Introduction
Macrophages are abundant in diverse tissues and
organs where they can function as immune effectors, immune
regulators, tissue remodelers, or quiescent scavengers. External
stimuli can cause macrophages to undergo a marked and coordinated
change in the expression of multiple gene products, changing the
functional capacity of the cell. The diversity of environments
surrounding macrophages in different tissues corresponds to an
equally diverse constellation of macrophage phenotypes in the host
(1). In the context of specific
immune response, the cytokine milieu compels mononuclear phagocytes
to express specialized and polarized functional properties. Since
they mirror the Th1/Th2 nomenclature, polarized macrophages are
regarded as classical macrophage activation (known as M1
activation) and alternative activation (known as M2 activation)
cells (2–5). Classically polarized activated M1
macrophages have long been known to be induced by IFN-γ alone or in
combination with microbial stimuli as LPS, or cytokines as TNF-α
and GM-CSF. M1 cells have an IL-12high,
IL-23high, IL-10low phenotype, are proficient
producers of effector molecules (reactive oxygen and nitrogen
intermediates) and inflammatory cytokines (IL-1β, TNF-α and IL-6),
contribute as inducer and effector cells in polarized Th1
responses, and mediate resistance against intracellular parasites
and tumors (6–10). By contrast, the alternative M2
form of macrophage activation is a generic name used for various
forms of non-classically activated macrophages resulting from cell
exposure to IL-4 or IL-13, immune complexes, IL-10, glucocorticoid,
or secosteroid (vitamin D3) (3,9,11).
The various forms of M2 macrophages share an IL-12low
and IL-23low phenotype, generally exhibit high levels of
scavenger, mannose (12), and
galactose-type receptors (3),
while arginine metabolism is shifted to the production of ornithine
and polyamines via arginase (13). However, the regulatory mechanisms
controlling the expression of the constellation of genes in
macrophages responding to activating conditions are not fully
defined.
microRNAs (miRNAs) are small, endogenous non-coding
RNA molecules (18–25 nucleotides) that post-transcriptionally
regulate gene expression (14).
miRNAs identify target mRNA through 5′-seed sequence interactions
with miRNA regulatory elements located in the 3′-untranslated
region of target mRNA (15).
Evidence has indicated that miRNAs play a critical role in
regulating cell processes such as cell proliferation, apoptosis,
and differentiation (16–18), leading to the hypothesis that
miRNAs are partially responsible for the coordinated changes in
gene expression occurring during macrophage polarization. This
hypothesis is supported by a number of published studies suggesting
different miRNAs in the human monocyte/macrophage response to
inflammatory stimuli (19–23).
In this study, we employed a miRNA microarray-based
profiling assay to document changes in the abundance of miRNAs
induced by the activation of primary bone marrow-derived
macrophages (BMDMs) with two distinct polarizing conditions to span
the spectrum of described activation patterns (M1 and M2). Our data
revealed that a number of miRNAs were consistently altered under
distinct polarizing conditions. Thus, the present study may be
useful for exploring the precise roles of miRNAs in macrophage
differentiation and polarized activation processes in the
future.
Materials and methods
Isolation and cultivation of murine
BMDMs
Bone marrow cells were obtained by flushing the
femurs from Balb/c mice with Dulbecco’s modified Eagle’s medium
(DMEM)-HEPES medium (Gibco, Eggenstein, Germany). Cells were
collected in 50 ml tubes and centrifuged for 10 min at 100 × g. The
supernatant was removed, and cells were suspended in DMEM (10% FCS;
20% L929 supernatant). Cells (1×106) were cultured in
6-well plates at 37°C and 5% CO2 for 7 days (M0).
Macrophage polarization was obtained by removing the culture medium
and culturing cells for an additional 18 h in RPMI-1640
supplemented with 5% FCS and 100 ng/ml LPS plus 20 ng/ml IFN-γ (for
M1 polarization) or 20 ng/ml IL-4 (for M2 polarization).
