Introduction
Prostate cancer has the second highest incidence and
fifth highest mortality rates among all malignancies affecting
males worldwide. According to the global cancer data reported in
2018, the number of new prostate cancer cases worldwide was
estimated to be ~1,276,106 annually, accounting for 13.5% of all
new cases of male cancer in with a corresponding mortality rate of
6.7% (1). The incidence rate of
prostate cancer in China has also been increasing annually and this
disease has become one of the problems affecting the health of
males in China. In recent years, the incidence rate of prostate
cancer is 6th among malignant tumors affecting males, whereas it is
mainly distributed in the age group >60 years and reaches a peak
in the age group of 80 years (2). The 5-year survival rate of patients
with advanced prostate cancer is ~30% compared with the 5-year
survival rate of >99% for localized or locally advanced prostate
cancer (3).
Long non-coding RNAs (lncRNAs) have been extensively
investigated with regard to their involvement in tumor progression.
Recently, it was reported that multiple lncRNAs play an important
role in prostate cancer (4-8).
lncRNA MIR4435-2HG, also known as LINC00978, has been shown to be
highly expressed in hepatocellular carcinoma, bladder, lung and
gastric cancer (9-12). The knockdown of MIR4435-2HG
expression has been shown to inhibit the proliferation, invasion
and migration of lung and bladder cancer cells (9,10). A previous study demonstrated that
lncRNA MIR4435-2HG induced β-catenin expression in gastric cancer
cells by combining with desmoplakin to promote their proliferation,
invasion and migration (11).
MIR4435-2HG has been shown to promote the proliferation and
metastasis of hepatocellular carcinoma cells (12).
Starbase (http://starbase.sysu.edu.cn/index.php) has predicted
that MIR4435-2HG targets ST8SIA1. ST8SIA1 expression has been found
to be increased in breast cancer tissues and cells, and its
inhibition completely suppresses tumor growth in vivo and
the metastasis of triple-negative breast cancer cells (13). ST8SIA1 has been shown to be
highly expressed in colorectal cancer tissues and HCT-8/5-FU cells
and it can promote the proliferation, invasion and migration of
colon cancer cells (14).
However, the expression levels and role of ST8SIA1 in prostate
cancer cells remain unknown. In addition, ST8SIA1 activates the
FAK/AKT/mammalian target of rapamycin (mTOR) signaling pathway to
promote the progression of breast cancer (13). The AKT/β-catenin signaling
pathway is a classic cancer-promoting signaling pathway (15-17).
Therefore, the aim of the present study was to
examine the effects of miR4435-2HG binding to ST8SIA1 on the
proliferation, invasion and migration of prostate cancer cells and
to explore the associated mechanism of action mediated by the
activation of the FAK/AKT/β-catenin signaling pathway.
Materials and methods
Cells and cell culture
The prostate cell line, WPMY-1 (CRL-2854), and the
prostate cancer cell lines, VCaP (CRL-2876), LNCaP (CRL-1740),
DU145 (HTB-81) and PC-3 (CRL-1435), were purchased from the
American Type Culture Collection (ATCC). The prostate cell line was
cultured in KSLM medium containing 10% FBS (Gibco; Thermo Fisher
Scientific, Inc.) and the prostate cancer cell lines were cultured
in RPMI-1640 medium containing 10% FBS in an incubator at 37°C in
the presence of 5% CO2 and saturated humidity.
Cell transfection
When the cells reached 80% confluency, they were
transfected with the plasmids containing pcDNA-negative control
(NC) (5 nM) and pcDNA-miR4435-2HG (5 nM) (Invitrogen; Thermo Fisher
Scientific, Inc.) and the sequences short hairpin (sh)RNA-NC (5
nM), shRNA-miR4435-2HG-1 (5 nM), shRNA-miR4435-2HG-2 (5 nM),
shRNA-ST8SIA1-1 (5 nM) and shRNA-ST8SIA1-2 (5 nM) (Guangzhou
RiboBio Co., Ltd.) using Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.). After 24 h of
transfection, subsequent experiments were conducted.
