining results, HUVECs were fluorescently stained
using the cell senescence detection kit SPiDER-βGal, and the
fluorescence intensity was measured by flow cytometry. The average
SA-β-GAL fluorescence intensity of nonirradiated HUVECs was
approximately 59,000, whereas the average SA-β-GAL fluorescence
intensity of 4 Gy-irradiated HUVECs was approximately 150,000,
which is a statistically substantial increase (Figs. 4E and S1). Because the expression of
CDC25A, which is involved in the cell cycle, was decreased,
we analyzed the cell cycle of HUVECs cultured for 10 days after 4
Gy irradiation. There were fewer S-phase cells in the 4
Gy-irradiated group compared with that in the nonirradiated group
(Figs. 4F and S2). This suggests that 4 Gy irradiation
induces senescence in HUVECs and affects the S phase of the cell
cycle through increased expression of miR-21-5p.
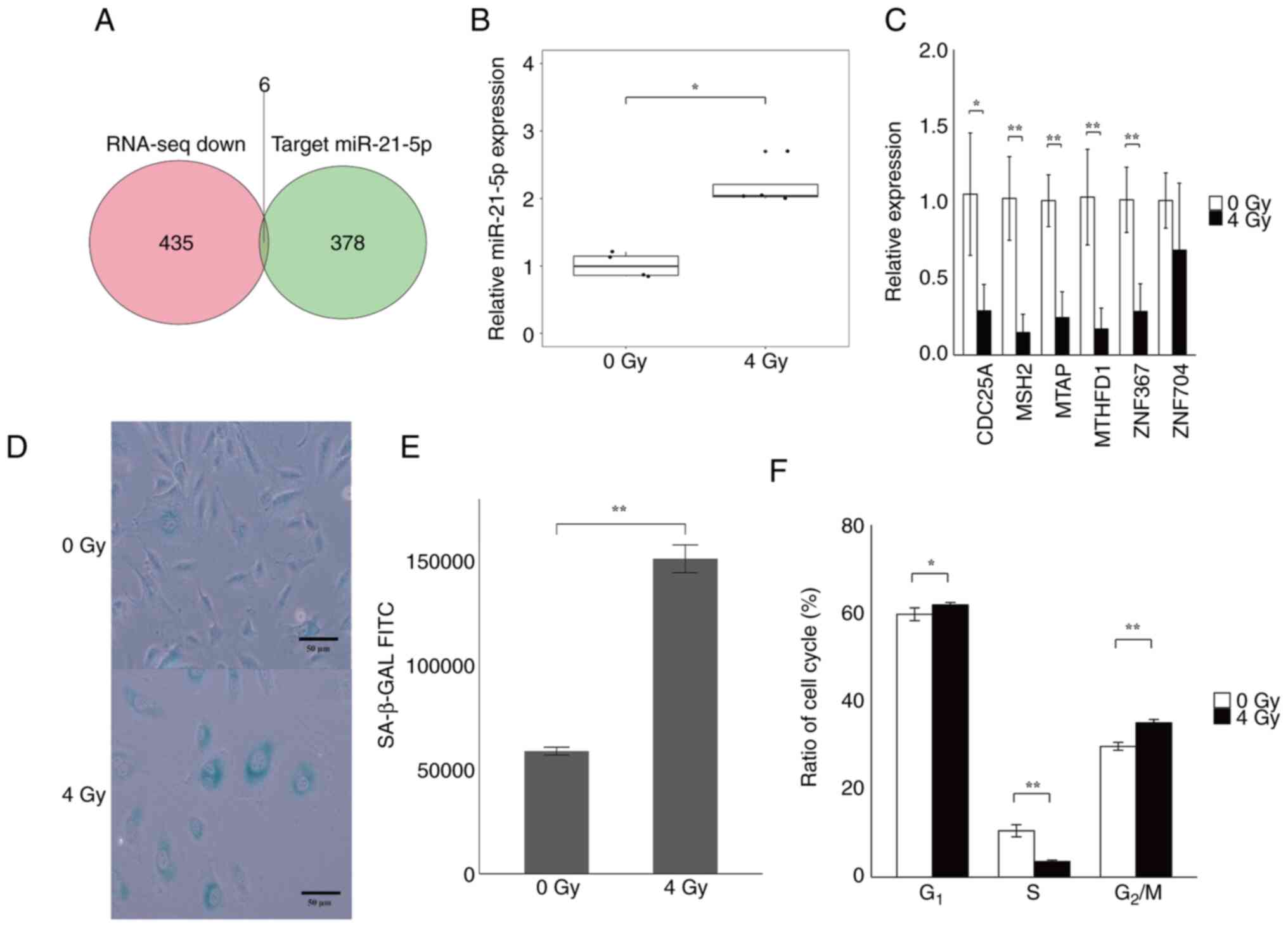 | Figure 4Quantitation of gene expression and
cell cycle analysis of senescence-induced HUVECs. Total RNA was
extracted from HUVECs cultured for 10 days after 4 Gy irradiation
and the expression levels of miR-21-5p and candidate target genes
were quantified by RT-qPCR. Senescence and cell cycle analysis of
HUVECs was performed. (A) Venn diagram of downregulated genes and
miR-21-5p target candidate genes based on the RNA-sequencing
reanalysis data. Candidate target genes were downloaded from
TargetScan (https://www.targetscan.org/vert_72/). A total of six
common genes, including CDC25A, MSH2, MTAP,
MTHFD1, ZNF367 and ZNF704, were detected. (B)
