Introduction
Curcumin
(1,7-bis(4-hydroxy-3-methoxyphenyl)-1,6-heptadiene-3,5-dione) is
the major yellow pigment extracted from turmeric, which is a
commonly used spice derived from the rhizome of the Curcuma
longa plant (1). An
increasing number of studies have recently supported its use in
cancer prevention therapy due to its anti-proliferative and
anti-carcinogenic properties (2).
Curcumin not only suppresses carcinogenesis of the skin, stomach,
colon and breast in vivo but also inhibits the growth of a
wide variety of tumor cells in vitro (3,4).
Prohibitin (PHB), also known as inhibin, is widely
distributed in bacteria, plants, yeast, protozoa and mammals and is
important in cell proliferation, differentiation and apoptosis
(5). This protein localizes to
the inner membrane of mitochondria, where it acts as a chaperone
protein (6,7), and is found in the nucleus, where it
negatively regulates transcription (8). The Phb1 gene, which is a
member of the PHB family, is located beside the tumor-suppressor
gene BRCA1 in the chromosome, rendering Phb1 highly
relevant in breast cancer (9).
Findings of a recent study revealed that the aberrant expression of
PHB was clearly overexpressed in gastric, liver and uterine cancer,
whereas another study showed that PHB is capable of promoting cell
apoptosis by interacting with a specific tumor-suppressor protein
(10–12). This finding appears to contradict
the tumor-suppressive role of PHB. At present, the mechanism
underlying its subcellular localization and the mechanisms through
which it regulates cell proliferation and differentiation remain to
be elucidated.
In the present study, we investigated the existence,
localization and alteration of the expression of PHB in HaCaT cells
in response to treatment with curcumin. We also examined the
interaction between PHB and proteins related to oncogenes and
tumor-suppressor genes during curcumin-induced apoptosis. Thus,
results of this study provided new insight on the functions and
mechanism of action of PHB as an antitumor target during cell
apoptosis.
Materials and methods
Materials
Immortalized human epidermal HaCaT cells were
obtained from the China Center for Type Culture Collection (Wuhan,
China). Goat anti-mouse HRP-IgG, goat anti-rabbit HRP-IgG, the
mouse anti-human PHB antibody, and the rabbit anti-human p53, Rb,
Fas and c-Myc antibodies were all obtained from Santa Cruz
Biotechnology, Inc. (Santa Cruz, CA, USA). RPMI-1640 was purchased
from Gibco-BRL (Carlsbad, CA, USA) and newborn calf serum was
obtained from Hangzhou Sijiqing Biological Engineering Material
Co., Ltd. (Hangzhou, China). Curcumin was obtained from the
National Institute of the Control Pharmaceutical and Biological
Products (NICPBP) (Beijing, China).
Cell culture and induction
Immortalized human epidermal HaCaT cells (China
Center for Type Culture Collection) were cultured in RPMI-1640
medium supplemented with 10% heat-inactivated fetal calf serum, 100
U/ml penicillin, and 100 mg/ml streptomycin (pH 7.2) at 37°C in air
with 5% CO2. Twenty-four hours after seeding, the HaCaT
cells were maintained in RPMI-1640 with 7.5 mg/l curcumin for 72 h
to induce differentiation. HaCaT cells cultured in RPMI-1640 medium
were used as the control.
Cell-selective extraction and sample
preparation for light microscopy
The cells were selectively extracted as described in
a previous study (13). After
selective extraction, the cells were prefixed in 2% glutaraldehyde
at 4°C for 30 min, and the nuclear matrix-intermediate filament
(NM-IF) samples were coverslipped and rinsed in phosphate-buffered
saline (PBS) at pH 7.4. The samples were then stained with 0.2%
Coomassie Brilliant Blue for 20 min, washed in distilled water,
air-dried, clarified by xylene, enveloped in a resin and observed
by Olympus BH-2 microscopy.
Purification of the nuclear matrix
protein
HaCaT cells were washed twice with cold PBS and
extracted using a cytoskeleton (CSK100) buffer (100 mM NaCl, 3 mM
MgCl2, 10 mM PIPES, 300 mM sucrose, 0.5% Triton X-100, 1
mM EGTA, and 1 mM PMSF, pH 6.8) for 10 min at 0°C. After
centrifugation at 1,000 × g for 5 min, the pellets were washed with
cold PBS to remove soluble cytoplasmic proteins and then
re-centrifuged and suspended in the digestion buffer CSK50
(identical to CSK100 buffer, except with 50 mM NaCl instead of KCl)
containing 400 U/ml DNase I for 30 min at room temperature. Cold
ammonium sulfate was added to a final concentration of 0.25 M to
precipitate the proteins. After centrifugation at 1,000 × g for 5
min, the pellets were washed with CSK50 buffer and dissolved in
lysis buffer [7 M urea, 2 M thiourea, 4% CHAPS, 1.5% Triton X-100
and 1% pharmalyte (pH 3–10; Amersham Biosciences), 65 mM DTT, 40 mM
Tris, 5 mg/ml aprotinin, 1 mg/ml leupeptin, 1 mg/ml pepstatin, 2 mM
PMSF and 4 mM EDTA]. The sample was sonicated at 0°C for 30 min and
centrifuged at 10,000 × g for 1 h. The protein concentrations of
the control and treated groups were determined using the Bradford
assay, and the proteins were stored at −80°C.
Western blot analysis
The protein lysates were electrophoretically
separated in 12% polyacrylamide gels and transferred onto PVDF
membranes. The membranes were incubated in 5% non-fat milk for 1.5
h at room temperature to block any non-specific binding and then
incubated with mouse PHB nucleophosmin antibody (1:1,000 dilution;
Santa Cruz Biotechnology, Inc.) in TBST for 1 h. The membranes were
then washed once with TBST for 10 min, incubated with horseradish
peroxidase-conjugated goat anti-mouse IgG (1:10,000 dilution; Santa
Cruz Biotechnology, Inc.) as the secondary antibody for 1 h at room
temperature, and washed three times with TBST for 30 min.
Immunoreactive bands were identified using an enhanced
chemiluminescence (ECL) detection system (Pierce Biotechnology,
Inc., Rockford, IL, USA). Samples incubated with 5% non-fat milk
instead of the primary antibodies were used as negative controls.
In addition, β-actin was used as an internal control.
Sample preparation for fluorescent
microscopy
The NM-IF samples on the cover slip were prefixed in
4% paraformaldehyde at 4°C for 10 min, rinsed twice in TPBS
(containing 0.5% Triton X-100) for 10 min, and blocked with 5% BSA
at room temperature for 1 h. The samples were incubated with mouse
anti-PHB antibody (1:300 dilution) at room temperature for 30 min,
incubated overnight at 4°C, and then washed three times with TPBS.