Reverse transcription-quantitative PCR
(RT-qPCR) analysis
The TRIzol® kit was used to extract total
RNA from the prostate cancer cells and the reverse transcription
kit (TransGen Biotech Co., Ltd.) was used to reverse transcribe
this into cDNA. The AceQ qPCR SYBR-Green Master Mix (Bio-Rad
Laboratories, Inc.) was used for the qPCR reaction. The
thermocycling conditions were as follows: Initial denaturation at
96°C for 4 min; followed by 40 cycles of denaturation at 95°C for
20 sec, annealing at 60°C for 30 sec and extension at 72°C for 30
sec. The primer sequences were as follows: lncRNA MIR4435-2HG
forward, 5′-GGAAGTGGTGGCTATGAGTCAG-3′ and reverse,
5′-TGTCAATTTGAAACTTAAAAAGCAG-3′; ST8SIA1 forward,
5′-TACTCTCTCTTCCCACAGG-3′ and reverse, 5′-GACAAAGGAGGGAGATTGC-3′;
GAPDH forward, 5′-CATGAGAAGTATGACAACAGCCT-3′ and reverse,
5′-AGTCCTTCCACGATACCAAAGT-3′. GAPDH was used as an internal refer-
ence to calculate the expression levels of miR4435-2HG and ST8SIA1
mRNA using the 2−ΔΔCq method (18).
Cell Counting kit (CCK)-8 assay
Following transfection for 24 h, the prostate cancer
cells were seeded into 96-well plates at a density of
4×103 cells/well and cultured for 24, 48 and 72 h. A
total of 10 μl CCK-8 solution was added to each well and the
plate was incubated at 37°C for 4 h. The absorbance value was
measured at 450 nm using a microplate reader (Multiskan MK3; Thermo
Fisher Scientific, Inc.).
Colony formation assay
Following transfection for 24 h, the prostate cancer
cells were collected, resuspended in medium and seeded into
single-cell suspension. Subsequently, the cells were counted and
their density was adjusted to 500 cells per well. Following
seeding, the KSLM medium was added to each well to a total volume
of 300 μl. Following 14 days of cell culture, the culture
medium was removed and the cells were rinsed with PBS twice. A
total of 200 μl and 1% crystal violet dye solution (Beyotime
Institute of Biotechnology) was added to each well, which was fully
covered at the bottom of the hole. Following 20 min of incubation
at room tempera- ture, the plate was washed using tap water and
dried for assay detection.
Wound healing assay
The prostate cancer cells were transfected, cultured
for 24 h and collected. The cells were resuspended in serum-free
medium and incubated in a 6-well plate (5×104) (the
incubated cells were covered with a layer of the culture plate the
following day) for 24 h. Following incuba- tion at room temperature
for 24 h, the cells were scratched with a 200-μl sterile
pipette tip and washed with PBS thrice. The migration of the cells
was observed and images were captured at 0 and 24 h using a light
microscope (magnification, ×100; Olympus Corporation).
Transwell assay
Matrigel was dissolved overnight at 4°C and diluted
in pre-cooled serum-free medium. A total of 40 μl Matrigel
was added to a pre-cooled Transwell chamber, which was allowed to
solidify following incubation for 2 h at 37°C. The transfected
cells (1×105) were mixed with 100 μl serum-free
DMEM and added to the upper chamber. A total of 600 μl
complete medium was added to the lower chamber. Following
incubation of the cells at 37°C with 5% CO2 for 24 h,
the cells in the upper chamber were wiped using a cotton swab,
fixed with 4% paraformaldehyde for 15 min, washed with PBS once,
stained with crystal violet for 10 min at room temperature, washed
with PBS again, imaged and counted under a light microscope
(Olympus Corporation).
Western blot analysis
The transfected cells and tumor tissues were lysed
in RIPA lysis buffer (CWBio) on ice for 30 min and the mixture was
centrifuged for 15 min (4°C, 14,000 × g) to obtain the supernatant.
Following quantitative analysis using a BCA kit (Beyotime Institute
of Biotechnology), the proteins were separated on 10% sodium
dodecyl sulfate polyacrylamide gels by electrophoresis, transferred
to polyvinylidene difluoride membranes and incubated with 5%
non-fat milk for 2 h at room temperature. Subsequently, the
membranes were incubated with antibodies against matrix
metalloproteinase (MMP)2 (ab92536; dilution, 1:1,000), MMP9
(ab76003; dilution, 1:1,000), Ki67 (ab92742; dilution, 1:5,000),
survivin (ab208938; dilution, 1:1,000), β-catenin (ab6302;
dilution, 1:4,000), c-MYC (ab32072; dilution, 1:1,000), cyclin D1
(ab16663; dilution, 1:200), phosphorylated (p)-FAK (ab81298;
dilution, 1:1,000), p-AKT (ab81298; dilution, 1:1,000) and GAPDH
(ab9485; 1:1,000) (all from Abcam) at 4°C overnight. The following
day, the membranes were incubated with a horseradish
peroxidase-labeled secondary antibody (#7074; 1:1,000; Cell
Signaling Technology, Inc.) at room temperature for 1 h and
visualized using ECL reagents (Research-bio). Relative protein
expression was quantified using Image-Pro Plus software (version
6.0; Media Cybernetics, Inc.).