Arginase activity assay
To prepare cell lysates for the arginase activity
assay, cells were first rinsed with ice-cold DPBS twice after each
specified treatment and then scraped into 300 ml of lysis buffer
containing 50 mM Tris-HCl (pH 7.5), 0.1 mM EDTA, 0.1 mM EGTA, 1
mg/ml leupeptin, 1 mg/ml aprotinin, and 0.1 mM phenylmethylsulfonyl
fluoride (PMSF; Sigma, St. Louis, MO, USA). Cells were then lysed
by sonication at the frequency of 20 kHz (Sonic and Materials,
Inc., Danbury, CT, USA) for 30 sec (10 sec/cycle). Arginase
activity in the cell lysates was measured as previously described
(24,25). Briefly, cell lysate (50 ml) was
added to 50 ml of Tris-HCl (50 mM; pH 7.5) containing 10 mM
MnCl2. Macrophage arginase was then activated by heating
this mixture at 55–60°C for 10 min. The hydrolysis reaction of
L-arginine by arginase was carried out by incubating the mixture
containing activated arginase with 50 ml of L-arginine (0.5 M; pH
9.7) at 37°C for 1 h and was stopped by adding 400 ml of the acid
solution mixture
(H2SO4:H3PO4:H2O,
51:3:7). For the colorimetric determination of urea,
a-isonitrosopropiophenone (25 ml, 9% in absolute ethanol) was then
added, and the mixture was heated at 100°C for 45 min. Samples were
placed in the dark for 10 min at room temperature, and the urea
concentration was determined spectrophotometrically by the
absorbance at 550 nm measured with a microplate reader (Molecular
Devices, Sunnyvale, CA, USA). The amount of urea produced was used
as an index for arginase activity.
ELISA assay
Cell culture supernatant was collected and stored at
−80°C until assayed. IL-12 and IL-10 concentrations were measured
with specific ELISA kits according to the manufacturer’s
instructions (R&D Systems, USA).
FACS analysis
BMDMs were stained with the following monoclonal
antibodies diluted in 1% FBS in PBS: FITC anti-F4/80 and PE
anti-F4/80 (both from eBioscience Inc., USA), purified
anti-inducible nitric oxide synthase (iNOS) (Santa Cruz
Biotechnology, Inc., Santa Cruz, CA, USA); PE anti-CD16/32, PE
anti-CD206 and PE anti-CD301. Cell fluorescence was measured using
FACS analysis and data were analyzed using CellQuest software (all
from BD Biosciences, San Jose, CA, USA).
miRNA microarray analysis
The miRNA microarray analysis was performed by the
Phalanx Biotech Group (Hsinchu, Taiwan). Total RNA was extracted
from BMDMs using TRIzol reagent (Invitrogen, Burlington, ON, USA)
according to the manufacturer’s instructions. Total RNA (2.5 μg)
was labeled with Cy5 fluorescent dyes using a miRNA ULSTM labeling
kit (Kreatech Diagnostics, Amsterdam, The Netherlands). Labeled
miRNA targets enriched by NanoSep 100K (Pall Corporation, Port
Washington, NY, USA) were hybridized to the MRmiOA-mmu-r1–3.0,
which contains triplicate 1111 unique miRNA probes from mouse
(miRBase Release 17.0), in technical replicates. Following
overnight hybridization at 37°C, non-specific binding targets were
washed away, and the slides were dried by centrifugation and
scanned using an Axon 4000B scanner. The Cy5 fluorescent
intensities of each spot were analyzed by GenePix4.1 software (both
from Molecular Devices). The signal intensity of each spot was
processed by the R program (version 2.12.1). The fine signals
(flag=0) were extracted and processed by log2 transformation, and
the quantile normalization method and ANOVA test. Experiment data
were saved as Microsoft Excel files.
qRT-PCR for miRNAs
Small RNA was extracted from BMDMs as described
above. RNA (2.5 μg) was then subjected to cDNA synthesis, using
either miRNA specific primers or U6 snRNA. Reactions were performed
using a TaqMan® MicroRNA reverse transcription kit
following the manufacturer’s instructions. qRT-PCR was performed
using TaqMan® Universal Master Mix on an Applied
Biosystems 7500 Real-Time PCR Systems. The 20 μl PCR reaction mix
included 1 μl RT products, 10 μl TaqMan® Universal PCR
Master Mix, No AmpErase® UNG (all from Applied
Biosystems, USA), and 1 μl TaqMan probe mix. The reactions were
incubated in 96-well plates at 95°C for 10 min, followed by 40
cycles of 95°C for 15 sec and 60°C for 1 min. Calculations of miRNA
expression levels (M1 vs. M2) were performed using the comparative
CT (ΔΔCT) method and normalized against U6 snRNA levels. Reactions
were run in triplicate.
Statistical analysis
Data were shown as the mean ± SEM. Statistical
analysis of the data was performed using the two-tailed independent
Student’s t-test or the ANOVA analysis using the GraphPad Prism
(version 4.0) statistical program. P<0.05 was considered
statistically significant.