The cells were then incubated with goat anti-mouse secondary
antibody labeled with the fluorescent dye FITC, washed in water,
and air-dried. Then, 90% glycerol in PBS was applied, and the cells
were observed by fluorescence microscopy. All of the steps after
incubation with the secondary antibody were performed in the dark,
and samples incubated with 5% BSA instead of the primary antibody
were used as the negative control.
Sample preparation for LSCM
The cells on the cover slips in the curcumin and
control groups were rinsed three times in PBS for 15 min and
submerged in 0.1 M TBS (containing 0.5% Triton X-100) for 20 min at
room temperature. The cells were fixed in 4% paraformaldehyde at pH
7.2 for 10 min, washed three times with PBS (pH 7.2) for 15 min,
blocked with 5% BSA at room temperature for 1 h, and then incubated
with dual primary antibodies at room temperature for 30 min and at
4°C overnight. The dual sets of primary antibodies were: PHB
(1:50)/Fas (1:30), PHB (1:50)/c-Myc (1:30), PHB (1:50)/P53 (1:30),
PHB (1:50)/Bax (1:30), and PHB (1:50)/Bcl-2 (1:30). After washing
with TBS, the cells were incubated with different secondary
antibody sets (goat anti-mouse and goat anti-rabbit, both diluted
at 1:200) and incubated at room temperature for 3 h in the dark.
The cells were then washed three times with PBS for 30 min,
enveloped with an anti-fluorescence quencher after drying, blocked
with nail polish, and observed under TSC-SP2-MP LSCM.
GST pull-down assay
The samples were inoculated with non-carrier
bacterial plasmids, and the constructed prokaryotic expressive
strains were induced through soluble expression. The GST protein
and GST combined protein were refined using glutathione sepharose
4B following the procedure described above. RIPA lysis buffer was
used to dissolve the HaCaT cells, and the cells were centrifuged at
12,000 × g and 4°C for 5 min. The centrifuged cell lysates were
incubated with GST and GST-PHB combined with beads at 4°C for 1 h
in a silent mixer. The cells were then washed three times using the
GST washing buffer. Then, 5X SDS loading buffer was added for
SDS-PAGE, and the same amount was added to the cell lysates as
described previously (16). The
interaction between the prey and bait protein was verified by
western blot analysis.
Results
Detection of PHB in the whole cells and
nuclear matrix by western blot analysis
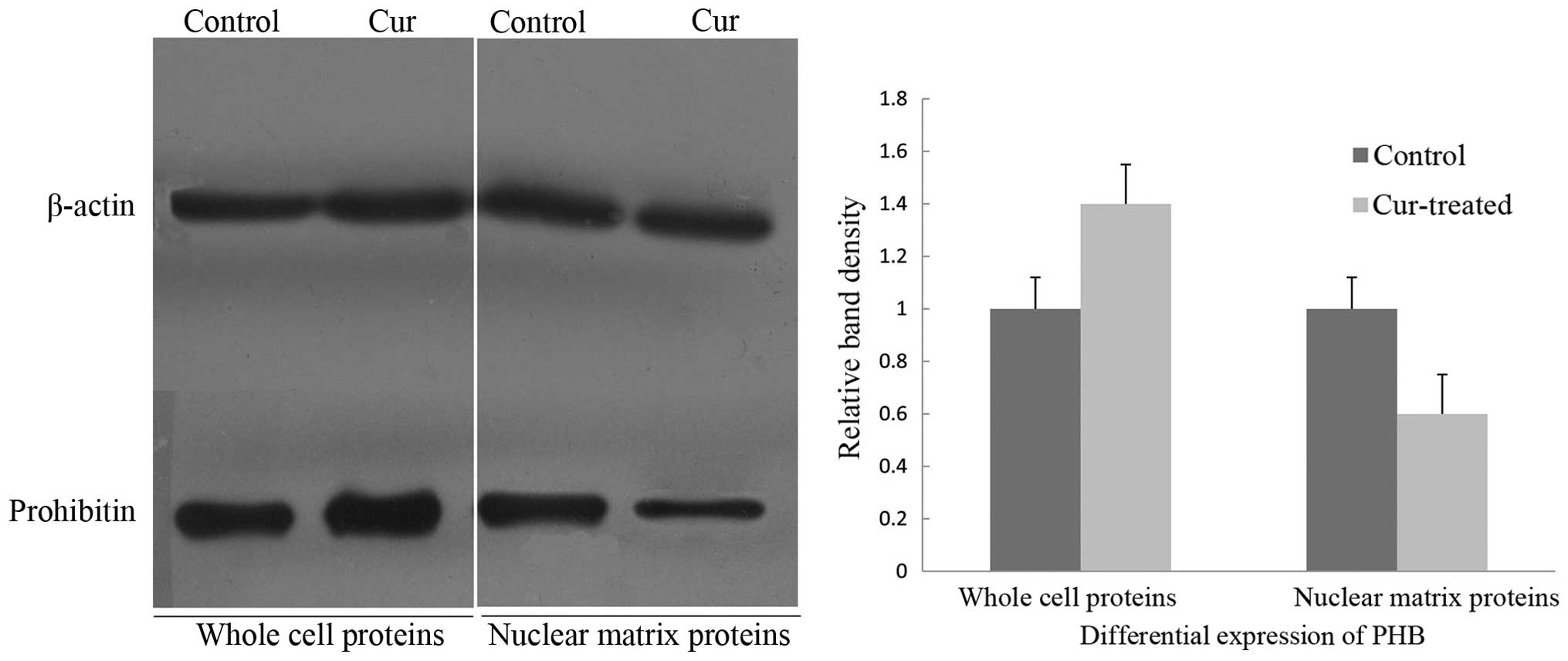
To verify the aberrant changes in PHB, western
immunoblotting was employed to confirm the expression levels of PHB
in the whole cells and nuclear matrix prior to and following
curcumin treatment, and the intensities of the protein bands were
densitometrically quantified as described by Sheffield (14). The expression level of PHB in
whole cells after exposure to curcumin for 48 h was increased,
whereas the level of the PHB was significantly decreased in the
nuclear matrix protein (Fig. 1).
Thus, the PHB expression in whole cells and the nuclear matrix of
HaCaT cells was inversely correlated.