Xenograft model
A total of 20 male nude mice were grown for 6-8
weeks with a weight of 18±2 g and were provided from Shanghai
Jesijie Experimental Animal Co., Ltd. These mice were divided (n=5)
randomly into the following 4 groups: shRNA-MIR4435-2HG,
pcDNA-miR4435-2HG, pcDNA-mi R4435-2HG + shRNA-ST8SIA1 and the
control. PC-3 cells were suspended with PBS. A total of 200
μl (1×104 cells/μl) cell suspension was
transplanted subcutaneously into the back of right forelimb of the
mice. The body weight and tumor volume were recorded on days 1, 5,
10, 15 and 20. Tumor volume was calculated as follows: Tumor volume
= (length × width × width)/2. At the end of the experiment (day
20), the 20 mice were euthanized with pentobarbital sodium (165
mg/kg, i.p.) and death was confirmed by monitoring the heartbeat.
The tumors were weighed, and the maximum tumor diameter and volume
obtained were 1.1 cm and 800 mm3, respectively. All
procedures were approved by the Animal Care and Use Committee of
the First Affiliated Hospital of Naval Medical University.
RNA binding protein immunoprecipitation
(RIP)
The binding sites of MIR4435-2HG and ST8SIA1 are
predicted by Starbase (http://starbase.sysu.edu.cn/index.php). RIP assays
were performed using the Magna RIP Kit (Millipore, Bedford, MA,
USA) according to manufacturer's instructions. The prostate cancer
cells were lysed with RIPA lysis buffer on ice. The lysate was
incubated with magnetic beads conjugated with IgG antibody, which
were pre-cleaned with RIP washing buffer twice. RNA-protein
complexes were immunoprecipitated with the Pierce Magnetic
RNA-Protein Pull-Down kit (Pierce; Thermo Fisher Scientific, Inc.).
Total RNA was extracted using TRIzol® reagent and
RT-qPCR was performed to detect the expression levels of
miR4435-2HG.
Statistical analysis
SPSS 22.0 (IBM Corp.) software was used for
statistical analyses. All the data are presented as the means ±
standard deviation. Two-group comparisons were analyzed with a
Student's t-test, while comparisons between multiple groups were
analyzed by one-way ANOVA with Tukey's post hoc test. A value of
P<0.05 was considered to indicate a statistically significant
difference.
Results
MIR4435-2HG expression levels are
increased in prostate cancer cell lines
The MI4435-2HG expression levels were increased in
the prostate cancer cell lines (VCaP, LNCaP, DU145 and PC-3 cells)
compared with those in the WPMY-1 cells. miR4435-2HG expression was
highest in the PC-3 cells, and these cells were thus selected for
use in subsequent experiments (Fig.
1).
Knockdown of MIR4435-2HG expression
inhibits the proliferation of prostate cancer cells
The MIR4435-2HG expression levels were decreased in
the PC-3 cells transfected with shRNA-MIR4435-2HG-1/2. The
MIR4435-2HG expression levels in the shRNA-MIR4435-2HG-1 group were
lower than those in the shRNA-MIR4435-2HG-2 group and
shRNA-MIR4435-2HG-1 was thus selected for use in subsequent
experiments (Fig. 2A). The
knockdown of MIR4435-2HG expression suppressed the viability
(Fig. 2B) and the clone
formation ability (Fig. 2C) of
the PC-3 cells.
Knockdown of MIR4435-2HG expression
inhibits the invasion and migration of prostate cancer cells
The knockdown of MIR4435-2HG expression inhibited
the migration (Fig. 3A and B)
and invasion (Fig. 3C and D) of
PC-3 cells. In addition, the knockdown of MIR4435-2HG expression
led to the downregulation of the expression levels of MMP2 and MMP9
in PC-3 cells (Fig. 3E).
