Introduction
Renal fibrosis is characterized by an enhancement
fibroblast proliferation and an increase in matrix protein
expression along with the loss of functioning nephrons. The final
common outcome of renal fibrosis is generally tubulointerstitial
fibrosis. It has been demonstrated that epithelial-to-mesenchymal
transition (EMT) is involved in the pathogenesis of
tubulointerstitial fibrosis (1).
In this process, tubular epithelial cells acquire a mesenchymal
phenotype which results in their transition into myofibroblasts
(2,3).
It is currently well known that EMT plays an
important role in embryonic development, as well as in wound
healing, tissue regeneration, cancer progression and organ
fibrosis, including renal fibrosis and is regulated by a series of
transcription factors and signaling pathways (4–7).
It has been demonstrated that the EMT-mediated renal fibrosis is
affected by a number of factors, including transforming growth
factor (TGF)-β1, connective tissue growth factor (CTGF),
Angiotensin II (ANG II), fibroblast growth factor (FGF)-2,
interleukin (IL)-1, hepatocyte growth factor (HGF) and bone
morphogenetic protein 7 (BMP-7) (8). The zinc-finger transcription factor,
Krüppel-like factor 4 (KLF4), performs divergent functions in a
number of physiological or pathological process. Recent studies
have demonstrated that KLF4 expression is decreased or lost in
cancer and the inhibition of its expression induces EMT by
regulating the expression of EMT-related genes, such as E-cadherin,
N-cadherin, vimentin, β-catenin, vascular endothelial growth factor
(VEGF)-A, endothelin-1 and Jnk1 in breast cancer (9) or through crosstalk with the TGF-β,
Notch and Wnt signaling pathways in gastrointestinal cancer
(10). Moreover, it has been
reported that KLF4 is epigenetically silenced by promoter
methylation and functions as a tumor suppressor gene in renal cell
carcinoma (11). However, the
expression pattern and role of KLF4 during EMT in renal fibrosis
remain unclear.
Epigenetic modificaions are essential for the
development and function of the kidneys (12), and aberrant methylation (13), histone modifications or the
dysregulation of microRNAs may cause chronic kidney disease
(14–16). It has been demonstrated that the
hypermethylation of RASAL1, encoding an inhibitor of the Ras
oncoprotein, is mediated by DNA methyltransferase 1 (Dnmt1) and is
associated with fibrogenesis in the kidneys (17). This prompted us to hypothesize
that the epigenetic repression of KLF4, similar to its role in
renal cell carcinoma (11), may
promote EMT in renal fibrogenesis.
In the present study, the expression, methylation
status and function of KLF4 in renal fibrosis were investigated. To
the best of our knowledge, we demonstrate for the first time that
the hypermethylation of KLF4 is associated with EMT in renal
fibrosis and that this hypermethylation is regulated by Dnmt. Thus,
our findings provide a new molecular basis for EMT in epithelial
cells in renal fibrosis.
Materials and methods
Cell culture
Human renal proximal tubule epithelial cells (HK-2
cells, ATCC® CRL-2190) were cultured in RPMI-1640 medium
containing 2,000 mg/l NaHCO3, supplemented with 10%
fetal bovine serum (FBS), 100 U/ml penicillin and 100 μg/ml
streptomycin in 5% CO2 at 37°C. The cells were then
stimulated for different periods of time with human recombinant
TGF-β1 (1 ng/ml; PeproTech, Rocky Hill, NJ, USA). Cell culture
reagents were obtained from Life Technologies Inc., Carlsbad, CA,
USA.
Animal model of unilateral ureteral
obstruction (UUO)
Male CD-1 mice were obtained from the Xiangya
Hospital of Central South University, Changsha, China. The mice in
the present study were divided into 3 groups as follows: i) the
mice in the control group (Con) were fed without surgical
manipulation; ii) the mice in the sham-operated group were
subjected to surgical manipulation without ureteral ligation; and
iii) the mice in the unilateral ureteral obstruction (UUO) group
were subjected to left ureteral ligation. UUO was performed as
previously described (18). All
mice were sacrificed 14 days following UUO and the kidney tissues
were collected for various analyses. The protocols for animal
experimentation and maintenance were approved by the Animal Ethics
Committee at our institute and carried out in accordance with
institutional guidelines.
Methylation-specific PCR (MSP)
MSP was carried out on bisulfate-treated DNA using
the EZ DNA Methylation-Gold™ kit (Zymo Research, Irvine, CA, USA)
according to the manufacturer’s instructions. Methylation-specific
primers for the KLF4 promoter were designed using the MethPrimer
program, as previously described (19). Bisulfite converted genomic DNA was
PCR-amplified using methylation-specific primers and SYBR-Green
reaction mix. The human primers used were unmethylated and were as
follows: KLF4 forward, 5′-tgtagggtttaaataggtgataatga-3′ and
reverse, 5′-aaataataaaaa ctcaaacaccaaa-3′; and methylated KLF4
forward, 5′-cgta gggtttaaataggtgataacg-3′ and reverse,
5′-aaataataaaaactcg aacaccgaa-3′. The mouse primers used were
unmethylated and were as follows: KLF4 forward,
5′-ttgtaaagagttttgagatttttgg-3′ and reverse,
5′-caaaattaaacacataatcatcatt-3′; and methylated KLF4 forward,
5′-gttcgtaaagagttttgagattttc-3′ and reverse,
5′-gaaattaaacacgtaatcatcgtt-3.
Epigenetic drug treatment of the
cells
The cells were treated with the demethylation drug,
5-Aza-2′-deoxycytidine (5-Aza), which was purchased from
Sigma-Aldrich (St. Louis, MO, USA; A3656) at 15.5 nM for 72 h.