Localization and expression of PHB in
HaCaT cells
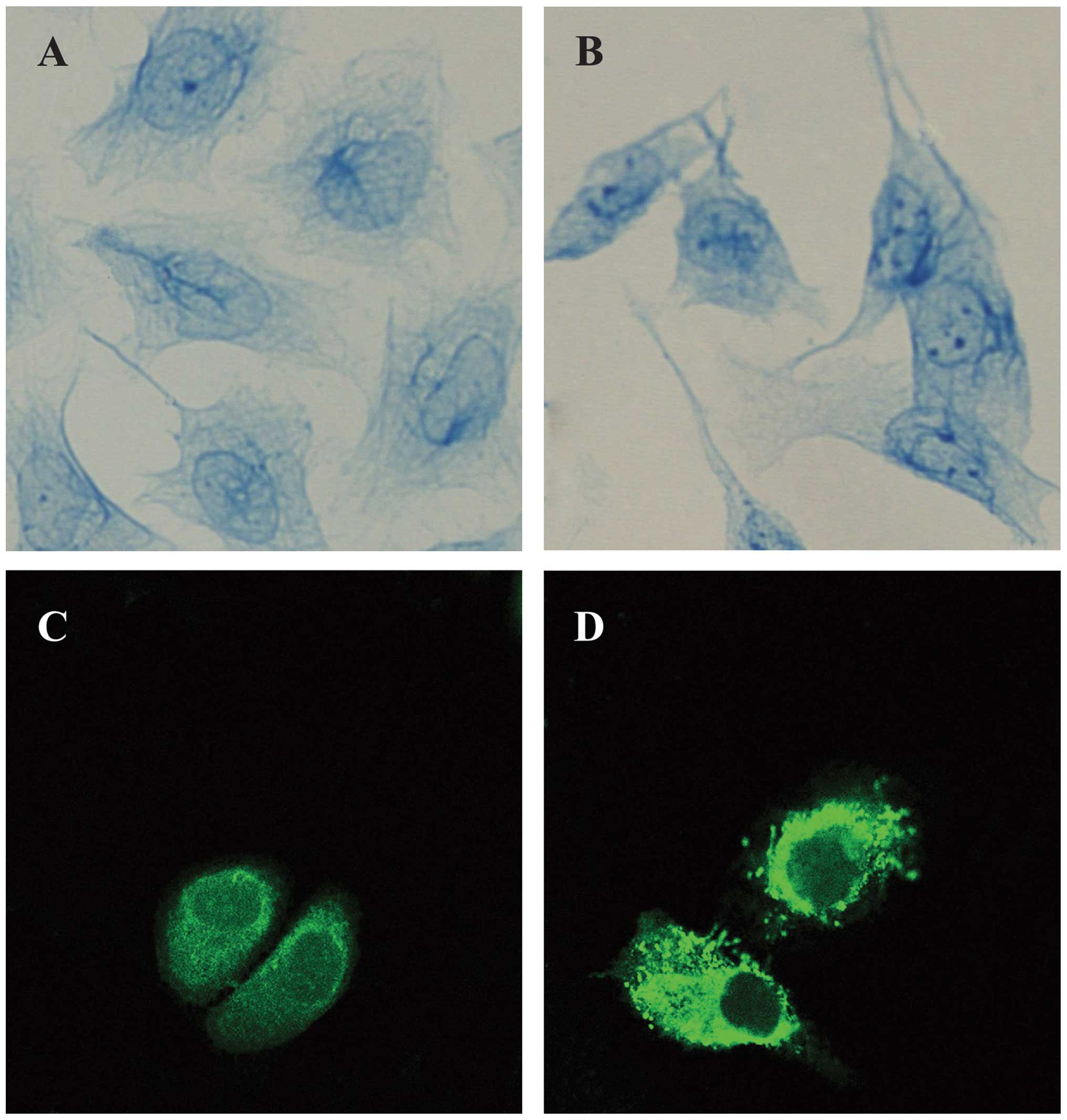
The light microscopy observations showed that the IF
in HaCaT cells was sparse and distributed irregularly in the
nucleus. In the curcumin-treated cells, the entire framework became
more widespread, whereas the NM-IF results showed characteristics
of uniform distribution. The evenly stained intermediate filaments
spread from the region around the nucleus to the edge of the cell
to form a uniform network throughout the cytoplasm (Fig. 2A and B).
The immunofluoresence analysis revealed the
localization and expression of PHB in HaCaT cells; the PHB protein
was labeled with FITC (green). The results revealed that PHB was
distributed throughout the cell. Its signal was strong in the
cytoplasm but weak in the nucleus, where it was distributed as
small particles, especially at the edge of nuclei; in particular,
the PHB signal was relatively strong in the nearby nuclear membrane
region. PHB was also evenly distributed in the cytoplasmic region
(Fig. 2C). The analysis of
curcumin-treated cells through microscopy showed that the
distribution and expression of PHB changed significantly. The
brightness of the fluorescence in the nucleus of curcumin-treated
HaCaT cells was obviously weakened, whereas the brightness of the
fluorescence of the cytoplasm of these cells was strengthened. This
finding indicates that PHB tended to move from the nuclear matrix
to the lamina and cytoplasm (Fig.
2D).
LSCM analysis of the co-localization of
PHB with oncogenes and tumor-suppressor genes during
curcumin-induced apoptosis
The localization of PHB and its related proteins,
c-Myc, Bax, p53 and Fas, was observed by LSCM. PHB was labeled with
FITC (green), and the other proteins were labeled with TRITC (red).
The co-localization fluorescence was yellow or orange, i.e., the
combination of the two different colors.
In HaCaT cells, PHB was distributed almost
throughout the entire cell. Compared with its distribution in the
cytoplasm, the amount and distribution of PHB in the karyoplasm was
lower and uneven, respectively. In response to curcumin treatment,
the level of PHB was enhanced and widely distributed in the
cytoplasm of HaCaT cells and decreased or even absent in the
karyoplasm.