Knockdown of MIR4435-2HG expression
inhibits the growth of prostate cancer cells in vivo
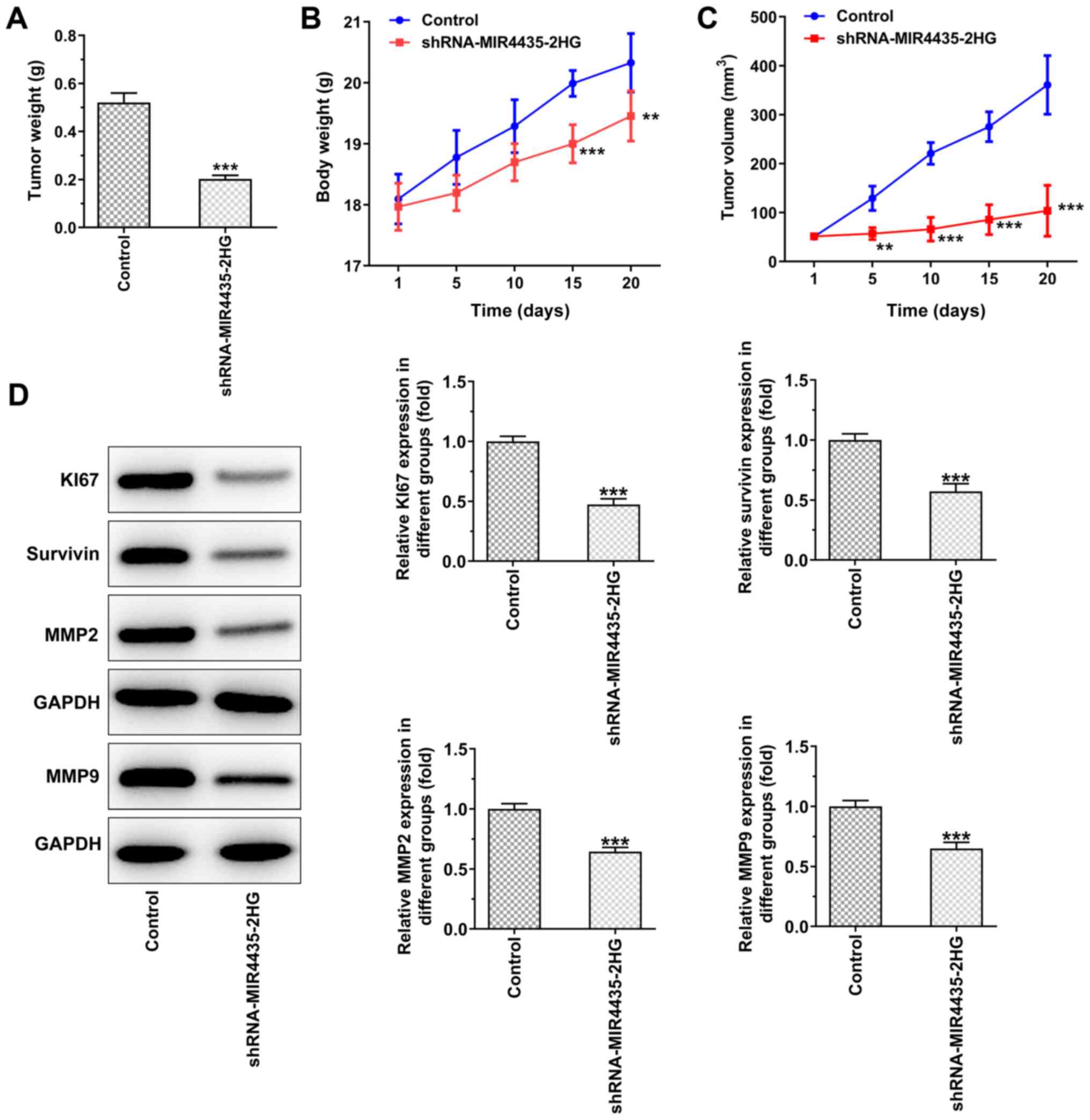
The visual investigation of euthanized mice and of
their corresponding tumors is presented in Fig. 4. The knockdown of MIR4435-2HG
expression decreased the tumor weight on the 20th day (Fig. 5A), whereas it also caused a
gradual decrease in the body weight of the mice resulting, in a
decrease in the tumor volume from the 1st to the 20th day (Fig. 5B and C). Moreover, the knockdown
of MIR4435-2HG expression suppressed the expression levels of Ki67,
survivin, MMP2 and MMP9 in the tumor tissues (Fig. 5D).