Reverse transcription-quantitative PCR
(RT-qPCR)
The total RNA was extracted from the cells using
TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the
manufacturer’s instructions, and the RNA was subsequently reverse
transcribed using the PrimeScript RT Master Mix Perfect Real-Time
kit (Takara, Dalian, China) to obtain the cDNA. Using the cDNA as a
template, a real-time PCR assay was performed using the following
pairs of primers: KLF-4 forward, 5′-ctgagcagcagggactg-3′ and
reverse, 5′-ttgagatgggaactctttgt-3′ in HK-2 cells, and KLF-4
forward, 5′-ttctccacgttcgcgtccgg-3′ and reverse,
5′-tctcgccaacggttagtcgggg-3′ in mice; and Dnmt1 forward,
5′-aaccttcacctagccccag-3′ and reverse, 5′-tgacaggtggt
cactcctcatg-3′; Dnmt3A forward, 5′-tattgatgagcgcacaagagagc-3′ and
reverse, 5′-gggtgttccagggtaacattgag-3′; Dnmt3B forward,
5′-tacacagacgtgtccaacatgggc-3′ and reverse, 5′-gaggaagctgct
aaggactagttc-3′; E-cadherin forward, 5′-aatgccgccatcgcttacac-3′ and
reverse, 5′-cgacgttagcctcgttctca-3′; α-smooth muscle actin (α-SMA)
forward, 5′-tcccttgagaagagttacgagtt-3′ and reverse,
5′-catgatgctgttgtaggtggtt-3′; zonula occludens-l (ZO-1) forward,
5′-ggaagttacgtggcgaag-3′ and reverse, 5′-ctctggcggacatcttgt-3′;
fibroblast-specific protein 1 (FSP-1) forward, 5′-cctctctacaa
ccctctct-3′ and reverse, 5′-ggacaccatcacatccag-3′; and β-actin
forward, 5′-aggggccggactcgtcatact-3′ and reverse, 5′-ggcggcacc
accatgtaccct-3′. The 20 μl real-time PCR reaction included
0.5 μl of cDNA template, 0.25 μl of Primer F, 0.25
μl of Primer R, 10 μl of RNase-free dH2O
and 8 μl of 2.5X Real-Time Master Mix (SYBR-Green I). The
reaction conditions included a pre-denaturation step at 95°C for 10
sec, and 40 cycles of 95°C for 15 sec and 60°C for 60 sec. After
the reaction, the data were subjected to statistical analysis.
Construction of lentivirus (LV) for KLF4
overexpression
To generate the KLF4-overexpression plasmid,
KLF4-CDS (NM_004235.4) was amplified by PCR using the following
primers: forward, 5′-gaattcatgaggcagccacctggc-3′ (including
the EcoRI site, shown in bold font) and reverse,
5′-ggatccttaaaaat gcctcttcatgtg-3′, (including the
BamHI site, shown in bold font); the sequence was then
cloned into the BamHI/EcoRI sites of the pLV-Neo
vector (Inovogen, Beijin, China). The pLV-Neo-KLF4 or pLV-Neo
vectors were co-transfected with packaging plasmids into 293T cells
using Lipofectamine 2000. The viral supernatant of pLV-Neo-KLF4 or
pLV-Neo was filtered through a 0.45-μm filter and renamed as
Lv-KLF4 and Lv-Con, respectively. All the supernatants were used to
infect the target cells with 10 μg/ml of polybrene
(Sigma-Aldrich). After the target cells were infected for 96 h,
KLF4 expression was confirmed by quantitative (real-time) PCR and
western blot analysis (Fig. 3).
The cells infected with Lv-KLF4 and Lv-Con were used for further
analyses.
Chromatin immunoprecipitation (ChIP)
assay
ChIP assay was carried out as previously described
(20). ChIP assays were performed
using the ChIP-IT kit (Active Motif, Carlsbad, CA, USA) according
to the manufacturer’s instructions. The HK-2 cells were
conventionally cultured to approximately 70–80% confluency, the
medium was decanted, and the medium containing 1% formaldehyde was
then added to fix the cells. The cells were washed with cold PBS,
and then glycine was added to terminate the fixation, and the cells
were washed again with cold PBS. After the cells were scaped from
the culture plate, the cold cell lysate was centrifuged to collect
the nuclei. The pelleted nuclei were resuspended, and chromatin was
cleaved into approximately 500-bp fragments by ultrasonication. The
cleaved chromatin was treated with RNase and proteinase K, and the
extent of cleavage was then assessed by 1% agarose gel
electrophoresis. Protein G beads were used to remove the
non-specific antibodies from the chromatin preparation,
Subsequently, an anti-Dnmt1 antibody (Cat. no. ab13537; Abcam,
Cambridge, UK), a positive control RNA polymerase (pol) II antibody
(Cat. no. ab5131; Abcam) and a negative control IgG antibody (Cat.
no. ab2410; Abcam) were added. The cells were incubated overnight
at 4°C. Protein G beads were added to the solution containing the
antibody/chromatin complexes, which were then washed and eluted,
de-cross-linked and the co-precipitated DNA was purified. Finally,
the co-precipitated DNA was used as a template in a PCR
amplification using the following primers: forward, 5′-cag
gagaatcgcttggacg-3′ and reverse, 5′-gaacaggcggaaagaggc-3′.