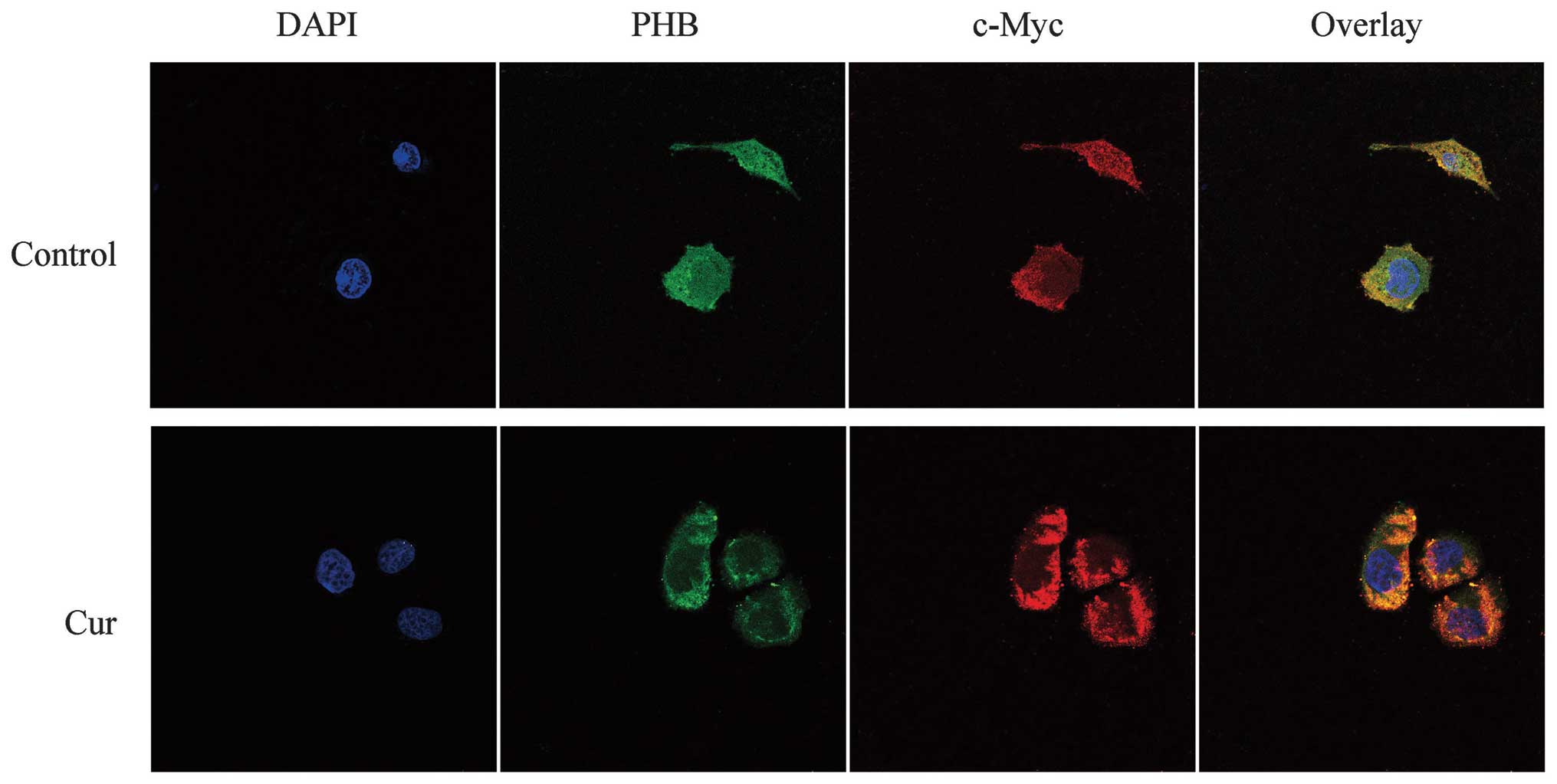
Co-localization of PHB with c-Myc in
HaCaT cells
Fluorescence microscopy revealed that the nuclei of
HaCaT cells stained with DAPI were blue in the control and
curcumin-treated group. The c-Myc protein, which was labeled by
TRITC, emitted a red fluorescence and was mainly distributed in the
cytoplasmic region. By contrast, the distribution of the green
fluorescence showed that PHB was distributed near the nuclear
membrane region, whereas other locations exhibited weaker
fluorescence. The yellow in the overlaid fluorescence image
indicated that the co-localization of PHB and c-Myc was found only
slightly in the nucleus edge region but at a higher amount in the
nucleolus. However, in the curcumin-treated HaCaT cells, both the
green fluorescence in the cytoplasm and the red color representing
c-Myc were strengthened. The co-localization between the two
proteins in the curcumin-treated cells was enhanced in the
cytoplasm, a result that most likely demonstrates that the
co-localization region of the two proteins was transferred from the
nucleus to the cytoplasm in response to curcumin treatment
(Fig. 3).
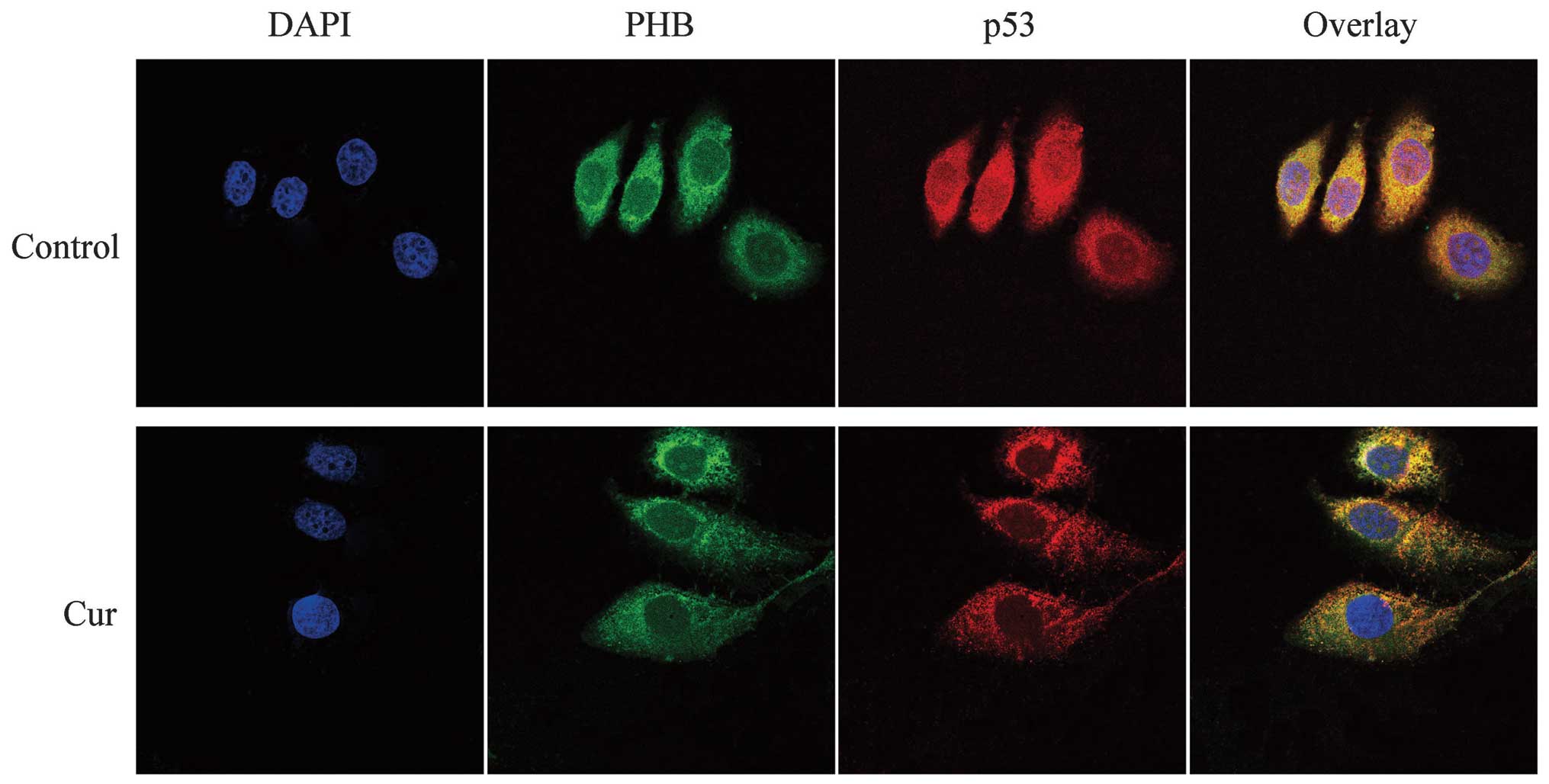
Co-localization of PHB with p53 in HaCaT
cells
In the control HaCaT cells, the green fluorescence
representing PHB was distributed throughout the cell, although it
was higher near the nuclear membrane in the cytoplasmic region and
weaker in the other cell regions. p53, which was labeled with
TRITC, exhibited red fluorescence that was distributed throughout
the cell, but the fluorescence intensity was higher in the
cytoplasm and nucleolus. The yellow fluorescence (overlaid signals)
indicated that the co-localization between PHB and p53 was high in
the nuclear membrane region. The curcumin-treated HaCaT cells
exhibited weaker fluorescence throughout the entire cell. The green