Knockdown of MIR4435-2HG expression
inhibits the activation of the FAK/AKT/β-catenin signaling pathway
in vitro and in vivo
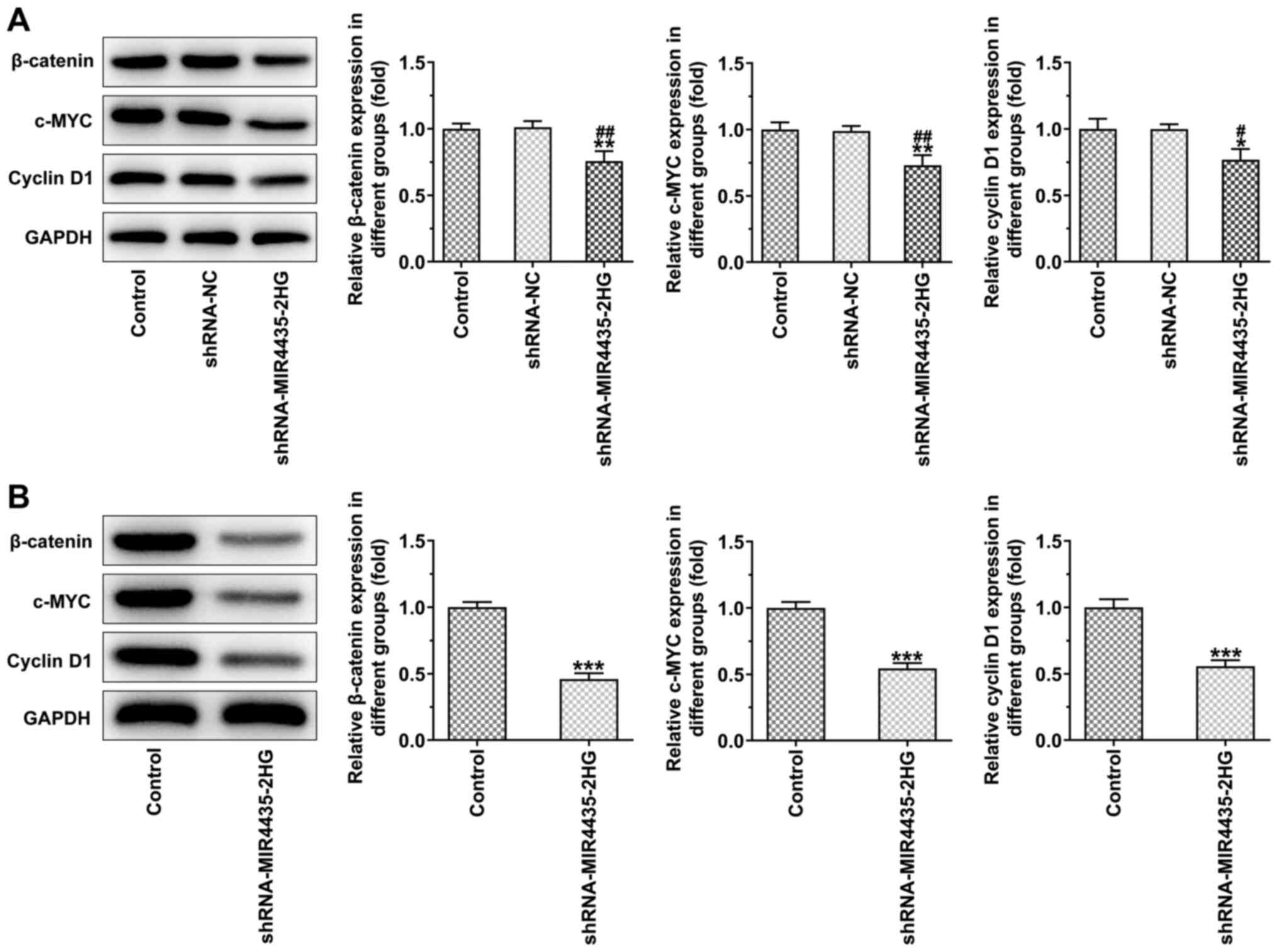
The knockdown of MIR4435-2HG expression in the
transfected PC-3 cells decreased the expression levels of
β-catenin, c-MYC and cyclin D1 (Fig.
6A). The expression levels of β-catenin, c-MYC and cyclin D1 in
the tumor tissues were also downregulated in the shRNA-MIR4435-2HG
group compared to those of the control group (Fig. 6B).