Results
Identification of ex vivo-programmed M1
and M2 macrophages
To screen for miRNAs whose abundance was altered
significantly following incubation of BMDMs in two distinct
polarizing conditions, we first generated M1 and M2 macrophages
in vitro. M1 macrophages induced by IFN-γ and LPS produced a
larger number of proinflammatory cytokines IL-12, while M2
macrophages, polarized by IL-4, showed increased IL-10 production
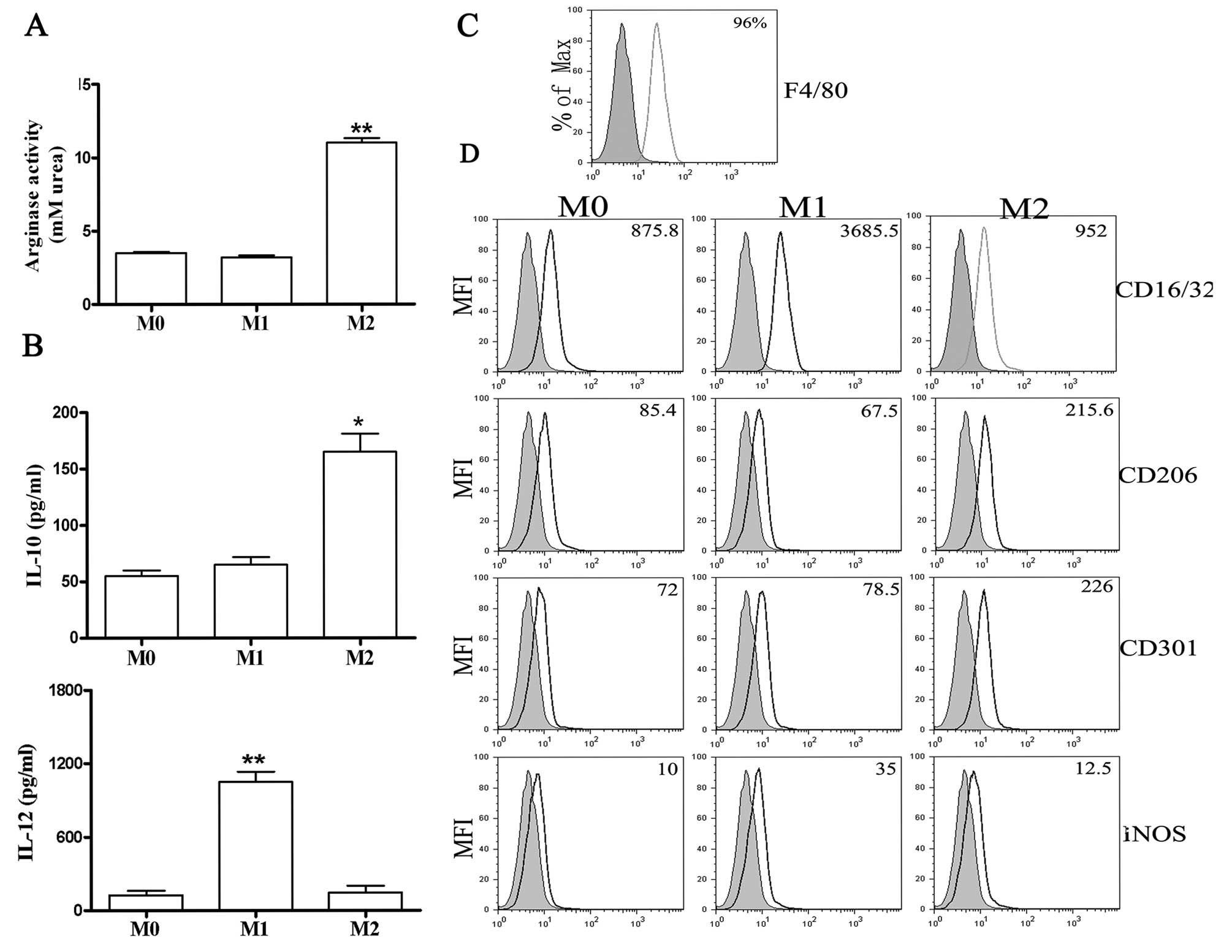
(Fig. 1B) and high levels of
arginase activity (Fig. 1A). By
assessing F4/80 expression, results of the FACS analysis showed
that the purity of BMDMs was ~96% (Fig. 1C). Moreover, FACS analysis showed
high levels of iNOS and CD16/32 expression in M1 macrophages and
increased CD206 and CD301 expression by M2 macrophages (Fig. 1D). These data confirmed that the
polarization conditions used in this study resulted in distinct
macrophage phenotypes.