Quantitation of gene expression by RT-qPCR of intracellular
miR-21-5p in HUVECs cultured for 10 days following 4 Gy
irradiation. The expression of intracellular miR-21-5p was
significantly increased in 4 Gy-irradiated HUVECs compared with in
nonirradiated HUVECs. (C) Quantitation of targeted-candidate genes
in the RNA-sequencing reanalysis data by RT-qPCR. The expression of
five genes was significantly decreased in 4 Gy-irradiated HUVECs
compared with nonirradiated HUVECs. (D) SA-β-GAL staining of HUVECs
cultured for 10 days after 4 Gy irradiation or nonirradiated cells.
Blue-stained cells are SA-β-GAL-positive cells. Scale bar, 50 µm.
(E) Quantitative analysis of fluorescence intensity by flow
cytometry using SPiDER-βGal. The fluorescence intensity was
significantly increased in HUVECs irradiated with 4 Gy compared
with nonirradiated HUVECs. (F) Cell cycle analysis of HUVECs
cultured for 10 days after irradiation with 4 Gy vs. nonirradiated
cells. The y-axis indicates the percentage of each cell cycle
phase. The 4 Gy-irradiated HUVECs exhibited a significantly lower
percentage of S phase cells, and a significant increase in
G1 and G2/M phase cells compared with
non-irradiated HUVECs. All experiments were performed with n=4.
*P<0.05, **P<0.01. CDC25A, cell
division cycle 25A; miR, microRNA; MSH2, mutS homolog 2;
MTAP, methylthioadenosine phosphorylase; MTHFD1,
methylenetetrahydrofolate dehydrogenase cyclohydrolase and
formyltetrahydrofolate synthetase 1; RNA-seq, RNA sequencing;
RT-qPCR, reverse transcription-quantitative PCR; SA-β-GAL,