MIR4435-2HG binds to ST8SIA1 in prostate
cancer cells
The binding sites of MIR4435-2HG and ST8SIA1 are
illustrated in Fig. 7A. RIP
assay indicated that MIR4435-2HG bound to ST8SIA1 in prostate
cancer cells (Fig. 7B). ST8SIA1
expression was also increased in the prostate cancer cells compared
with the WPMY-1 cells, whereas the highest expression was noted in
the PC-3 cells (Fig. 7C). The
knockdown of MIR4435-2HG expression suppressed the expression of
ST8SIA1 (Fig. 7D).
Knockdown of ST8SIA1 expression inhibits
the effects of MIR4435-2HG on the proliferation of prostate cancer
cells
MIR4435-2HG expression was increased in the PC-3
cells transfected with pcDNA-MIR4435-2HG (Fig. 8A), whereas ST8SIA1 expression was
decreased in the PC-3 cells transfected with shRNA-ST8SIA1-1/2.
ST8SIA1 protein expression was lower in the shRNA-ST8SIA1-1 group
compared with that in the shRNA-ST8SIA1-2 group (Fig. 8B), whereas ST8SIA1 mRNA
expression in the shRNA-ST8SIA1-2 group demonstrated similar
changes to those noted for the protein expression levels of the
ST8SIA1 (Fig. 8C). MIR4435-2HG
overexpression promoted the proliferation of the PC-3 cells, which
was reversed following the knockdown of ST8SIA1 expression
(Fig. 8D).
Knockdown of ST8SIA1 expression inhibits
the effects of MIR4435-2HG on the invasion and migration of
prostate cancer cells
The invasion and migration of the PC-3 cells was
observed by wound healing and the Transwell assays following cell
transfection (Fig. 9A and B).
MIR4435-2HG overexpression promoted the migration (Fig. 9C) and invasion (Fig. 9D) of PC-3 cells, which was
reversed following the knockdown of ST8SIA1 expression. MIR4435-2HG
overexpression also increased the levels of MMP2 and MMP9 in PC-3
cells, which were reversed following the knockdown of ST8SIA1
expression (Fig. 9E).
Knockdown of ST8SIA1 expression inhibits
the effects of MIR4435-2HG on the growth of prostate cancer cells
in vivo
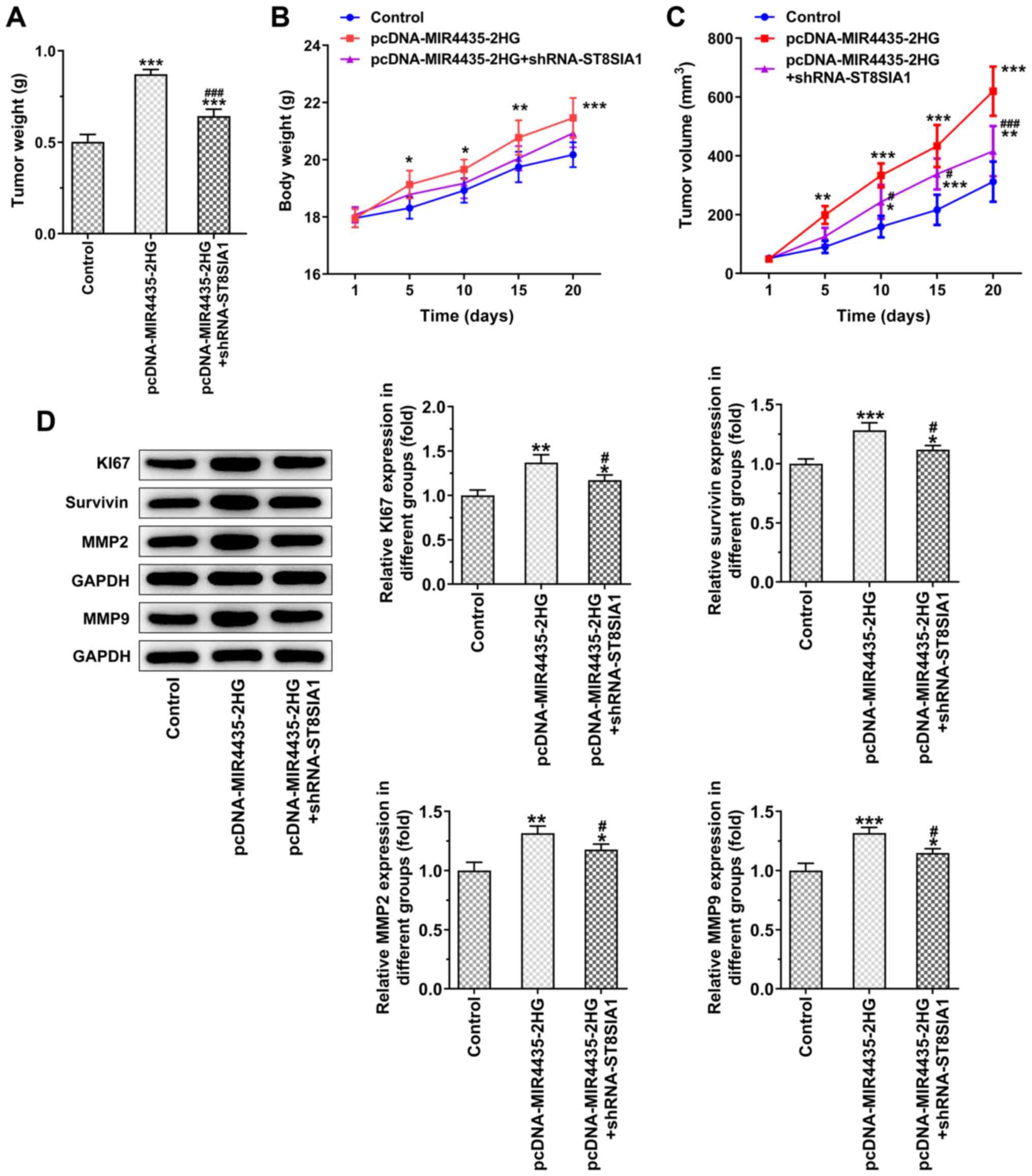
The visual investigation of euthanized mice and
their tumors is presented in Fig.
10. MIR4435-2HG overexpression increased tumor weight in the
mice and the knockdown of ST8SIA1 expression suppressed the effects
of MIR4435-2HG overexpression on cell proliferation on day 20
(Fig. 11A). As the time period