miRNA microarray results analysis
To identify miRNAs that are differentially expressed
among M0, M1 and M2, we prepared a mouse miRNA microarray
containing 1111 oligonucleotide probes complementary to known
mammalian miRNAs. Probes were repeated three times in each
microarray and each microarray contained controls. The miRNA
expression patterns for M1 and M2 were compared. Significance
analysis of microarray and a fold change criterion (M1/M2 ratio) of
>2 or <0.5 and P<0.05 were used to identify significant
differences. Using these criteria, we identified 109 miRNAs that
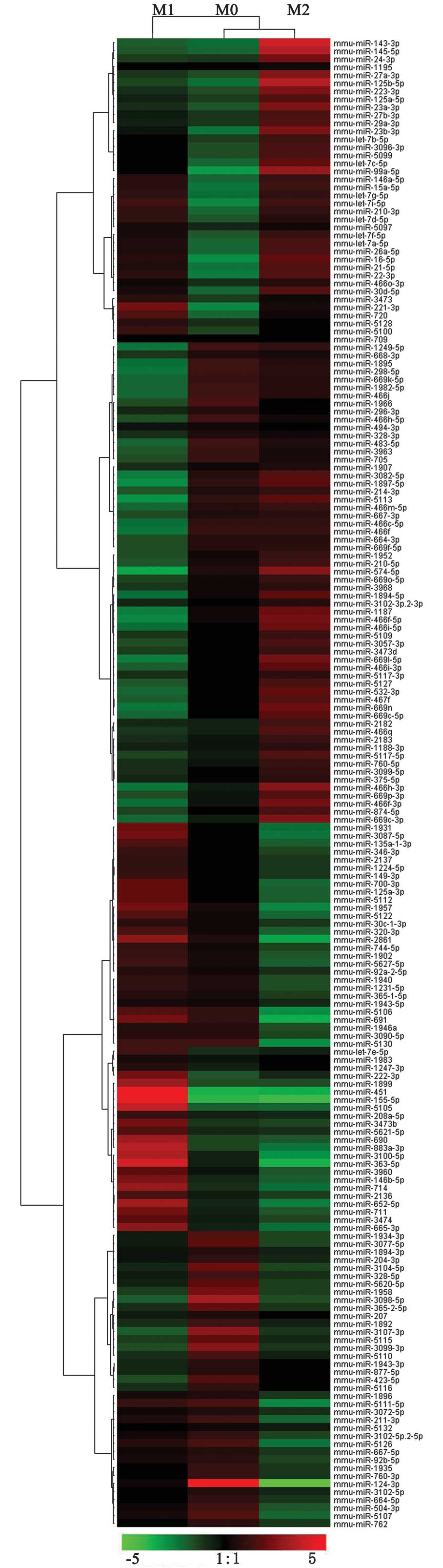
were differentially expressed between M1 and M2. Of these, 104
miRNAs were upregulated and 5 miRNAs were downregulated in M1
compared with M2 (Fig. 2,
Tables I and II).
 | Table IThe number of microRNAs differentially
expressed in polarized macrophages (expression fold of >2). |
Table I
The number of microRNAs differentially
expressed in polarized macrophages (expression fold of >2).
| No. | Comparison | Upregulation | Downregulation |
|---|
| 1 | M1 vs. M0 | 120 | 1 |
| 2 | M2 vs. M0 | 4 | 2 |
| 3 | M1 vs. M2 | 104 | 5 |
 | Table IImicroRNAs differentially expressed in
polarized macrophages (M1 vs. M2, expression fold >2). |
Table II
microRNAs differentially expressed in
polarized macrophages (M1 vs. M2, expression fold >2).