Western blot analysis
The cells were lysed in cell lysate, and then
centrifuged at 12,000 × g for 20 min at 4°C. The supernatant was
collected and denatured. Proteins were separated by 10% SDS-PAGE
and blotted onto polyvinylidene difluoride (PVDF) membranes. The
PVDF membranes were treated with TBST containing 50 g/l skimmed
milk at room temperature for 4 h, followed by incubation with the
primary antibodies, anti-KLF4 (Cat. no. BM0485; Abzoom Biolabs,
Dallas, TX, USA), anti-E-cadherin (Cat. no. BM0530; Abzoom
Biolabs), anti-α-SMA (Cat. no. YT5053; ImmunoWay Biotechnology
Company, Newark, DE, USA), anti-ZO-1 (1:1,000 dilution; Cat. no.
ab59720; Abcam), anti-FSP-1 (1:5,000 dilution; Cat. no. ab27957;
Abcam) and anti-β-actin (Cat. no. BM0272; Abzoom Biolabs),
respectively, at 37°C for 1 h. The membranes were rinsed and
incubated for 1 h with the corresponding peroxidase-conjugated
secondary antibodies. Chemiluminescent detection was performed
using the ECL kit (Pierce Chemical Co., Rockford, IL, USA). The
amount of the protein of interest, expressed as arbitrary
densitometric units, was normalized to the densitometric units of
β-actin.
Immunocytochemistry
The expression of E-cadherin, ZO-1, α-SMA and FSP-1
in the control HK-2 cells (infected with Lv-Con) and stimulated or
not with TGF-β1, and in the HK-2 cells infected with Lv-KLF4 and
stimulated with TGF-β1, was determined by immunocytochemistry. A
standard immunostaining procedure was performed using antibodies
against E-cadherin (1:500 dilutions), α-SMA (1:500 dilutions),
FSP-1 (1:200 dilutions), ZO-1 (1:100 dilutions). Protein expression
was graded on a scale of ‘±’ to ‘+++’ as previously described
(21).
Statistical analysis
Data are expressed as the means ± SD from at least 3
separate experiments. Statistical analysis was carried out using
SPSS 15.0 software. Differences between 2 groups were analyzed
using the Student’s t-test. A value of P<0.05 was considered to
indicate a statistically significant difference.
Results
The expression and methylation status of
KLF4 in mice subjected to UUO
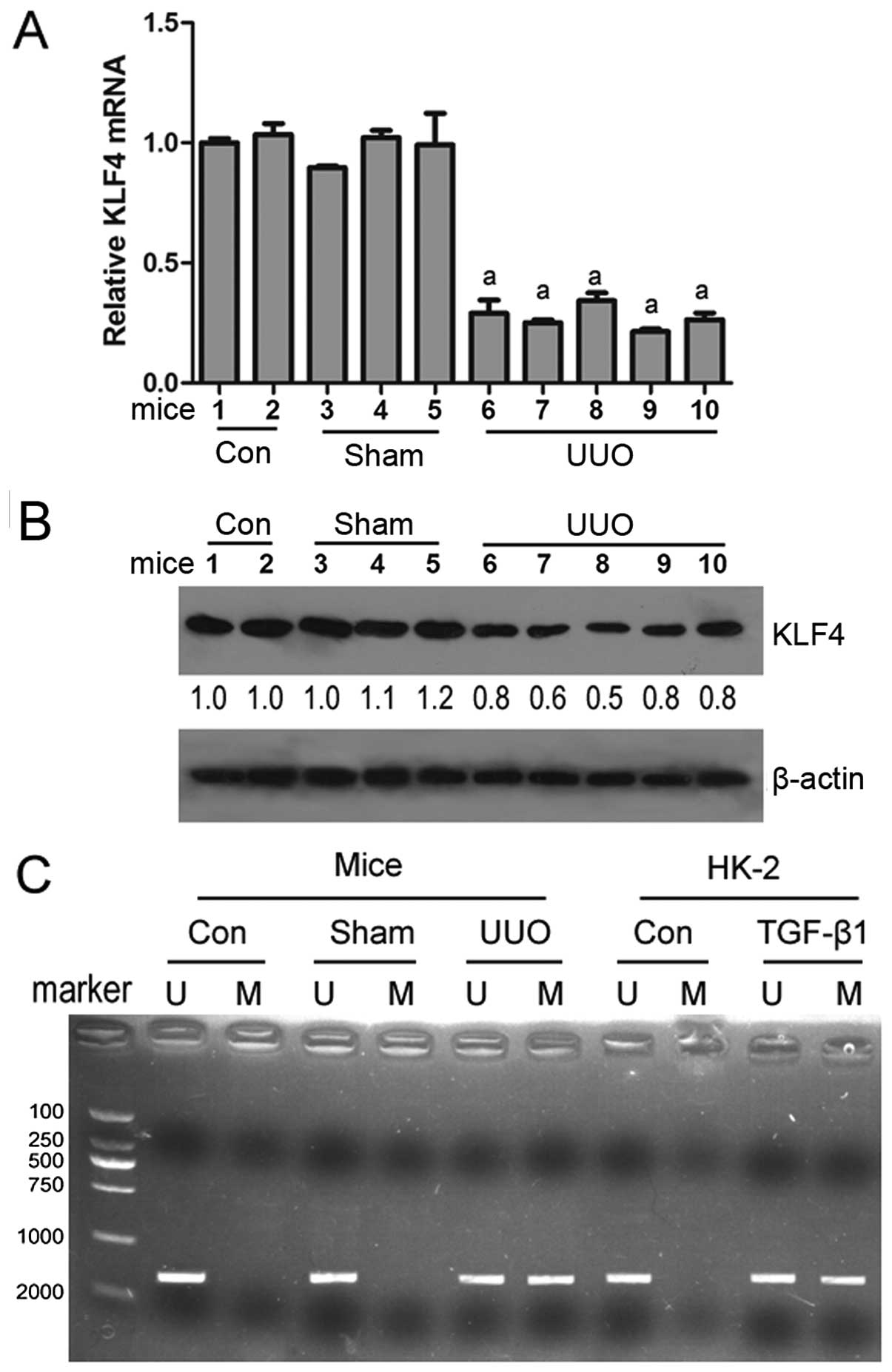
To determine whether KLF4 is involved in renal
fibrosis, an animal model of renal disease was created by
subjecting mice to UUO. RT-qPCR and western blot analysis revealed
that the KLF4 mRNA and protein levels were decreased in the renal
tissues from the mice subjected to UUO compared with the mice in
the control or sham-operated groups (Fig. 1A and B). MSP analysis revealed
that KLF4 methylation was observed in the renal tissues of mice
subjected to UUO, while KLF4 methylation was not observed in the
mice in the control group or sham-operated group (Fig. 1C). These results suggest that KLF4
is downregulated by hypermethylation, thus contributing to renal
fibrogenesis.
