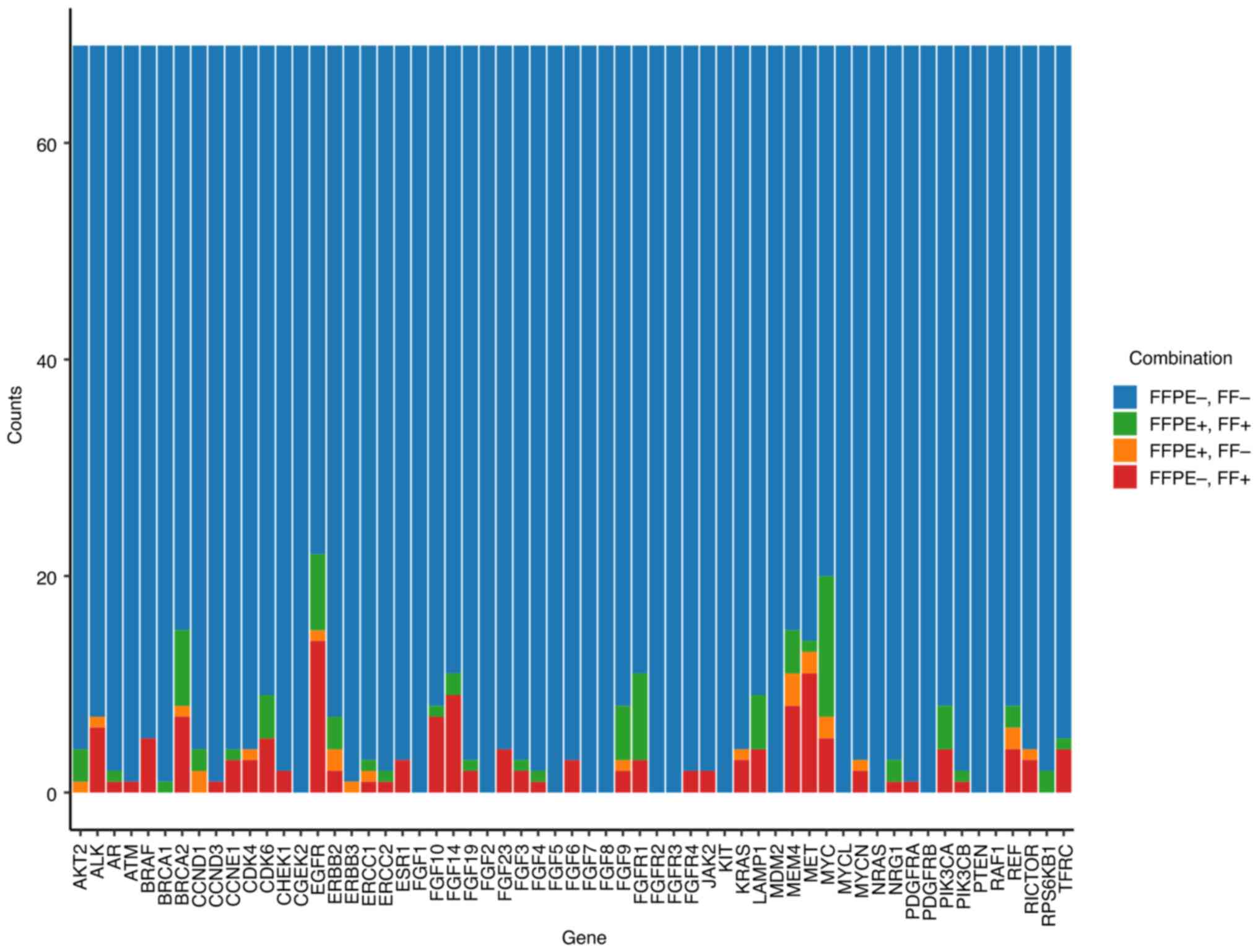
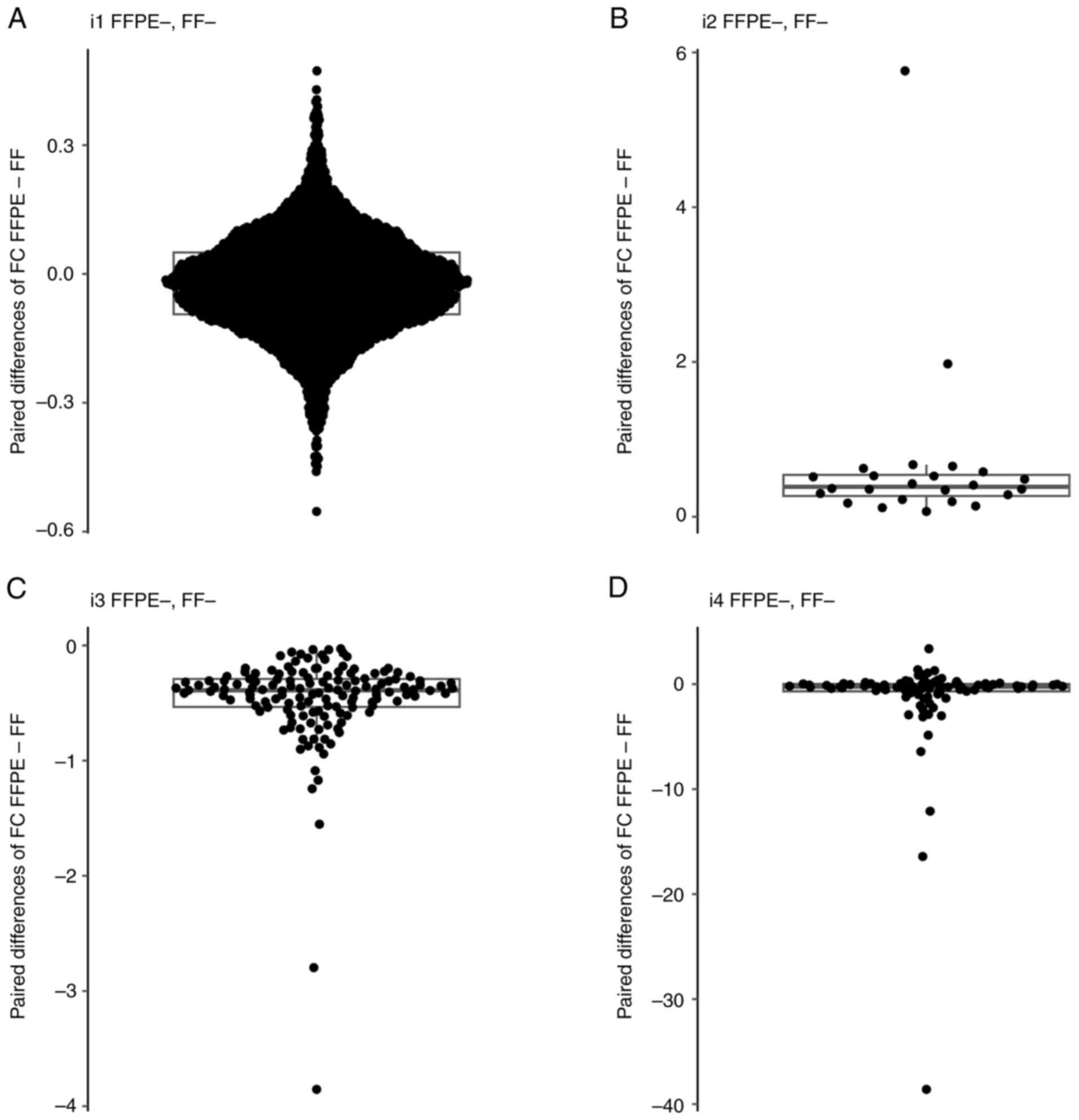
|
1
|
Giacò L, Palluzzi F, Guido D, Nero C,
Giacomini F, Duranti S, Bria E, Tortora G, Cenci T, Martini M, et
al: A computational framework for comprehensive genomic profiling
in solid cancers: The analytical performance of a high-throughput
assay for small and copy number variants. Cancers (Basel).
14(6152)2022.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Froyen G, Geerdens E, Berden S, Cruys B
and Maes B: Diagnostic validation of a comprehensive targeted panel
for broad mutational and biomarker analysis in solid tumors.
Cancers (Basel). 14(2457)2022.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Conroy JM, Pabla S, Glenn ST, Seager RJ,
Van Roey E, Gao S, Burgher B, Andreas J, Giamo V, Mallon M, et al:
A scalable high-throughput targeted next-generation sequencing
assay for comprehensive genomic profiling of solid tumors. PLoS
One. 16(e0260089)2021.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Ottestad AL, Huang M, Emdal EF, Mjelle R,
Skarpeteig V and Dai HY: Assessment of two commercial comprehensive
gene panels for personalized cancer treatment. J Pers Med.
13(42)2022.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Pestinger V, Smith M, Sillo T, Findlay JM,
Laes JF, Martin G, Middleton G, Taniere P and Beggs AD: Use of an