color was scattered in the cytoplasm and decreased in the nucleus;
however, the nucleolus exhibited green fluorescence. In addition,
p53 was distributed in the cytoplasmic region and exhibited a
weaker fluorescence in the nucleolus of these curcumin-treated
cells. The overlapped yellow fluorescent color demonstrated that
PHB and p53 are co-localized in the nuclear membrane region,
particularly in the cytoplasmic region. In addition, the
co-localization in the nucleolus was markedly lower compared with
that found in the control cells. The abovementioned evidence
suggests that the co-localization region of the two proteins moved
from the nucleus to the cytoplasmic region in response to curcumin
treatment (Fig. 4).
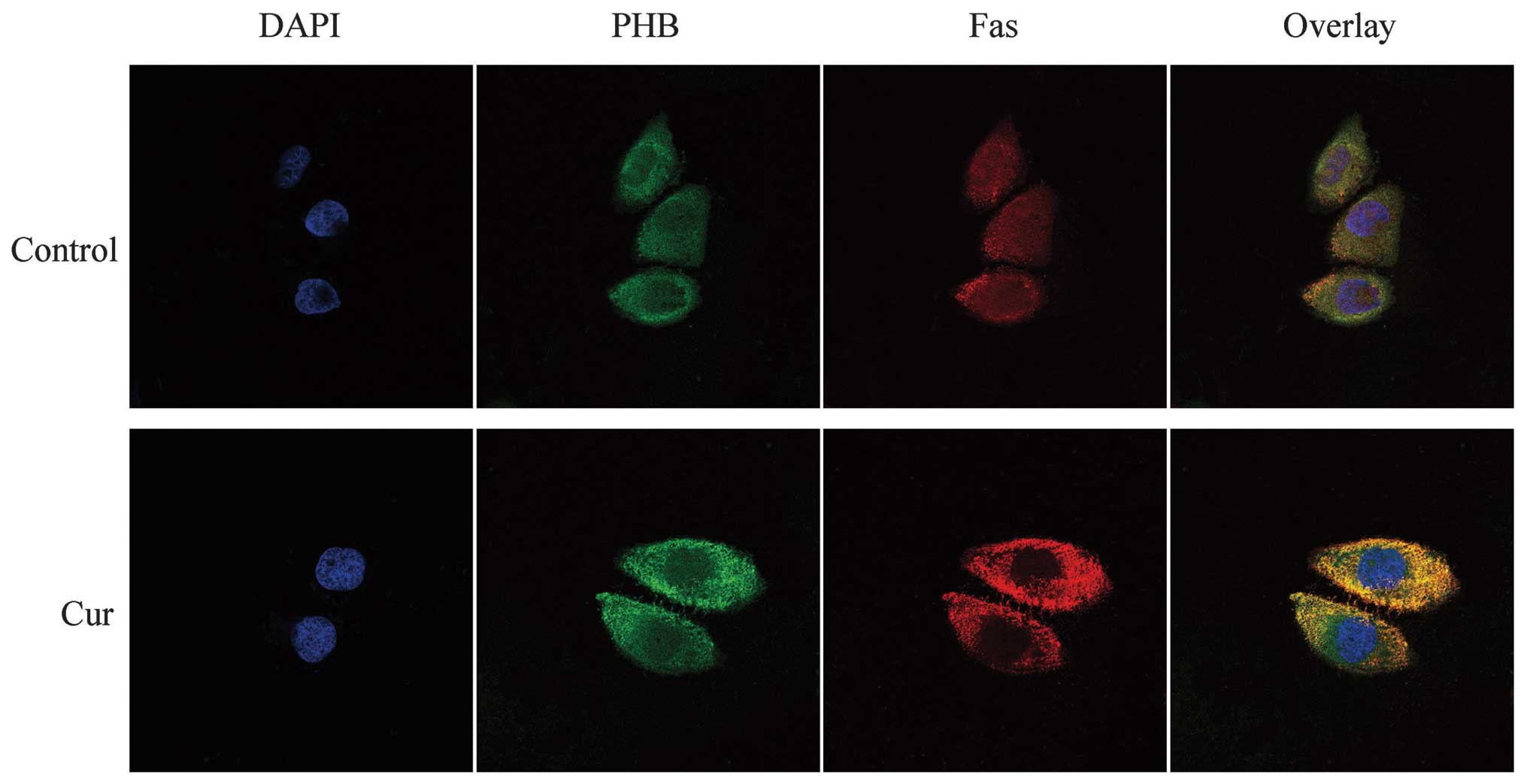
Co-localization of PHB with Fas in HaCaT
cells
According to the fluorescent staining results, the
red fluorescence, which represented Fas, was well distributed
throughout the cell. Compared with the fluorescence intensity in
the nucleus, the intensity increased near the nuclear membrane
region. The yellow fluorescence clearly revealed the
co-localization of PHB and Fas in the nucleolus and around the
nuclear membrane. The green fluorescence, which represented PHB,
was weaker in the HaCaT cell nucleus following treatment with
curcumin and was increased in the cytoplasm. In addition, after
curcumin treatment, Fas was well distributed in the cytoplasm and
was slightly weaker in the nucleus, although its fluorescence
remained strong. The co-localization of PHB and Fas after curcumin
treatment suggests that the two proteins were clearly co-localized
around the cytoplasm and the cytomembrane, although the
co-localization relationship in the nucleus was weakened or
completely absent. This finding suggests that the PHB and Fas
co-localization region shifted from the nucleus to the cytoplasm in
response to curcumin treatment (Fig.