senescence-associated β-galactosidase; ZNF367, zinc finger
protein 367; ZNF704, zinc finger protein 704. |
miR-21-5p expression is increased in
EVs secreted from 4 Gy-irradiated HUVECs
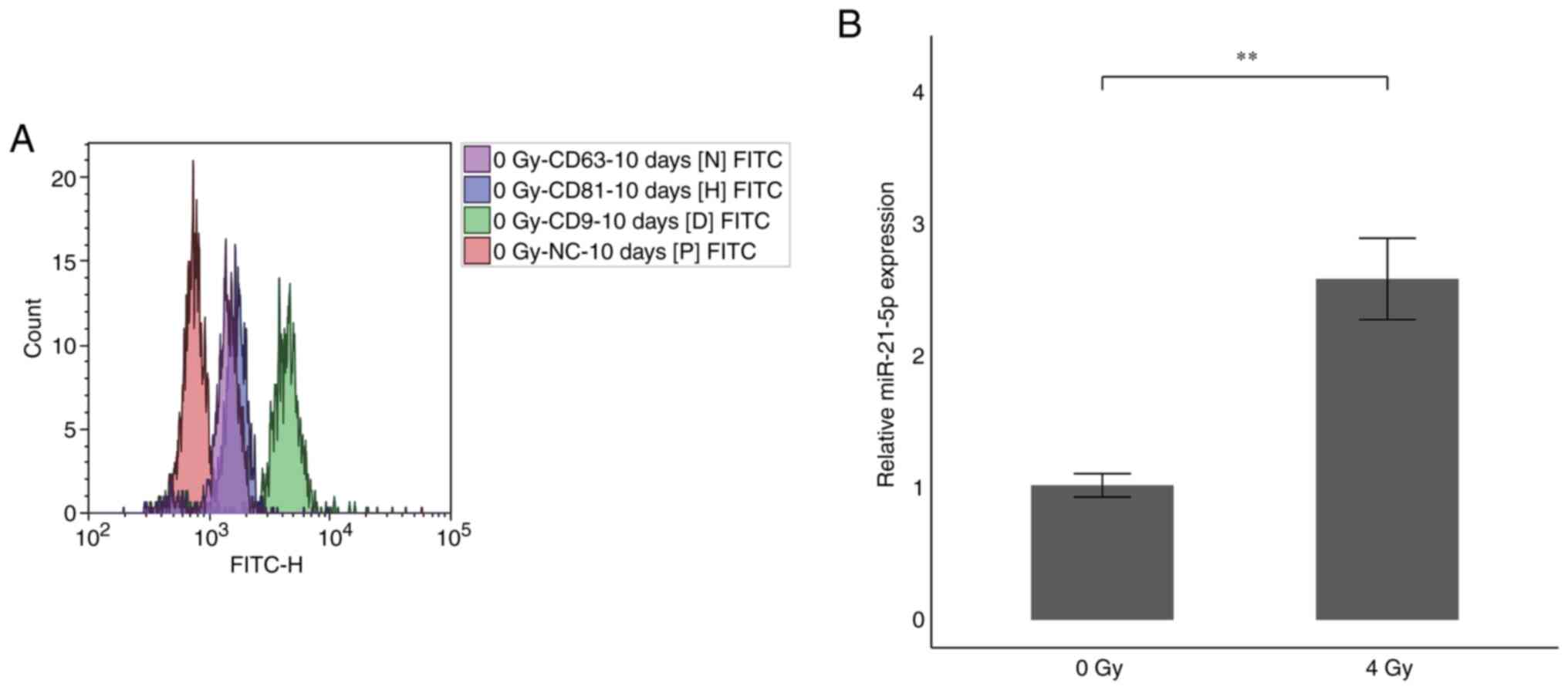
To identify tetraspanins, which are surface antigens
of EVs secreted from HUVECs cultured for 10 days after 4 Gy
irradiation, EVs were collected from 2 ml of culture supernatant
using PS capture and incubated with specific antibodies for each
FITC-labeled tetraspanin family member and fluorescence was
detected by flow cytometry. The results indicated that CD9 was
strongly expressed in EVs secreted from HUVECs, confirming that EVs
were collected (Fig. 5A). Next, we
measured the expression of miR-21-5p in the EVs. The miR-21-5p
expression in the supernatant of HUVECs cultured for 10 days after
4 Gy irradiation considerably increased by 2.7-fold compared with
the nonirradiated HUVECs (Fig.
5B). This indicates that miR-21-5p is increased not only
present intracellularly, but also in EVs of HUVECs irradiated with
4 Gy.
Discussion
In this study, we found that mmu-miR-21a-5p was
increased in blood EVs of aging mice. Increased expression of
miR-21-5p, with the same sequence as that in mice, was observed in
senescent HUVECs. Moreover, miR-21-5p was also increased in EVs in
the culture supernatant of senescence-induced HUVECs. Based on
these findings, the increased expression of EV miR-21-5p
extracellularly in senescence-induced HUVECs in vitro suggests
increased expression of miR-21-5p in the blood EVs of aging mice.