KLF4 is hypermethylated and downregulated
in HK-2 cells stimulated with TGF-β1
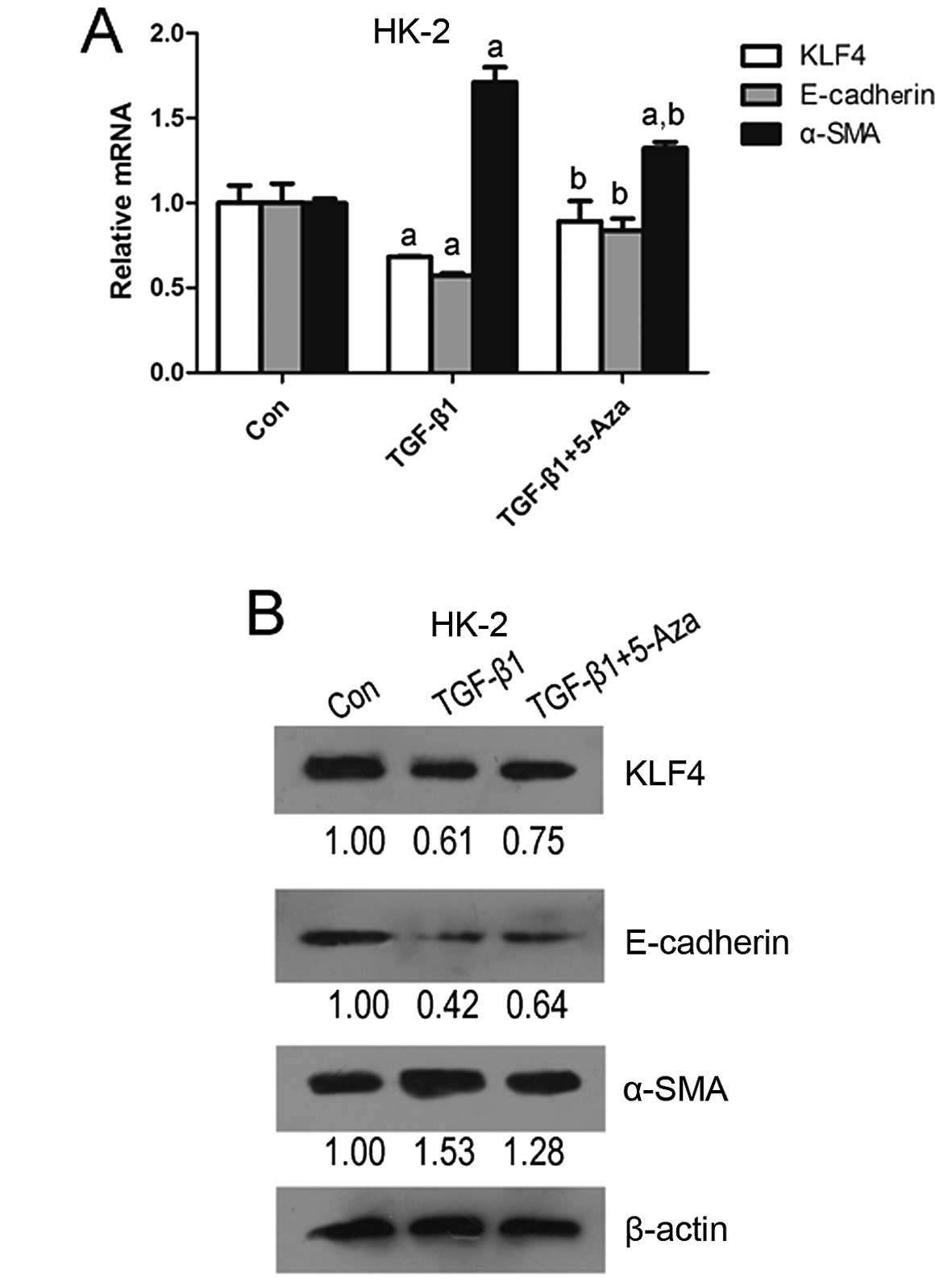
To further determine whether KLF4 is involved in
renal fibrosis by affecting EMT in renal tubular epithelial cells,
human renal proximal tubule epithelial cells (HK-2 cells) were
stimulated with TGF-β1, which induces EMT in epithelial cells.
Following stimulation for 36 h, a decrease in KLF4 and E-cadherin
expression accompanied by an increase in α-SMA expression were
observed in the HK-2 cells (Fig.