5).
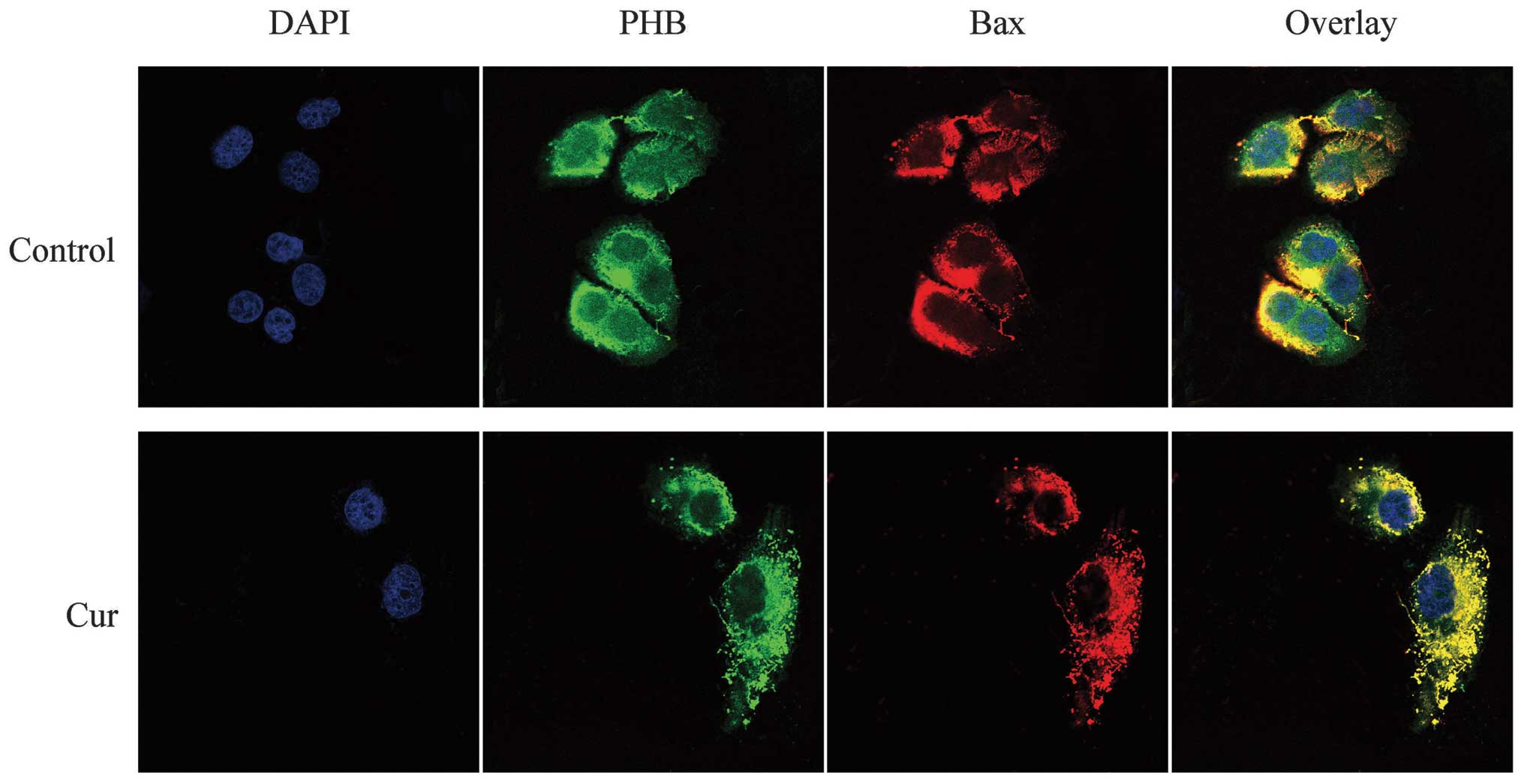
Co-localization of PHB with Bax in HaCaT
cells
Bax was labeled with TRITC and thus exhibited a red
fluorescence. The LSCM results showed that Bax was distributed
throughout the cell, although the fluorescence intensity was weaker
in the nucleus but stronger around the nucleolus. Similarly, PHB
exhibited green fluorescence that was well distributed in the
nucleus, although the fluorescence intensity was mostly clustered
near the nucleolus. The overlaid yellow fluorescence of the two
proteins was strongly co-localized near the nucleus and less
co-localized in the nucleolus. After treatment with curcumin, the
level of PHB decreased significantly in the nucleus, and the green
fluorescence was clearly enhanced in the cytoplasm. Following
curcumin treatment, the red fluorescence, which represents Bax,
decreased significantly or even disappeared in the nucleus and was
significantly enhanced in the cytoplasm. The yellow fluorescence
showed that the two proteins were clearly co-localized in the
cytoplasm and not co-localized in the nucleus. This finding
demonstrates that the co-localization of PHB and Bax shifted from
the nucleus to the cytoplasm in response to curcumin treatment
(Fig. 6).