| Upregulated | Downregulated |
|---|
|
|
|---|
| mmu-miR-155-5p | mmu-miR-3091-3p | mmu-miR-410-3p | mmu-miR-125b-5p |
| mmu-miR-451 | mmu-miR-301b-5p | mmu-miR-544-3p | mmu-miR-466f-5p |
| mmu-miR-363-5p | mmu-miR-153-5p | mmu-miR-675-5p | mmu-miR-466h-3p |
| mmu-miR-3100-5p | mmu-miR-33-3p | mmu-miR-1932 | mmu-miR-145-5p |
| mmu-miR-5617-3p | mmu-miR-542-5p | mmu-miR-1197-5p | mmu-miR-143-3p |
| mmu-miR-7b-5p | mmu-miR-1930-5p | mmu-miR-1969 | mmu-miR-574-5p |
| mmu-miR-448-3p | mmu-miR-3109-3p | mmu-miR-3970 | |
| mmu-miR-1947-5p | mmu-miR-1298-5p | mmu-miR-5709 | |
| mmu-miR-141-3p |
mmu-miR-344d-2-5p |
mmu-miR-3095-5p | |
| mmu-miR-1950 | mmu-miR-714 |
mmu-miR-5617-5p | |
| mmu-miR-2139 |
mmu-miR-3086-3p |
mmu-miR-1955-3p | |
|
mmu-miR-3093-3p |
mmu-miR-465a-5p | mmu-miR-223-5p | |
|
mmu-miR-883a-3p | mmu-miR-221-5p | mmu-miR-191-3p | |
|
mmu-let-7f-1-3p | mmu-miR-381-3p |
mmu-miR-181d-3p | |
| mmu-miR-2861 |
mmu-miR-551b-5p | mmu-miR-152-5p | |
| mmu-miR-202-3p | mmu-miR-212-5p | mmu-miR-683 | |
| mmu-miR-5105 |
mmu-miR-301a-5p |
mmu-miR-344b-5p | |
| mmu-miR-880-3p | mmu-miR-33-5p | mmu-miR-205-3p | |
| mmu-miR-1953 | mmu-miR-379-3p | mmu-miR-453 | |
| mmu-miR-691 |
mmu-miR-3067-3p | mmu-miR-432 | |
|
mmu-miR-146a-3p |
mmu-miR-1199-5p |
mmu-miR-291b-5p | |
| mmu-miR-876-3p | mmu-miR-592-3p |
mmu-miR-190b-3p | |
| mmu-miR-431-3p | mmu-miR-433-5p | mmu-miR-409-3p | |
|
mmu-miR-1948-5p | mmu-miR-491-5p | mmu-miR-652-5p | |
| mmu-miR-127-3p |
mmu-miR-3079-3p | mmu-miR-421-3p | |
|
mmu-miR-1897-3p |
mmu-miR-3104-3p |
mmu-miR-450a-2-3p | |
|
mmu-miR-291a-5p | mmu-miR-410-5p |
mmu-miR-3080-3p | |
|
mmu-miR-1298-3p |
mmu-miR-1955-5p | mmu-miR-876-5p | |
| mmu-miR-471-3p |
mmu-miR-200b-3p |
mmu-miR-181a-1-3p | |
| mmu-miR-204-5p | mmu-miR-804 |
mmu-miR-551b-3p | |
|
mmu-miR-5626-5p |
mmu-miR-3082-3p | mmu-miR-299-5p | |
| mmu-miR-23b-5p | mmu-miR-122-3p | mmu-miR-136-5p | |
| mmu-miR-124-3p | mmu-miR-205-5p |
mmu-miR-128-1-5p | |
|
mmu-miR-3106-3p |
mmu-miR-92a-1-5p |
mmu-miR-3094-3p | |
| mmu-miR-25-5p | mmu-miR-107-5p | | |
To evaluate the function categories of miRNAs, we
used the bioinformatics tool, TAM (http://202.38.126.151/hmdd/tools/tam.html) (26), to integrate differentially
expressed miRNAs into different sets according to various rules and
provide information to mine potential biological meaning behind the
list of miRNAs studied. Significantly enriched terms in the miRNAs
with increased or decreased expression were apoptosis, cell
differentiation, cell proliferation, immune response and
inflammation (Table III).
 | Table IIIFunctional categories of
differentially expressed microRNAs strictly associated with
macrophage polarization. |
Table III
Functional categories of
differentially expressed microRNAs strictly associated with
macrophage polarization.