Therefore, increased serum miR-21-5p may be a biomarker for
vascular endothelial cell senescence.
Reanalysis of RNA-sequence data from HUVECs
identified 1881 genes with altered expression (Fig. 2C). Enrichment analysis of these
altered genes revealed that many of the downregulated genes in 4
Gy-irradiated HUVECs were associated with the cell cycle and DNA
repair, suggesting that the downregulation of these genes may
directly affect these processes. Bouten et al (29) performed RNA-sequencing on human
lung microvascular endothelial cells irradiated with 10 Gy. In
their enrichment analysis, GO term and KEGG pathways also
identified terms or pathways related to the cell cycle and DNA
repair, which supports our reanalysis of the GSE130727 data
(Fig. 3). These molecular changes
are thought to result in the inability of cells to proliferate due
to reduced expression of genes related to DNA replication and the
cell cycle, resulting in cell senescence.
MSH2 is probably involved in processing biologically
substantial clustered DNA damages and in executing apoptosis
induced by ionizing radiation. The human DNA mismatch complex
MSH2-MSH3 recognizes small loops via a mechanism different from
that of MSH2-MSH6, which is specific for single-base mismatches
(37). This salvage pathway has
been implicated in cell apoptosis, proliferation, differentiation,
and inflammatory responses. MTAP catalyzes the reversible
phosphorylation of 5'-methylthioadenosine, a co-product of
polyamine biosynthesis (38). It
has been reported that loss of MTAP in hepatocellular carcinoma
cells results in cell cycle arrest (39,40).
Therefore, reduced expression of MTAP may be related to the cell
cycle. MTHFD1 plays a role in nucleotide synthesis and cell cycle
(41,42). MTHFD1 has been reported to be
related to serine metabolism, and it has been reported that when
serine metabolism ceases, the expression of folate
metabolism-related genes such as MTHFD1 declines and cells
senescence (43). It has been
reported that knockdown of ZNF704 in cancer cells halts the cell
cycle and induces apoptosis (44).
ZNF367 is involved in YAP signaling in carcinomas and is frequently
enriched in aged brain tissue (45,46).
In breast carcinomas, it negatively correlates with miR-21-5p,
which has been shown to inhibit cancer cell proliferation (47).
In colon adenocarcinomas, increased expression of
miR-21-5p reportedly increases cell proliferation (48). However, Li et al (49) reported that in chondrosarcoma,
increased expression of miR-21-5p induces apoptosis by causing
G0/G1 cell cycle arrest. Furthermore, Liu et al (50) compared kidney aging in freely-fed
3-month-old rats, 24-month-old rats, and calorie-restricted
24-month-old rats. They observed higher miR-21 expression in the
freely-fed 24-month-old rats than in the calorie-restricted
24-month-old rats, suggesting that the miR-21 expression increased
in the kidneys of the 24-month-old rats. This supports our results
that increased miR-21 expression is associated with kidney aging
(50). Mensà et al
(51) reported that miR-21-5p
expression is increased in EVs of aging HUVECs, which are secreted
by aging HUVECs as an aging signal. This suggests that miR-21-5p
expression increases as cells undergo senescence (51). Thus, the accumulation of miR-21-5p
contributes to the induction of cell senescence. Dellago et
al (52) reported that
overexpression of miR-21 in HUVECs causes cellular senescence,
suggesting that suppression of miR-21 expression may halt the
progression of cellular senescence. They also found that CDC25A, a
target gene of miR-21, inhibits cell proliferation through an
increase in cyclin-dependent kinase 2 (CDK2), a protein required
for the transition from the G1 to S phase of the cell cycle
(53). Cell cycle analysis
suggests that the S phase of the cell cycle is reduced and G1
arrest occurs because of the failure of the transition from the G1
to S phase caused by decreased CDC25A expression. Our data
also suggest that HUVECs with increased intracellular miR-21-5p and
SA-β-GAL upon senescence induction are involved in the cell cycle
and DNA repair by RNA-sequence enrichment analysis and cell cycle
analysis. This suggests that miR-21-5p is involved in the cell
cycle and DNA repair, which supports the results of our previous
study.