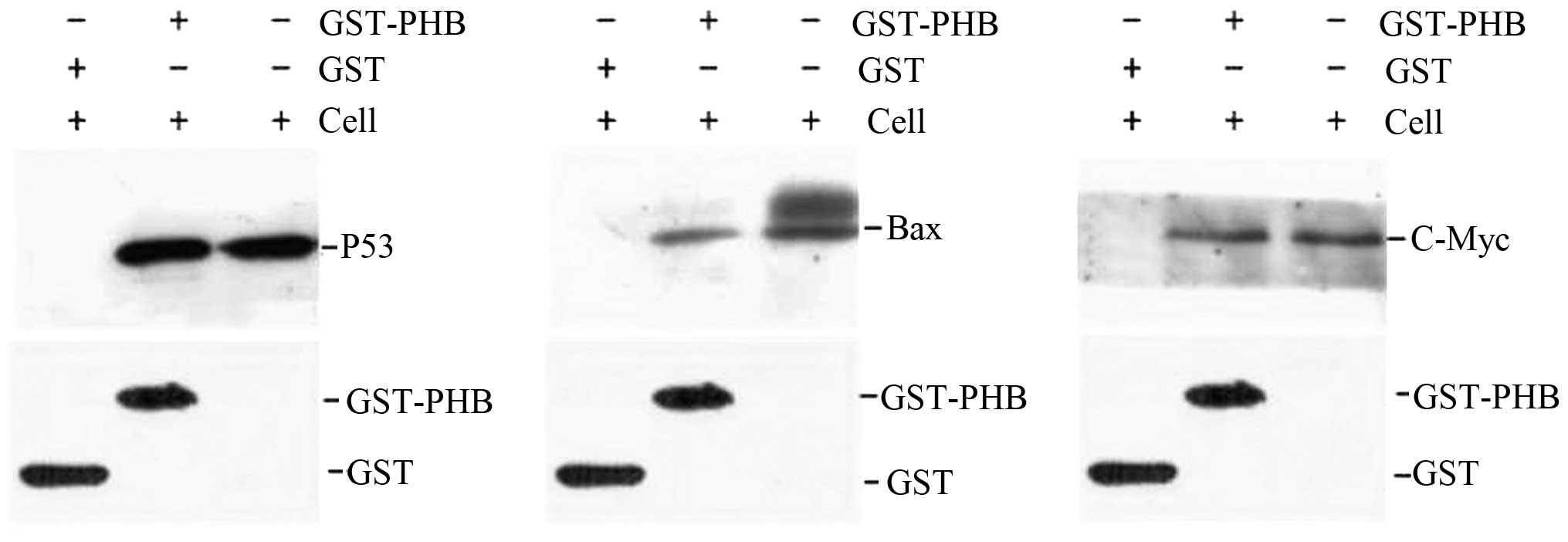
Interaction of PHB with p53, c-Myc, Bax
and Fas in HaCaT cells
To verify the interaction between PHB and the
oncogene proteins in HaCaT cells, a GST pull-down assay was
employed. GST-PHB and GST were expressed in bacteria and purified
to near homogeneity. After the recombinant proteins GST-PHB and GST
were incubated with the HaCaT cell lysate supernatant, the western
blotting results suggested the existence of a direct interaction
between GST-PHB and p53 in vitro, although no interaction
was identified between GST and p53 (Fig. 7). Identical results were obtained
for the analysis of GST-PHB with c-Myc and Bax, and no significant
interaction was detected between GST-PHB and Fas (data not shown).
As a result, we suggest that PHB directly interacts with p53, c-Myc
and Bax, and these results are consistent with the cellular
co-localization results (Fig.
7).
Discussion
Expression and subcellular localization
changes of PHB
The PHB protein is encoded by the PHB gene, which is
related to cell apoptosis, and is found to be diffused throughout
all eukaryotic cells, which means that it is highly conserved in
evolution. Results of a previous study have shown that PHB is
upregulated in tumor cells compared with normal cells (15). Results of the present study show
that PHB was upregulated in whole cells and markedly downregulated
in the nuclear matrix. Additionally, we found that PHB is
translocated from the nuclei to the nuclear membrane and cytoplasm
after curcumin treatment. These results indicate that PHB is found
not only in the mitochondria of the cytoplasm but also in the
nuclear matrix of HaCaT cells and that its decreased nuclear
expression and localization is associated with canceration and
reversion. These results are consistent with those of our previous
study, which suggests that PHB is capable of being downregulated
and transported from the cytoplasm to the nucleus in osteosarcoma
MG-63 cells (16). This ability
of PHB may be one of the molecular mechanisms associated with its
effectiveness in the prevention of cancer. There are conflicting
experimental results regarding the manner in which a microinjection
of PHB mRNA blocks cell proliferation (17) and in the manner in wihch PHB
functions as a tumor-suppressor protein and arrests G1-S cell
transition through a process involving the repression of
E2F-mediated transcription (18).
We hypothesized that PHB has different functions in different cell
lines, that its expression levels are related to the tumor clinical
stage, and that it may act as a molecular switch that controls cell
proliferation.
Alteration of the co-localization and
interaction between PHB and other oncogenes and tumor-suppressor
genes
The immunofluorescence microscopy and LSCM results
revealed that PHB co-localizes with the products of p53, c-Myc, Bax
and Fas genes in HaCaT cells and that the location of this
co-localization is altered by curcumin treatment. In addition, the
GST pull-down assay showed that PHB directly interacts with p53,
c-Myc and Bax in HaCaT cells.
The tumor-suppressor gene p53 is a type of specific
transcription factor and is highly conserved in evolution, similar
to PHB. p53 is important in the inhibition of the cell cycle, the
promotion of cell apoptosis and aging (19). DNA damage cannot be repaired in
time if the p53 gene is in an inactive state, and the accumulation
of DNA damage contributes to the conversion of a cell into a tumor
cell. Our LSCM results indicate that the co-localization of p53 and
PHB is unevenly distributed, mainly in the nuclear region. However,
the co-localization location of the two proteins was altered
following treatment with curcumin. This phenomenon shows that the
interaction between the two proteins may regulate cell apoptosis,
and these results were also verified through the GST pull-down
experiment.
Additionally, the c-Myc oncogene encodes the c-Myc
protein. The c-Myc protein is a type of transcription factor, and
its expression has been found to be increased in most human tumor
cells. In addition, c-Myc has the dual effect of promoting cell
proliferation and apoptosis (20). Findings of a previous study have
shown that c-Myc is important in mammalian cell apoptosis (21). Therefore, c-Myc is the main factor
that regulates cell proliferation, differentiation, apoptosis,
canceration and metastasis. Our LSCM results revealed that the PHB
and c-Myc co-localization is found in the nucleolus and near the
nucleus in control cells and is mainly found in the cytoplasm
following curcumin treatment. This change in the co-localization
location suggests that there is a close relationship between the
interaction of the two proteins and cell apoptosis, as was
confirmed by the GST pull-down results.
Bax, which is located in the cytoplasm, belongs to
the Bcl-2 family and is another factor that controls cell
apoptosis. Homodimers of Bax or the combination of Bax and Bcl-2
are capable of inducing cell apoptosis (22). However, the Bax/Bac-2 dipolymer
may exhibit a rivalry with Bcl-2 in the inhibition of apoptosis.
These proteins regulate cell apoptosis mainly by controlling the
transfer of Bax to the mitochondria (23). The LSCM results obtained in this
study suggest that the location of the co-localization between Bax
and PHB is altered from the nucleus to the cytoplasm following
treatment with curcumin. This finding suggests the hypothesis that
the expression of Bax is increased via its interaction with PHB,
which prevents Bax from translocating to the mitochondrion and
promotes the induction of the apoptosis pathway. The findings
obtained in our study suggest that the interaction of Bax with PHB
after PHB is transported out of the nucleus leads to the activation
of an apoptosis signal factor, which contributes to the death of
HaCaT cells.