2), suggesting that KLF4 functions as a suppressor of EMT
induced by TGF-β1 in tubular epithelial cells. Moreover,
5-aza-2′-deoxycytidine, as a demethylating agent, attenuated the
TGF-β1-induced downregulation in KLF4 expression (Fig. 2). MSP analysis indicated that both
methylated and unmethylated KLF4 were detected in the HK-2 cells
stimulated with TGF-β1; however, only unmethylated KLF4 was
detected in the controls (untreated cells) (Fig. 1C). These results demonstrate that
the downregulation of KLF4 is at least partly induced by
hypermethylation alternation, and that this may promote EMT in
tubular epithelial cells in the process of renal fibrogenesis.
Dnmt1 is directly involved in the
hypermethylation of KLF4 in HK-2 cells stimulated with TGF-β1
The results from RT-qPCR revealed that the Dnmt1 and
Dnmt3A expression levels were significantly increased in the HK-2
cells stimulated with TGF-β1; however, Dnmt3B expression was
decreased (Fig. 3A). ChIP assay
indicated that after Dnmt1 protein-KLF4 promoter complexes were
captured with Dnmt1-specific antibodies, KLF4-promoter-specific
segments were detected (Fig. 3B).
These data demonstrate that Dnmt1 plays a direct role in the
TGF-β1-mediated hypermethylation of KLF4 in HK-2 cells.
KLF4 functions as a suppressor of EMT in
renal epithelial cells
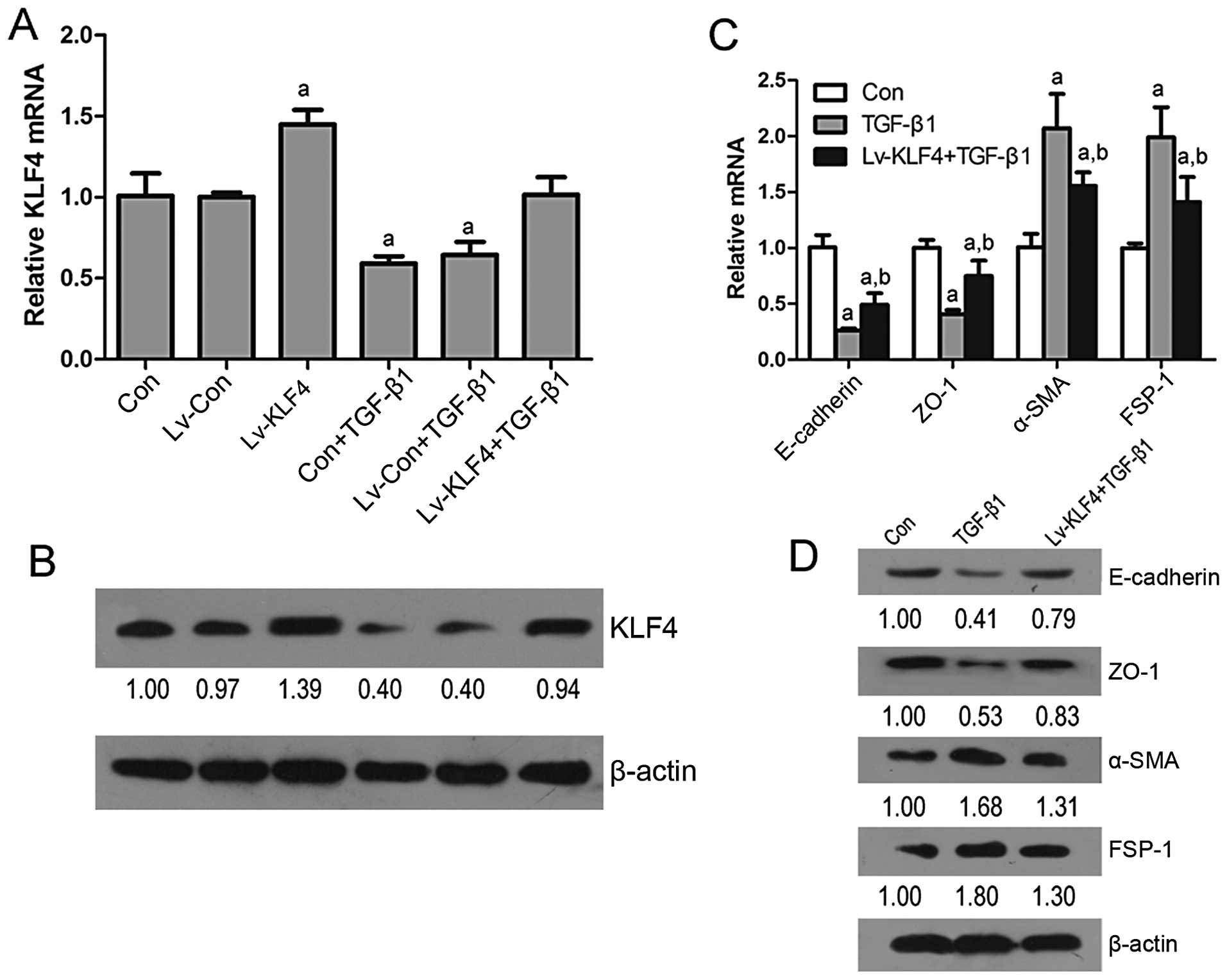
To validate whether KLF4 plays a role in EMT in
renal epithelial cells, the expression of KLF4 was enforced in the
HK-2 cells. The results from RT-qPCR and western blot analysis
revealed that the transfection of the HK-2 cells with Lv-KLF4
induced an increase in the expression of KLF4, E-cadherin and ZO-1,
and a decrease in the expression of α-SMA and FSP-1, and that this
attenuated the TGF-β1-induced downregulation in KLF4, E-cadherin
and ZO-1 expression and the upregulation in α-SMA and FSP-1
expression (Fig. 4). In addition,
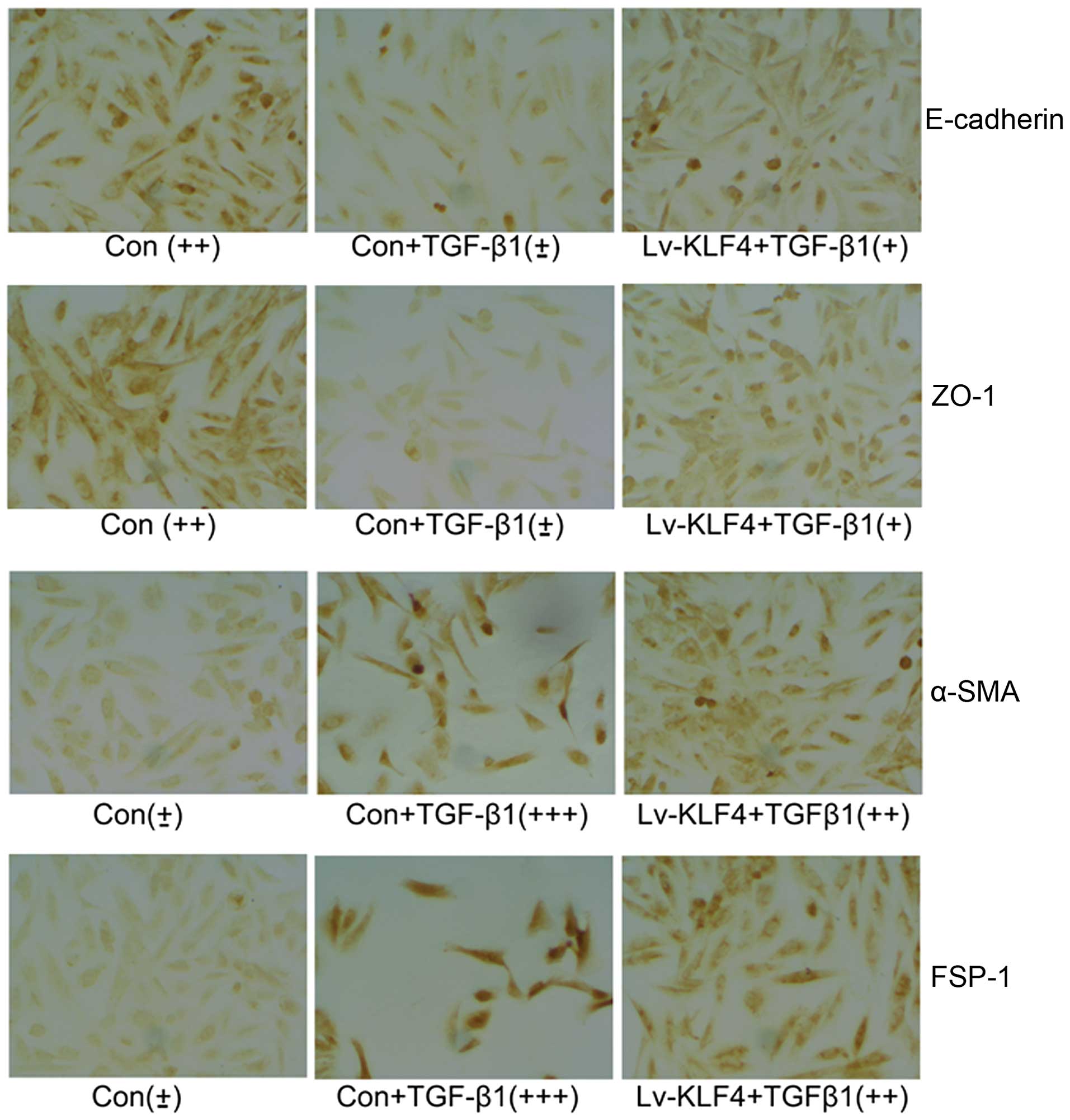