integrated pan-cancer oncology enrichment next-generation
sequencing assay to measure tumour mutational burden and detect
clinically actionable variants. Mol Diagn Ther.
24(505)2020.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Meireles SI, Cruz MV, de Godoy CD and de
Testagrossa L: Performance of non-formalin fixed paraffin embedded
samples in hybrid capture and amplicon next-generation sequencing
panels. Diagn Cytopathol. 52:171–182. 2024.PubMed/NCBI View
Article : Google Scholar
|
|
7
|
Betge J, Kerr G, Miersch T, Leible S,
Erdmann G, Galata CL, Zhan T, Gaiser T, Post S, Ebert MP, et al:
Amplicon sequencing of colorectal cancer: Variant calling in frozen
and formalin-fixed samples. PLoS One. 10(e0127146)2015.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Robbe P, Popitsch N, Knight SJL, Antoniou
P, Becq J, He M, Kanapin A, Samsonova A, Vavoulis DV, Ross MT, et
al: Clinical whole-genome sequencing from routine formalin-fixed,
paraffin-embedded specimens: Pilot study for the 100,000 Genomes
Project. Genet Med. 20:1196–1205. 2018.PubMed/NCBI View Article : Google Scholar
|
|
9
|
De Paoli-Iseppi R, Johansson PA, Menzies
AM, Dias KR, Pupo GM, Kakavand H, Wilmott JS, Mann GJ, Hayward NK,
Dinger ME, et al: Comparison of whole-exome sequencing of matched
fresh and formalin fixed paraffin embedded melanoma tumours:
implications for clinical decision making. Pathology. 48:261–266.