We previously showed increased expression of
mmu-miR-21a-5p in serum in cognitively impaired 58-week-old mice
(28). Because the Aging mice used
in the present study were even older, it suggests that the
expression of mmu-miR-21a-5p persistently increases with
senescence. Moreover, Accardi et al (54) found that plasma miR-21-5p was
increased in individuals aged 51 to 99 years. They also
demonstrated that passaging HUVECs induced senescence, resulting in
increased intracellular miR-21-5p expression (29), which supports our results. However,
they did not analyze EV miR-21-5p. Thus, our results are novel as
we found miR-21-5p secretion from senescence-induced HUVECs. This
suggests that the senescent vascular endothelial cells secrete
miR-21-5p, which accumulates in the blood. Based on our results and
those of other groups, there is a close relationship between
cognitive function, aging, and miR-21-5p expression. EV miR-21-5p
may be a biomarker for the aging of vascular endothelial cells. By
integrating our results with those of other researchers, we
demonstrate a close relationship among miR-21-5p expression,
cognitive function, and aging. In particular, miR-21-5p in EVs is a
promising biomarker for vascular endothelial cell aging,
potentially serving as a starting point for new studies exploring
the connection between age-related cognitive decline and vascular
degeneration. Future studies are required to further elucidate the
functional role and mechanisms of miR-21-5p to enhance our
understanding of the pathophysiology of aging. The limitation of
this study is the extent to which the accumulation of endothelial
cell senescence and miR-21-5p expression influence individual
aging. Future research should increase the number of individual
mice for detailed analysis and quantify the relationship between
individual aging and the accumulation of endothelial cell
senescence.
Supplementary Material
SA-β-GAL analysis of HUVECs exposed to
4 Gy irradiation. The FITC fluorescence intensity of HUVECs stained
with SPiDER-βGal working solution was detected using a CytoFLEX
flow cytometer. SA-β -GAL, senescence-associated
β-galactosidase.
Cell cycle analysis of HUVECs exposed
to 4 Gy irradiation. PI-stained HUVECs were detected using the
CytoFLEX flow cytometer, and the Kaluza 2.2 software was used to
calculate the percentage of cells in the G1, S and
G2/M phases.
Primer sequences for miRNA reverse
transcription-quantitative PCR.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by The JSPS KAKENHI
(grant no. 21H04844) and JST SPRING (grant no. JPMJSP2152).
Availability of data and materials
The microarray data generated in the present study
may be found in the Gene Expression Omnibus database under
accession number GSE274943 or at the following URL: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE274943.
The other data generated in the present study may be requested from
the corresponding author.
Authors' contributions
KY and MC were major contributors in performing the
experiments and writing the manuscript. KY and MC confirm the
authenticity of all the raw data. All authors read and approved the
final version of the manuscript.
Ethics approval and consent to
participate
All animal experiments were performed in accordance
with The Guideline for Animal Experimentation of Hirosaki
University. The Animal Research Committee of Hirosaki University
(approval no. AE01-2023-097-1; Hirosaki, Japan) approved and
monitored the procedures. The Ethics Committee of Hirosaki
University Graduate School of Health Sciences (Hirosaki, Japan)
confirmed that ethical review is not required for research using
commercially available, frequently used cultured cells.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Guo J, Huang X, Dou L, Yan M, Shen T, Tang
W and Li J: Aging and aging-related diseases: From molecular
mechanisms to interventions and treatments. Signal Transduct Target
Ther. 7(391)2022.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Coppé JP, Desprez PY, Krtolica A and
Campisi J: The senescence-associated secretory phenotype: The dark
side of tumor suppression. Annu Rev Pathol. 5:99–118.
2010.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Wong PF, Tong KL, Jamal J, Khor ES, Lai SL
and Mustafa MR: Senescent HUVECs-secreted exosomes trigger
endothelial barrier dysfunction in young endothelial cells. EXCLI.