immunocytochemistry of the HK-2 cells infected with Lv-KLF4 or
Lv-KLF4 + TGF-β1 revealed similar results as regards the expression
of these factors (Fig. 5). These
data confirm that KLF4 inhibits the progression of EMT induced by
TGF-β1 in renal epithelial cells.
Discussion
EMT in the adult kidneys is one of the key events in
renal tubulointerstitial fibrosis (22). The transition of tubular
epithelial cells into myofibroblasts through EMT finally
contributes to renal fibrogenesis. Strutz et al (23) discovered that tubular epithelial
cells expressed FSP-1, a cytoskeleton-associated, calcium-binding
protein that is normally expressed in fibroblasts, but not in
epithelial cells, in a mouse model of anti-tubular basement
membrane disease and firstly demonstrated the presence of EMT in
renal fibrosis using FSP-1 as a marker. Subsequently, Iwano et
al (24) demonstrated that up
to 36% of all FSP-1-positive fibroblasts within the interstitial
space originate from renal proximal tubules following UUO, clearly
confirming the significant contribution of EMT to the pathogenesis
of kidney fibrosis in the model of UUO. Studies have demonstrated
that tubular epithelial cells in vitro undergo phenotypic
conversion after being incubated with TGF-β1 and that tubular
epithelial cells transdifferentiate into myofibroblasts (25,26). EMT can be regulated by a number of
factors in different ways; however, TGF-β1 is the most potent
inducer that is capable of initiating and completing the entire EMT
course (8). Thus, the mouse model
of UUO and the tubular epithelial cell model stimulated by TGF-β1
are classic in vivo and in vitro renal EMT models
(24,27). In addition, EMT in renal fibrosis
is generally identified by the loss of epithelial proteins,
including E-cadherin, ZO-1 and cytokeratin (28,29), and the acquisition of new
mesenchymal markers, including vimentin, α-SMA and FSP-1 (30–32).