| Functions | microRNAs |
|---|
| Apoptosis | miR-181a, miR-155,
miR-204, miR-92a, miR-146a, miR-221 |
| Cell
differentiation | miR-145, miR-155,
miR-143, miR-127 |
| Cell
proliferation | miR-125b, miR-451,
miR-145, miR-143, miR-124, miR-221, miR-92a |
| Immune
response | miR-25, miR-125b,
miR-181a, miR-223, miR-155, miR-143, miR-146a, miR-92a |
| Inflammation | miR-25, miR-125b,
miR-181a, miR-223, miR-155, miR-146a, miR-143 |
qRT-PCR confirmation of the miRNA
microarray results
According to the function classification of
differentially expressed miRNAs, sets of miRNAs were selected and
qRT-PCR was used to confirm the results of the miRNA microarray
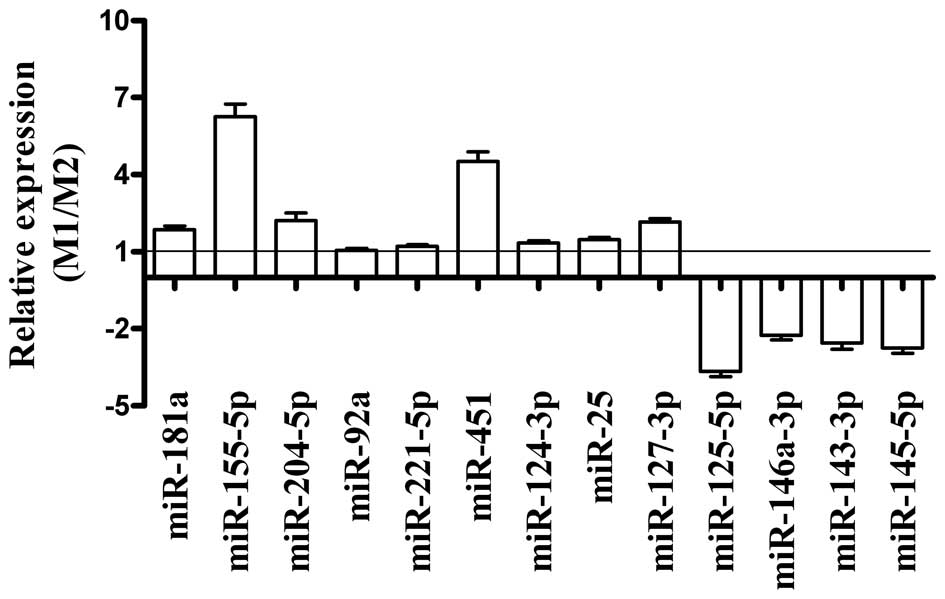
analysis. Of the nine miRNAs identified by the microarray as being
the most overexpressed in M1 compared to M2 (miR-155-5p, miR-181a,
miR-204-5p, miR-92a, miR-221-5p, miR-451, miR-124-3p, miR-25 and
miR-127-3p), qRT-PCR confirmed that five (miR-181a, miR-155-5p,
miR-204-5p, miR-451 and miR-127-3p) were overexpressed (Fig. 3). Of the four miRNAs identified as
being underexpressed in M1 by the microarray (miR-125-5p,
miR-146a-3p, miR-143-3p and miR-145-5p), the qRT-PCR analysis
confirmed that all four were underexpressed (Fig. 3). Overall, the qRT-PCR analysis
showed that the miRNA microarray results had some small errors,
however, it confirmed that a significant number of miRNAs were
differentially regulated in macrophages that responded to M1 and M2
polarizing conditions.
Discussion
Mammalian macrophages are induced to adopt a
spectrum of widely divergent phenotypes in response to diverse
external stimuli. The present study was based upon the hypothesis
that miRNAs are regulators that coordinate, in part, the global
changes in the expression of a number of genes that occur after
macrophage exposure to different activating conditions.
Expression profiling experiments have reported
changes in miRNA expression in human and murine monocytic cells
responding to selected inflammatory conditions (19–23,27–33). In these experiments, a subset of
miRNAs was repeatedly found to be induced following
inflammation-inducing stimuli, which caused induced differentiation
towards the M1 phenotype. We hypothesized that miRNAs are involved,
not only in macrophage responses to inflammatory conditions, but
also in the modifications of gene expression required to generate a
spectrum of macrophage activation patterns.
The aim of this study was to identify miRNAs that
respond to stimuli inducing two different patterns of macrophage
activation (M1 and M2). We profiled the expression of a number of
miRNAs using miRNA microarray and demonstrated that the expression
of 109 miRNAs was significantly different in M1 compared with M2.
Using function classification, we were able to correlate the
upregulation and downregulation of miRNAs to the
inflammation-related processes, such as apoptosis, cell
differentiation, cell proliferation, immune response, and
inflammatory response. However, few regulatory pathways have been
experimentally validated, the functional confirmation of which is
to be confirmed in subsequent studies.