of treatment increased, the body weights (Fig. 11B) of the mice and their tumor
volume (Fig. 11C) gradually
increased in the pcDNA-MIR4435-2HG group. The increase noted in the
pcDNA-MIR4435-2HG + shRNA-ST8SIA1 group was lower than that in the
pcDNA-MIR4435-2HG group. MIR4435-2HG overexpression also promoted
the expression of KI67, survivin, MMP2 and MMP9 in the tumor
tissues, whereas the knockdown of ST8SIA1 expression attenuated the
effects of MIR4435-2HG overexpression (Fig. 11D).
MIR4435-2HG overexpression activates the
FAK/AKT/β- catenin signaling pathway, which is inhibited following
the knockdown of ST8SIA1 expression
MIR4435-2HG overexpression caused an upregulation in
the expression levels of p-FAK, p-AKT, β-catenin, c-MYC and cyclin
D1, which was inhibited following the knockdown of ST8SIA1
expression in PC-3 cells (Fig.
12A) and in tumor tissues (Fig.
12B).
Discussion
Prostate cancer is a malignant disease, which
affects the health of male patients. It is not easily diagnosed due
to the lack of specific tumor markers present in the early stages
of the disease. Therefore, effective treatment is not usually
applied at an early stage and the disease rapidly progresses
(19). The proliferation and
invasion of the cells in malignant tumors remains a major obstacle
in the treatment process of this disease. The reduction in the
proliferative and invasive rate is conducive to the remission of
the disease. Therefore, research on the targets genes that can
reduce the proliferation and invasion of the cells may result in
the treatment of this disease (20).
lncRNAs affect the proliferation, invasion and
migration of multiple cancer types. MIR4435-2HG expression has been
shown to be upregulated in gastric cancer (21), hepatocellular carcinoma (22), glioblastoma (23), colorectal cancer (24) and oral squamous cell carcinoma
(25). This elevated expression
promotes the proliferation, invasion and migration of the
aforementioned cancer cell types. In the present study, MIR4435-2HG
expression was also found to be elevated in prostate cancer cells,
and the knockdown of its expression inhibited the proliferation,
clone formation ability, invasion and migration of PC-3 cells in
vitro as well as their tumor growth in vivo.