Li et al (11) reported that the hypermethylation
of the KLF4 promoter mainly resulted in the inhibition of its
expression in renal cancer and the overexpression of KLF4
suppressed renal cancer cell migration and invasion by altering
EMT-related factors. In the present study, we investigated the
expression and methylation status of KLF4 in renal EMT models in
vivo and in vitro. We found that the KLF4 mRNA and
protein levels were decreased in the renal tissues of mice
subjected to UUO and in tubular epithelial HK-2 cells stimulated
with TGF-β1, and illustrated that the reduced expression of KLF4 in
renal EMT models in vivo or in vitro was accompanied
by the hypermethylation of the KLF4 promoter that may lead to lower
transcript levels of KLF4. In vitro, the reduced KLF4
expression accompanied by a decrease in E-cadherin expression and
an increase in α-SMA expression were observed in the HK-2 cells
stimulated with TGF-β1. However, the demethylating agent,
5-aza-2′-deoxycytidine, attenuated the TGF-β1-induced
downregulation of KLF4 and E-adherin and upregulation of E-cadherin
in the HK-2 cells. It is thus verified that KLF4 is hypermethylated
and downregulated in the renal EMT process. These data also suggest
that KLF4 functions as a suppressor of EMT induced by TGF-β1 in
tubular epithelial cells.
It has been demonstrated that epienetic alterations
of functional genes are associated with DNA methyltransferase
(17) or histone deacetylase
(14). Our results revealed that
an increased Dnmt1 and Dnmt3 expression was detected in the HK-2
cells stimulated with TGF-β1. It was confirmed that Dnmt1 directly
participates in the TGF-β1-mediated hypermethylation of the KLF4
promoter in HK-2 cells. It has been demonstrated that Dnmt1
catalyzes DNA methylation and subsequently leads to the
transcriptional repression of genes associated with fibrogenesis;
for example, the induction of RASAL1 hypermethylation contributes
to renal fibroblast activation (17), the mediation of PTEN
hypermethylation (33) and the
epigenetic repression of Smad7 (34) in liver fibrosis. Our data provide
evidence supporting the hypothesis that an increased Dnmt1
expression induced by TGF-β contributes to the epigenetic
repression of KLF4 in EMT in tubular epithelial HK-2 cells.
To confirm that KLF4 functions as a suppressor of
EMT in renal fibrosis, the expression of KLF4 was enforced in the
HK-2 cells. The results revealed that the overexpression of KLF4
induced an increase in E-cadherin and ZO-1 expression and a
decrease in a-SMA and FSP-1 expression, and attenuated the
TGF-β1-induced downregulation of E-cadherin and ZO-1 expression and
the upregulation of α-SMA and FSP-1. These data confirm that KLF4
inhibits the TGF-β1-induced EMT progression in renal epithelial
cells, and suggest that KLF4 plays a suppressive role in EMT in
renal fibrosis.
In conclusion, in this study, we demonstrate that
KLF4 is downregulated in a renal EMT model in vivo and in
vitro, and that KLF4 functions as suppressor of renal
fibrogenesis and the hypermethylation of KLF4 mediated by Dnmt1.
The downregulatin of KLF4 contributes to the progression of EMT in
renal epithelial cells. Future studies are required to elucidate
the utility of methylated KLF4 as a diagnostic marker or
therapeutic target in renal fibrosis.
References
|
1
|
Carew RM, Wang B and Kantharidis P: The
role of EMT in renal fibrosis. Cell Tissue Res. 347:103–116. 2012.
View Article : Google Scholar
|
|
2
|
Zeisberg M and Kalluri R: The role of
epithelial-to-mesenchymal transition in renal fibrosis. J Mol Med
(Berl). 82:175–181. 2004. View Article : Google Scholar
|
|
3
|
Yang J and Liu Y: Dissection of key events
in tubular epithelial to myofibroblast transition and its
implications in renal interstitial fibrosis. Am J Pathol.
159:1465–1475. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Caramel J, Papadogeorgakis E, Hill L, et
al: A switch in the expression of embryonic EMT-inducers drives the
development of malignant melanoma. Cancer Cell. 24:466–480. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Simic P, Williams EO, Bell EL, Gong JJ,
Bonkowski M and Guarente L: SIRT1 suppresses the
epithelial-to-mesenchymal transition in cancer metastasis and organ
fibrosis. Cell Rep. 3:1175–1186. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Guarino M, Tosoni A and Nebuloni M: Direct
contribution of epithelium to organ fibrosis:
epithelial-mesenchymal transition. Hum Pathol. 40:1365–1376. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Liu YN, Abou-Kheir W, Yin JJ, et al:
Critical and reciprocal regulation of KLF4 and SLUG in transforming
growth factor beta-initiated prostate cancer epithelial-mesenchymal
transition. Mol Cell Biol. 32:941–953. 2012. View Article : Google Scholar :
|
|
8
|
Liu Y: Epithelial to mesenchymal
transition in renal fibrogenesis: pathologic significance,
molecular mechanism, and therapeutic intervention. J Am Soc
Nephrol. 15:1–12. 2004. View Article : Google Scholar
|
|
9
|
Tiwari N, Meyer-Schaller N, Arnold P, et
al: Klf4 is a transcriptional regulator of genes critical for EMT,
including Jnk1 (Mapk8). PLoS One. 8:e573292013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cui J, Shi M, Quan M and Xie K: Regulation
of EMT by KLF4 in gastrointestinal cancer. Curr Cancer Drug
Targets. 13:986–995. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li H, Wang J, Xiao W, et al: Epigenetic
alterations of Kruppel-like factor 4 and its tumor suppressor
function in renal cell carcinoma. Carcinogenesis. 34:2262–2270.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Dressler GR: Epigenetics, development, and
the kidney. J Am Soc Nephrol. 19:2060–2067. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Smyth LJ, McKay GJ, Maxwell AP and
McKnight AJ: DNA hypermethylation and DNA hypomethylation is
present at different loci in chronic kidney disease. Epigenetics.