18:764–776. 2019.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Mongiardi MP, Merolle M, Fustaino V, Levi
A and Falchetti ML: Gene expression profiling of hypoxic response
in different models of senescent endothelial cells. Aging Clin Exp
Res. 33:1993–2001. 2001.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Ridzuan N, Al Abbar A, Yip WK, Maqbool M
and Ramasamy R: Characterization and expression of senescence
marker in prolonged passages of rat bone marrow-derived mesenchymal
stem cells. Stem Cells Int. 2016(8487264)2016.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Xu J, Liu D, Zhao D, Jiang X, Meng X,
Jiang L, Yu M, Zhang L and Jiang H: Role of low-dose radiation in
senescence and aging: A beneficial perspective. Life Sci.
302(120644)2022.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Tanaka Y and Takahashi A:
Senescence-associated extracellular vesicle release plays a role in
senescence-associated secretory phenotype (SASP) in age-associated
diseases. J Biochem. 169:147–153. 2021.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Hallal S, Tűzesi Á, Grau GE, Buckland ME
and Alxander KL: Understanding the extracellular vesicle surface
for clinical molecular biology. J Extracell Vesicles.
11(e12260)2022.PubMed/NCBI View Article : Google Scholar
|
|
9
|
O'Brien K, Breyne K, Ughetto S, Laurent LC
and Breakefield XO: RNA delivery by extracellular vesicles in
mammalian cells and its applications. Nat Rev Mol Cell Biol.
21:585–606. 2020.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Nowicka G: Extracellular vesicles in the
diagnosis and treatment of cardiovascular disease. What's behind?
What do we need to implement them into clinical practice? Int J
Biochem Cell Biol. 172(106600)2024.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Adduri RSR, Cai K, Velasco-Alzate K,
Vasireddy R, Miller JW, de Frías SP, de Frías FP, Horimasu Y,
Iwamoto H, Hattori N, et al: Plasma extracellular vesicle proteins
as promising noninvasive biomarkers for diagnosis of idiopathic
pulmonary fibrosis. J Extracell Biol. 2(e98)2023.PubMed/NCBI View
Article : Google Scholar
|
|
12
|
García-Silva S, Gallardo M and Peinado H:
DNA-loaded extracellular vesicles in liquid biopsy: Tiny players
with big potential? Front Cell Dev Biol. 8(622579)2021.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Ekström K, Valadi H, Sjöstrand M, Malmhäll
C, Bossios A, Eldh M and Lötvall J: Characterization of mRNA and
microRNA in human mast cell-derived exosomes and their transfer to
other mast cells and blood CD34 progenitor cells. J Extracell
Vesicles. 1(18389)2012.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Lagos-Quintana M, Rauhut R, Lendeckel W
and Tuschl T: Identification of novel genes coding for small
expressed RNAs. Science. 294:853–858. 2001.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Das K and Rao LV: The role of microRNAs in
inflammation. Int J Mol Sci. 23(15479)2022.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Nunes AD, Weigl M, Schneider A, Noureddine
S, Yu L, Lahde C, Saccon TD, Mitra K, Beltran E, Grillari J, et al:
miR-146a-5p modulates cellular senescence and apoptosis in visceral
adipose tissue of long-lived Ames dwarf mice and in cultured
pre-adipocytes. Geroscience. 44:503–518. 2022.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Suh N: MicroRNA controls of cellular
senescence. BMB Rep. 51:493–499. 2018.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Chan JA, Krichevsky AM and Kosik KS:
MicroRNA-21 is an antipoptotic factor in human glioblastoma cells.
Cancer Res. 65:6029–6033. 2005.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Zhang X, Azhar G and Wei JY: The
expression of microRNA and microRNA clusters in the aging heart.
PLoS One. 7(e34688)2012.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Wang P, Zou F, Zhang X, Li H, Dulak A,
Tomko RJ Jr, Lazo JS, Wang Z, Zhang L and Yu J: microRNA-21
negatively regulates Cdc25A and cell cycle progression in colon
cancer cells. Cancer Res. 69:8157–8165. 2009.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Zhang Y, Liu Y, Liu H and Tang WH:
Exosomes: Biogenesis, biologic function and clinical potential.