According to the function analysis of differentially
expressed miRNAs, 13 of the 109 miRNAs were selected and qRT-PCR
was used to confirm the results of the miRNA microarray analysis.
In total, 9 of the 13 miRNAs tested were identified as being
differentially expressed in M1 by the microarray and qRT-PCR.
Similarities between our microarray data and those of previous
studies have been identified. Previous studies have shown that
several miRNAs, including miR-29b, miR-146a, miR-155, miR-193b,
miR-222 and miR-125b, were elevated during monocytic cell
differentiation towards macrophages (21,23,33–35). These miRNAs appeared to be
regulated in monocyte-derived macrophages (MDMs). In our study, in
addition to miR-146a, miR-155 and miR-125b, high levels of
miR-181a, miR-204-5p, miR-451 and miR-127-3p in M1 cells, and
miR-143-3p and miR-145-5p in M2 cells were detected. Although there
were discrepancies in the results of the microarray and the qRT-PCR
analyses, the miRNA microarray provided a rapid method for
identifying a large number of differentially expressed miRNAs in
M1, which was confirmed by qRT-PCR.
This study has examined the global expression
patterns of miRNAs in macrophage polarization and contributed to
the growing understanding of the role of miRNAs in macrophage
exposure to different activating conditions. Thus, miRNA profiling
reveals novel molecules and signatures associated with
differentiation of BMDMs and polarized activation which may be
candidate targets in pathophysiology.
Acknowledgements
This study was financially supported by the Natural
Science Foundation of the Anhui Higher Education Institutions of
China (KJ2011B191, KJ2011B198) and the Foundation of Introducing
Talents of Yijishan Hospital (YJRC2009003).
References
|
1
|
Rosa A, Ballarino M, Sorrentino A, et al:
The interplay between the master transcription factor PU1 and
miR-424 regulates human monocyte/macrophage differentiation. Proc
Natl Acad Sci USA. 104:19849–19854. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Androulidaki A, Iliopoulos D, Arranz A, et
al: The kinase Akt1 controls macrophage response to
lipopolysaccharide by regulating microRNAs. Immunity. 31:220–231.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bazzoni F, Rossato M, Fabbri M, et al:
Induction and regulatory function of miR-9 in human monocytes and
neutrophils exposed to proinflammatory signals. Proc Natl Acad Sci
USA. 106:5282–5287. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Du CS, Liu C, Kang JH, et al: MicroRNA
miR-326 regulates TH-17 differentiation and is associated with the
pathogenesis of multiple sclerosis. Nat Immunol. 10:1252–1259.
2009. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chang CI, Liao JC and Kuo L: Arginase
modulates nitric oxide production in activated macrophages. Am J
Physiol. 274:H342–H348. 1998.PubMed/NCBI
|
|
6
|
Chen T, Huang Z, Wang L, Wang Y, Wu F,
Meng S and Wang C: MicroRNA-125a-5p partly regulates the
inflammatory response lipid uptake and ORP9 expression in
oxLDL-stimulated monocyte/macrophages. Cardiovas Res. 83:131–139.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Cheng Y, Kuang WH, Hao YH, et al:
Downregulation of miR-27a* and miR-532-5p and upregulation of
miR-146a and miR-155 in LPS-induced RAW2647 macrophage cells.
Inflammation. 35:1308–1313. 2012.
|
|
8
|
Coley W, Van Duyne R, Carpio L, et al:
Absence of DICER in monocytes and its regulation by HIV-1. J Biol
Chem. 285:31930–31943. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Dalton DK, Pitts-Meek S, Keshav IS, Figari
A, Bradley A and Stewart TA: Multiple defects of immune cell
function in mice with disrupted interferon-gamma genes. Science.
259:1739–1742. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Forrest AR, Kanamori-Katayama M, Tomaru Y,
et al: Induction of microRNAs mir-155 mir-222 mir-424 and mir-503
promotes monocytic differentiation through combinatorial
regulation. Leukemia. 24:460–466. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Goerdt S and Orfanos CE: Other functions
other genes: alternative activation of antigen-presenting cells.
Immunity. 10:137–142. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Goerdt S, O Politz K, Schledzewski R, et
al: Alternative versus classical activation of macrophages.
Pathobiology. 67:222–226. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gordon S: Alternative activation of
macrophages. Nat Rev Immunol. 3:23–35. 2003. View Article : Google Scholar
|
|
14
|
Graff JW, Dickson AM, Gwendolyn C,
McCaffrey AP and Wilson ME: Identifying functional microRNAs in
macrophages with polarized phenotypes. J Biol Chem.