The interaction of Fas with its natural ligand FasL
activates the apoptosis signal for transmembrane delivery and is
thus termed the ‘dead ligand’. The Fas protein is expressed in
numerous tumor cells, and the Fas and FasL compounds may activate
the cascade reaction of the caspase family, thereby inducing the
apoptosis of target cells (24,25). The LSCM results of the present
study reveal that Fas is found mainly in the cytoplasm and the
nucleus and that its co-localization with PHB is mainly found in
the nucleolus and cell cytoplasm. Following treatment with
curcumin, the location of the co-localization of the two proteins
was altered in the cytoplasm, which demonstrates the relationship
between the interaction of the two proteins and HaCaT cell
apoptosis. However, these results were not confirmed through our
GST pull-down experiment. Therefore, additional investigation on
the Fas protein is required as there are no studies on the
relationship between the interaction of PHB and Fas and cell
apoptosis.
In this study, we confirmed the activity of PHB as a
significant regulatory factor in the proliferation and apoptosis of
HaCaT cells. This conclusion was based on the findings after
curcumin treatment that its expression was decreased in the nucleus
and increased in the whole cell and that PHB is co-localized and
translocated with oncogene proteins and tumor-suppressor proteins.
We provide evidence for additional investigation to be conducted
into the function of PHB during the proliferation of human
epidermal HaCaT cells. Additionally, the described interactions of
PHB with the proteins encoded by oncogenes and tumor-suppressor
genes offer new insight into the mechanism underlying the
development of canceration and its reversion.
Acknowledgements
This study was supported by National Natural Science
Foundation of China (grant nos. 81071669, 81272921 and 81272245)
and Natural Science Foundation of Fujian Province (grant no.
2011J01256).
References
|
1
|
Jana NR, Dikshit P, Goswami A and Nukina
N: Inhibition of proteasomal function by curcumin induces apoptosis
through mitochondrial pathway. J Biol Chem. 279:11680–11685. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Boucher BJ: Curcumin and diabetes: a role
for the vitamin D receptor? Br J Nutr. 108:21042012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Rodwell C: Curcumin curries favour? Nat
Rev Cancer. 12:3762012. View
Article : Google Scholar
|
|
4
|
Watson JL, Hill R, Yaffe PB, et al:
Curcumin causes superoxide anion production and p53-independent
apoptosis in human colon cancer cells. Cancer Lett. 297:1–8. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mishra S, Ande SR and Nyomba BL: The role
of prohibitin in cell signaling. FEBS J. 277:3937–3946. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Merkwirth C and Langer T: Prohibitin
function within mitochondria: essential roles for cell
proliferation and cristae morphogenesis. Biochim Biophys Acta.
1793:27–32. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhou P, Qian L, D’Aurelio M, et al:
Prohibitin reduces mitochondrial free radical production and
protects brain cells from different injury modalities. J Neurosci.
32:583–592. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gamble SC, Odontiadis M, Waxman J, et al:
Androgens target prohibitin to regulate proliferation of prostate
cancer cells. Oncogene. 23:2996–3004. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jupe ER, Badgett AA, Neas BR, et al:
Single nucleotide polymorphism in prohibitin 3′ untranslated region
and breast-cancer susceptibility. Lancet. 357:1588–1589. 2001.
|
|
10
|
Fusaro G, Dasgupta P, Rastogi S, Joshi B
and Chellappan S: Prohibitin induces the transcriptional activity
of p53 and is exported from the nucleus upon apoptotic signaling. J
Biol Chem. 278:47853–47861. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhu B, Zhai J, Zhu H and Kyprianou N:
Prohibitin regulates TGF-beta induced apoptosis as a downstream
effector of smad-dependent and -independent signaling. Prostate.
70:17–26. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wong PF, Cheong WF, Shu MH, Teh CH, Chan
KL and AbuBakar S: Eurycomanone suppresses expression of lung
cancer cell tumor markers, prohibitin, annexin 1 and endoplasmic
reticulum protein 28. Phytomedicine. 19:138–144. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li QF: Effect of retinoic acid on the
changes of nuclear matrix in termediate filament system in gastric
carcinoma cells. World J Gastroenterol. 5:417–420. 1999.PubMed/NCBI
|
|
14
|
Sheffield JB: ImageJ, a useful tool for
biological image processing and analysis. Microsc Microanal.
13:200–201. 2007. View Article : Google Scholar
|
|
15
|
Kang X, Zhang L, Sun J, et al: Prohibitin:
a potential biomarker for tissue-based detection of gastric cancer.
J Gastroenterol. 43:618–625. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Shi SL, Li QF, Liu QR, et al: Nuclear
matrix protein, prohibitin, was down-regulated and translocated
from nucleus to cytoplasm during the differentiation of
osteosarcoma MG-63 cells induced by ginsenoside Rg1, cinnamic acid,
and tanshinone IIA (RCT). J Cell Biochem. 108:926–934. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Nuell MJ, Stewart DA, Walker L, et al:
Prohibitin, an evolutionarily conserved intracellular protein that
blocks DNA synthesis in normal fibroblasts and HeLa cells. Mol Cell
Biol. 11:1372–1381. 1991.PubMed/NCBI
|
|
18
|
McClung JK, Jupe ER, Liu XT and Dell’Orco
RT: Prohibitin: potential role in senescence, development, and
tumor suppression. Exp Gerontol. 30:99–124. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yu J and Zhang L: The transcriptional
targets of p53 in apoptosis control. Biochem Biophys Res Commun.
331:851–858. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ninomiya I, Yonemura Y, Matsumoto H, et
al: Expression of c-myc gene product in gastric carcinoma.
Oncology. 48:149–153. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cao X, Bennett RL and May WS: c-Myc and
caspase-2 are involved in activating Bax during cytotoxic
drug-induced apoptosis. J Biol Chem. 283:14490–14496. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li X, Ye H, Cai L, et al: Millimeter wave
radiation induces apoptosis via affecting the ratio of Bax/Bcl-2 in
SW1353 human chondrosarcoma cells. Oncol Rep. 27:664–672.
2012.PubMed/NCBI
|
|
23
|
Yang D, Liu X, Zhang R, et al: Increased
apoptosis and different regulation of pro-apoptosis protein bax and
anti-apoptosis protein bcl-2 in the olfactory bulb of a rat model
of depression. Neurosci Lett. 504:18–22. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Xu B, Xu Z, Xia T, et al: Effects of the
Fas/Fas-L pathway on fluoride-induced apoptosis in SH-SY5Y cells.
Environ Toxicol. 26:86–92. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang XY, Zhang R and Lian S: Aberrant
expression of Fas and FasL pro-apoptotic proteins in basal cell and
squamous cell carcinomas. Clin Exp Dermatol. 36:69–76. 2011.
View Article : Google Scholar : PubMed/NCBI
|