Cell Biosci. 9(19)2019.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Iglesias MJ, Kruse LD, Sanchez-Rivera L,
Enge L, Dusart P, Hong MG, Uhlén M, Renné T, Schwenk JM, Bergstrom
G, et al: Identification of endothelial proteins in plasma
associated with cardiovascular risk factors. Arterioscler Thromb
Vasc Biol. 41:2990–3004. 2021.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Berezin AE and Berezin AA: Extracellular
endothelial cell-derived vesicles: Emerging role in cardiac and
vascular remodeling in heart failure. Front Cardiovasc Med.
15(47)2020.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Casella G, Munk R, Kim KM, Piao Y, De S,
Abdelmohsen K and Gorospe M: Transcriptome signature of cellular
senescence. Nucleic Acids Res. 47:7294–7305. 2019.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Chiba M, Uehara H, Niiyama I, Kuwata H and
Monzen S: Changes in miRNA expressions in the injured small
intestine of mice following high-dose radiation exposure. Mol Med
Rep. 21:2452–2458. 2020.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Yamamoto K and Chiba M: Examination and
comparison of the RNA extraction methods using mouse serum. Biomed
Rep. 20(51)2024.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43(e47)2015.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Yamamoto K, Miyano K, Fujita M, Kurata W,
Ohta H, Matsumoto K and Chiba M: Changes in cognitive ability and
serum microRNA levels during aging in mice. Exp Ther Med.
27(120)2024.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Bouten RM, Dalgard CL, Soltis AR, Slaven
JE and Day RM: Transcriptomic profiling and pathway analysis of
cultured human lung microvascular endothelial cells following
ionizing radiation exposure. Sci Rep. 11(24214)2021.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Chen S, Zhou Y, Chen Y and Gu J: fastp: An
ultra-fast all-in-one FASTQ preprocessor. Bioinformatics.
34:i884–i890. 2018.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Patro R, Duggal G, Love MI, Irizarry RA
and Kingsford C: Salmon provides fast and bias-aware quantification
of transcript expression. Nat Methods. 14:417–419. 2017.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Soneson C, Love MI and Robinson MD:
Differential analyses for RNA-seq: Transcript-level estimates
improve gene-level inferences. F1000Res. 4(1521)2015.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Chen Y, Lun AT and Smyth GK: From reads to
genes to pathways: Differential expression analysis of RNA-Seq
experiments using Rsubread and the edgeR quasi-likelihood pipeline.
F1000Res. 5(1438)2016.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Yu G, Wang LG, Han Y and He QY:
clusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Yu G and He QY: ReactomePA: An
R/Bioconductor package for reactome pathway analysis and
visualization. Mol Biosyst. 12:477–479. 2016.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Edelbrock MA, Kaliyaperumal S and Williams
KJ: Structural, molecular and cellular functions of MSH2 and MSH6
during DNA mismatch repair, damage signaling and other noncanonical
activities. Mutat Res. 743:53–66. 2013.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Fan N, Zhang Y and Zou S:
Methylthioadenosine phosphorylase deficiency in tumors: A
compelling therapeutic target. Front Cell Dev Biol.
5(1173356)2023.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Marjon K, Cameron MJ, Quang P, Clasquin
MF, Mandley E, Kunii K, McVay M, Choe S, Kernytsky A, Gross S, et
al: MTAP deletions in cancer create vulnerability to targeting of
the MAT2A/PRMT5/RIOK1 axis. Cell Rep. 15:574–587. 2016.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Kryukov GV, Wilson FH, Ruth JR, Paulk J,
Tsherniak A, Marlow SE, Vazquez F, Weir BA, Fitzgerald ME, Tanaka
M, et al: MTAP deletion confers enhanced dependency on the PRMT5
arginine methyltransferase in cancer cells. Science. 351:1214–1218.
2016.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Galbiatti AL, da Silva LM, Ruiz-Cintra MT,
Raposo LS, Maníglia JV, Pavarino ÉC and Goloni-Bertollo EM:
Association between 11 genetic polymorphisms in folate-metabolising
genes and head and neck cancer risk. Eur J Cancer. 48:1525–1531.