287:21816–21825. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing often flanked by adenosines indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liu G, Friggeri A, Yang Y, Park YJ,
Tsuruta Y and Abraham E: miR-147 a microRNA that is induced upon
Toll-like receptor stimulation regulates murine macrophage
inflammatory responses. Proc Natl Acad Sci USA. 106:15819–15824.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lu M, Shi B, Wang J, Cao Q and Cui QH:
TAM: a method for enrichment and depletion analysis of a microRNA
category in a list of microRNAs. BMC Bioinformatics. 11:419–426.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lumeng CN, Bodzin JL and Saltiel AR:
Obesity induces a phenotypic switch in adipose tissue macrophage
polarization. J Clin Invest. 117:175–184. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Mallory AC and Vaucheret H: MicroRNAs:
something important between the genes. Curr Opin Plant Biol.
7:120–125. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mantovani A, Sica A and Locati M:
Macrophage polarization comes of age. Immunity. 23:344–346. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Mantovani A, Sica A, Sozzani S, Allavena
P, Vecchi A and Locati M: The chemokine system in diverse forms of
macrophage activation and polarization. Trends Immunol. 25:677–686.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mantovani A, Sozzani S, Locati M, Allavena
P and Sica A: Macrophage polarization: tumor-associated macrophages
as a paradigm for polarized M2 mononuclear phagocytes. Trends
Immunol. 23:549–555. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Mills CD, Kincaid K, Alt JM, Heilman MJ
and Hill AM: M-1/M-2 macrophages and the Th1/Th2 paradigm. J
Immunol. 164:6166–6173. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Monk CE, Hutvagner G and Arthur JS:
Regulation of miRNA transcription in macrophages in response to
Candida albicans. PLoS One. 5:e136692010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Mosser DM: The many faces of macrophage
activation. J Leukoc Biol. 73:209–212. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Mosser DM and Edwards JP: Exploring the
full spectrum of macrophage activation. Nat Rev Immunol. 8:958–969.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
27
|
Munder M, Eichmann K and Modolell M:
Alternative metabolic states in murine macrophages reflected by the
nitric oxide synthase/arginase balance: competitive regulation by
CD4+ T cells correlates with Th1/Th2 phenotype. J
Immunol. 160:5347–5354. 1998.
|
|
28
|
Nathan CF, Murray HW, Wiebe ME and Rubin
BY: Identification of interferon-gamma as the lymphokine that
activates human macrophage oxidative metabolism and antimicrobial
activity. J Exp Med. 158:670–689. 1983. View Article : Google Scholar
|
|
29
|
O’Connell RM, Taganov KD, Boldin MP, Cheng
G and Baltimore D: MicroRNA-155 is induced during the macrophage
inflammatory response. Proc Natl Acad Sci USA. 104:1604–1609.
2007.PubMed/NCBI
|
|
30
|
O’Connell RM, Kahn D, Gibson WS, et al:
MicroRNA-155 promotes autoimmune inflammation by enhancing
inflammatory T cell development. Immunity. 33:607–619.
2010.PubMed/NCBI
|
|
31
|
Ruggiero T, Trabucchi M, De Santa F, et
al: LPS induces KH-type splicing regulatory protein-dependent
processing of microRNA-155 precursors in macrophages. FASEB J.
23:2898–2908. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Schulte LN, Eulalio A, Mollenkopf HJ,
Reinhardt R and Vogel J: Analysis of the host microRNA response to
Salmonella uncovers the control of major cytokines by the let-7
family. EMBO J. 30:1977–1989. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Taganov KD, Boldin MP, Chang KJ and
Baltimore D: NF-kappaB-dependent induction of microRNA miR-146 an
inhibitor targeted to signaling proteins of innate immune
responses. Proc Natl Acad Sci USA. 103:12481–12486. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Tili E, Michaille JJ, Cimino A, et al:
Modulation of miR-155 and miR-125b levels following
lipopolysaccharide/TNF-alpha stimulation and their possible roles
in regulating the response to endotoxin shock. J Immunol.
179:5082–5089. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Tserel L, Runnel T, Kisand K, et al:
MicroRNA expression profiles of human blood monocyte-derived
dendritic cells and macrophages reveal miR-511 as putative positive
regulator of Toll-like receptor 4. J Biol Chem. 286:26487–26495.
2011. View Article : Google Scholar
|