9:366–376. 2014. View Article : Google Scholar :
|
|
14
|
Sanders YY, Tollefsbol TO, Varisco BM and
Hagood JS: Epigenetic regulation of thy-1 by histone deacetylase
inhibitor in rat lung fibroblasts. Am J Respir Cell Mol Biol.
45:16–23. 2011. View Article : Google Scholar :
|
|
15
|
Denby L, Ramdas V, Lu R, et al:
MicroRNA-214 antagonism protects against renal fibrosis. J Am Soc
Nephrol. 25:65–80. 2014. View Article : Google Scholar :
|
|
16
|
Zarjou A, Yang S, Abraham E, Agarwal A and
Liu G: Identification of a microRNA signature in renal fibrosis:
role of miR-21. Am J Physiol Renal Physiol. 301:F793–F801. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bechtel W, McGoohan S, Zeisberg EM, et al:
Methylation determines fibroblast activation and fibrogenesis in
the kidney. Nat Med. 16:544–550. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang Y, Kong J, Deb DK, Chang A and Li
YC: Vitamin D receptor attenuates renal fibrosis by suppressing the
reninangiotensin system. J Am Soc Nephrol. 21:966–973. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li LC and Dahiya R: MethPrimer: designing
primers for methylation PCRs. Bioinformatics. 18:1427–1431. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sun Y, Wang L, Zhou Y, Mao X and Deng X:
Cloning and characterization of the human trefoil factor 3 gene
promoter. PLoS One. 9:e955622014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wei S, Xiong M, Zhan DQ, et al: Ku80
functions as a tumor suppressor in hepatocellular carcinoma by
inducing S-phase arrest through a p53-dependent pathway.
Carcinogenesis. 33:538–547. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Galichon P and Hertig A: Epithelial to
mesenchymal transition as a biomarker in renal fibrosis: are we
ready for the bedside? Fibrogenesis Tissue Repair. 4:112011.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Strutz F, Okada H, Lo CW, et al:
Identification and characterization of a fibroblast marker: FSP1. J
Cell Biol. 130:393–405. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Iwano M, Plieth D, Danoff TM, Xue C, Okada
H and Neilson EG: Evidence that fibroblasts derive from epithelium
during tissue fibrosis. J Clin Invest. 110:341–350. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lei X, Wang L, Yang J and Sun LZ: TGFbeta
signaling supports survival and metastasis of endometrial cancer
cells. Cancer Manag Res. 2009:15–24. 2009.PubMed/NCBI
|
|
26
|
Chen YX, Li Y, Wang WM, et al:
Phosphoproteomic study of human tubular epithelial cell in response
to transforming growth factor-beta-1-induced
epithelial-to-mesenchymal transition. Am J Nephrol. 31:24–35. 2010.
View Article : Google Scholar
|
|
27
|
Yang J and Liu Y: Blockage of tubular
epithelial to myofibroblast transition by hepatocyte growth factor
prevents renal interstitial fibrosis. J Am Soc Nephrol. 13:96–107.
2002.
|
|
28
|
Kanlaya R, Sintiprungrat K and
Thongboonkerd V: Secreted products of macrophages exposed to
calcium oxalate crystals induce epithelial mesenchymal transition
of renal tubular cells via RhoA-dependent TGF-beta1 pathway. Cell
Biochem Biophys. 67:1207–1215. 2013. View Article : Google Scholar
|
|
29
|
Forino M, Torregrossa R, Ceol M, et al:
TGFbeta1 induces epithelial-mesenchymal transition, but not
myofibroblast transdifferentiation of human kidney tubular
epithelial cells in primary culture. Int J Exp Pathol. 87:197–208.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Huang B, Pi L, Chen C, et al: WT1 and Pax2
re-expression is required for epithelial-mesenchymal transition in
5/6 nephrectomized rats and cultured kidney tubular epithelial
cells. Cells Tissues Organs. 195:296–312. 2012. View Article : Google Scholar
|
|
31
|
Gu L, Gao Q, Ni L, Wang M and Shen F:
Fasudil inhibits epithelial-myofibroblast transdifferentiation of
human renal tubular epithelial HK-2 cells induced by high glucose.
Chem Pharm Bull (Tokyo). 61:688–694. 2013. View Article : Google Scholar
|
|
32
|
Han WQ, Zhu Q, Hu J, Li PL, Zhang F and Li
N: Hypoxia-inducible factor prolyl-hydroxylase-2 mediates
transforming growth factor beta 1-induced epithelial-mesenchymal
transition in renal tubular cells. Biochim Biophys Acta.
1833:1454–1462. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Bian EB, Huang C, Ma TT, et al:
DNMT1-mediated PTEN hypermethylation confers hepatic stellate cell
activation and liver fibrogenesis in rats. Toxicol Appl Pharmacol.
264:13–22. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Bian EB, Huang C, Wang H, et al:
Repression of Smad7 mediated by DNMT1 determines hepatic stellate
cell activation and liver fibrosis in rats. Toxicol Lett.
224:175–185. 2014. View Article : Google Scholar
|