2016.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Kerick M, Isau M, Timmermann B, Sültmann
H, Herwig R, Krobitsch S, Schaefer G, Verdorfer I, Bartsch G,
Klocker H, et al: Targeted high throughput sequencing in clinical
cancer settings: Formaldehyde fixed-paraffin embedded (FFPE) tumor
tissues, input amount and tumor heterogeneity. BMC Med Genomics.
4(68)2011.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Fielding DI, Dalley AJ, Singh M,
Nandakumar L, Lakis V, Chittoory H, Fairbairn D, Patch AM, Kazakoff
SH, Ferguson K, et al: Evaluating diff-quik cytology smears for
large-panel mutation testing in lung cancer-predicting DNA content
and success with low-malignant-cellularity samples. Cancer
Cytopathol. 131:373–382. 2023.PubMed/NCBI View Article : Google Scholar
|
|
12
|
R Core Team (2021) R: A language and
environment for statistical computing. R Foundation for Statistical
Computing, Vienna, 2021.
|
|
13
|
tabulizer: Bindings for Tabula PDF Table
Extractor Library. R package version 0.2.2.
|
|
14
|
Ooms J: The jsonlite Package: A practical
and consistent mapping between JSON data and R Objects 2014.
|
|
15
|
Iannone R and Roy O: DiagrammeR:
Graph/Network Visualization. 2024.
|
|
16
|
Iannone R: DiagrammeRsvg: Export
DiagrammeR Graphviz Graphs as SVG. 2016.
|
|
17
|
Ooms J and Brüggemann S: Render SVG images
into PDF, PNG, (Encapsulated) PostScript, or Bitmap Arrays
2023.
|
|
18
|
Larsson J, Godfrey AJR, Gustafsson P,
Eberly DH, Huber E and Privé F: Area-proportional euler and venn
diagrams with ellipses, 2024.
|
|
19
|
Knaus BJ and Grünwald NJ: vcfr: A package
to manipulate and visualize VCF format data in R. Mol Ecol Resour.
17:44–53. 2017.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Local run manager trusight oncology
comprehensive (EU) analysis module. 2022.
|
|
21
|
Bonneville R, Krook MA, Kautto EA, Miya J,
Wing MR, Chen HZ, Reeser JW, Yu L and Roychowdhury S: Landscape of
microsatellite instability across 39 cancer types. JCO Precis
Oncol. 17(00073)2017.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Baum JE, Zhang P, Hoda RS, Geraghty B,
Rennert H, Narula N and Fernandes HD: Accuracy of next-generation
sequencing for the identification of clinically relevant variants
in cytology smears in lung adenocarcinoma. Cancer Cytopathol.
125:398–406. 2017.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Faber E, Grosu H, Sabir S, Lucas FAS,
Barkoh BA, Bassett RL, Luthra R, Stewart J and Roy-Chowdhuri S:
Adequacy of small biopsy and cytology specimens for comprehensive
genomic profiling of patients with non-small-cell lung cancer to
determine eligibility for immune checkpoint inhibitor and targeted
therapy. J Clin Pathol. 75:612–619. 2022.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Ramani NS, Chen H, Broaddus RR, Lazar AJ,
Luthra R, Medeiros LJ, Patel KP, Rashid A, Routbort MJ, Stewart J,
et al: Utilization of cytology smears improves success rates of
RNA-based next-generation sequencing gene fusion assays for
clinically relevant predictive biomarkers. Cancer Cytopathol.
129:374–382. 2021.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Pepe F, Pisapia P, Gristina V, Rocco D,
Micheli M, Micheli P, Iaccarino A, Tufano R, Gragnano G, Russo G,
et al: Tumor mutational burden on cytological samples: A pilot
study. Cancer Cytopathol. 129:460–467. 2021.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Sanghvi RV, Buhay CJ, Powell BC, Tsai EA,
Dorschner MO, Hong CS, Lebo MS, Sasson A, Hanna DS, McGee S, et al:
Characterizing reduced coverage regions through comparison of exome
and genome sequencing data across 10 centers. Genet Med.
20:855–866. 2018.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Wang Q, Shashikant CS, Jensen M, Altman NS
and Girirajan S: Novel metrics to measure coverage in whole exome
sequencing datasets reveal local and global non-uniformity. Sci
Rep. 7(885)2017.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Chou D, Chen X, Purdy A, Teng L, Luo B,
Zhao C, Ball L, Castaneda A, Clark K, Crain B, et al: Abstract
3732: Analytical performance of TruSight® Tumor 170 on
small nucleotide variations and gene amplifications using DNA from
formalin-fixed, paraffin-embedded (FFPE) solid tumor samples.
Cancer Res. 2017.
|