2012.PubMed/NCBI View Article : Google Scholar
|
|
42
|
He D, Yu Z, Liu S, Dai H, Xu Q and Li F:
Methylenetetrahydrofolate dehydrogenase 1 (MTHFD1) is
underexpressed in clear cell renal cell carcinoma tissue and
transfection and overexpression in Caki-1 cells inhibits cell
proliferation and increases apoptosis. Med Sci Monit. 21:8391–8400.
2018.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Zhou S, Cui J and Shi Y: Serine metabolism
regulates the replicative senescence of human dental pulp cells
through histone methylation. Curr Issues Mol Biol. 24:2856–2870.
2024.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Luo J, Li H, Xiu J, Zeng J, Feng Z, Zhao
H, Li Y and Wei W: Elevated ZNF704 expression is associated with
poor prognosis of uveal melanoma and promotes cancer cell growth by
regulating AKT/mTOR signaling. Biomark Res. 10(38)2023.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Lei T, Gao Y, Duan Y, Cui C, Zhang L and
Si M: Inhibition of zinc finger protein 367 exerts a tumor
suppressive role in colorectal cancer by affecting the activation
of oncogenic YAP1 signaling. Environ Toxicol. 36:2278–2290.
2021.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Baumgart M, Groth M, Priebe S, Savino A,
Testa G, Dix A, Ripa R, Spallotta F, Gaetano C, Ori M, et al:
RNA-seq of the aging brain in the short-lived fish N.
furzeri-conserved pathways and novel genes associated with
neurogenesis. Aging Cell. 13:965–974. 2014.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Du L, Tao X and Shen X: Human umbilical
cord mesenchymal stem cell-derived exosomes inhibit migration and
invasion of breast cancer cells via miR-21-5p/ZNF367 pathway.
Breast Cancer. 28:829–837. 2021.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Yu W, Zhu K, Wang Y, Yu H and Guo J:
Overexpression of miR-21-5p promotes proliferation and invasion of
colon adenocarcinoma cells through targeting CHL1. Mol Med.
24(36)2018.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Li G, Yang Y, Xu S, He M and Zhang Z:
mir-21-5p inhibits the progression of human chondrosarcoma by
regulating CCR7/STAT3/NF-κB pathway. Connect Tissue Res.
62:313–324. 2021.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Liu JR, Cai GY, Ning YC, Wang JC, Lv Y,
Guo YN, Fu B, Hong Q, Sun XF and Chen XM: Caloric restriction
alleviates aging-related fibrosis of kidney through downregulation
of miR-21 in extracellular vesicles. Aging. 27:18052–18072.
2020.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Mensà E, Guescini M, Giuliani A, Bacalini
MG, Ramini D, Corleone G, Ferracin M, Fulgenzi G, Graciotti L,
Prattichizzo F, et al: Small extracellular vesicles deliver miR-21
and miR-217 as pro-senescence effectors to endothelial cells. J
Extracell Ves. 18(1725285)2020.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Dellago H, Preschitz-Kammerhofer B,
Terlecki-Zaniewicz L, Schreiner C, Fortschegger K, Chang MW, Hackl
M, Monteforte R, Kühnel H, Schosserer M, et al: High levels of
oncomiR-21 contribute to the senescence-induced growth arrest in
normal human cells and its knock-down increases the replicative
lifespan. Aging Cell. 12:446–458. 2013.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Donzelli M and Draetta GF: Regulating
mammalian checkpoints through Cdc25 inactivation. EMBO Rep.
4:671–677. 2003.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Accardi G, Bono F, Cammarata G, Aiello A,
Herrero MT, Alessandro R, Augello G, Carru C, Colomba P, Costa MA,
et al: miR-126-3p and miR-21-5p as hallmarks of bio-positive
ageing; correlation analysis and machine learning prediction in
young to ultra-centenarian Sicilian population. Cells.
11(1505)2022.PubMed/NCBI View Article : Google Scholar
|