ST8SIA1 is also known as ganglioside GD3 synthase
(GD3s) (26,27). It has been reported that ST8SIA1
is highly expressed in tumors. Yamashiro et al (28) examined the expression levels of
ST8SIA1 in different tumor cell lines and found that it was highly
expressed in melanoma, neuroblastoma and glioma cells. ST8SIA1 has
also been shown to be highly expressed in triple-negative breast
cancer (29). Previous studies
have demonstrated that ST8SIA1 promotes the development of tumors
by modulating GD3 and GD2 and by various other mechanisms of
action. Cazet et al (30)
demonstrated that ST8SIA1 overexpression induced the accumulation
of GD2 and GD3 on the surface of MDA-MB-231 breast cancer cells,
while it activated the downstream MEK/ERK and PI3K/Akt signaling
pathways by activating C-MET, which in turn promoted the
proliferation of tumor cells. Moreover, ST8SIA1 stimulates the
proliferation of melanoma cells (31). In SK-MEL-2 melanoma cells, NF-κB
regulates the transcriptional activity of ST8SIA1 and affects the
concentration of GD3 acting as a tumor suppressor (32). In the present study, ST8SIA1
expression was increased in prostate cancer cells and the knockdown
of its expression impaired the role of MIR4435-2HG by suppressing
the proliferation, invasion and migration of PC-3 cells in
vitro and their tumor growth in vivo.
FAK signaling plays a role in cell adhesion,
migration and metastasis (33).
FAK regulates various cellular activities including cell survival,
adhesion, proliferation and migration by activating several signal
transducers, such as PI3K, AKT and mTOR (34,35). The knockdown of ST8SIA1
expression inhibits FAK/AKT/ERK/mTOR signaling in order to regulate
the activity of breast cancer stem-like cells in triple-negative
breast cancer (TNBC) (13).
ST8SIA1 inhibition has been shown to suppress the FAK/Akt/mTOR
pathway in chemoresistant TNBC cells and ST8SIA1 inhibition can
cause the β-catenin degradation and subsequent reduction of
Wnt/β-catenin activity. In addition, a previous study reported the
decreased phosphorylation of FAK at Y861, Akt at Ser473 and mTOR at
Ser2448 in ST8SIA1-depleted MDA-MB-231-dox-r and BT 549-pac-r cells
(36). In the present study, the
knockdown of ST8SIA1 also suppressed the FAK/Akt/β-catenin
signaling pathway. p-AKT activates its downstream molecules. It can
thus be hypothesized that the phosphorylation of FAK at Y861, Akt
at Ser473 and mTOR at Ser2448 is also regulated by ST8SIA1; the
authors aim to investigate this in future studies, as their
functions are closely associated with tumor invasion and
metastasis. The inhibition of AKT activation reduces the invasive
and metastatic activities of multiple types of tumor cells
(37). An increased AKT activity
may increase the expression of the β-catenin protein in tumor cells
and promote its nuclear entry and transcriptional activity, which
accelerates the migration and invasion of tumor cells (38). The present study demonstrated
that the in vitro proliferation, invasion and migration of
PC-3 cells and tumor growth in vivo were suppressed by the
inhibition of the FAK/AKT/β-catenin signaling pathway. It was
hypothesized that there may be other existing pathways through
which MIR4435-2HG regulates FAK-AKT-β-catenin. MIR4435-2HG has been
found to target miR-330; miR-330 has been demonstrated to regulate
the PKC expression (39).
miR-330 has been demonstrated to regulate the development of
prostate cancer (40,41). According to KEGG pathway
analysis, the PKC gene is the upstream gene of FAK.
MIR4435-2HG/miR-330/FAK upstream gene (PKC) may be the other
pathway through which MIR4435-2HG regulates FAK/AKT/β-catenin. In
future studies, the authors aim to verify the combination of
MIR4435-2HG and miR-330 and explore the mechanisms through which
MIR4435-2HG regulates miR-330 expression. In the future, the
authors also aim to investigate the effects of changes in miR-330
expression on the FAK/AKT/β-catenin pathway.
In conclusion, the present study demonstrated that
the knockdown of MIR4435-2HG expression inhibited the
proliferation, invasion and migration of prostate cancer cells
in vitro and in vivo, whereas the knockdown of
ST8SIA1 expression inhibited the effects of miR4435-2HG on the
in vitro proliferation, invasion and migration of PC-3 cells
and on the in vivo tumor growth by suppressing the
FAK/AKT/β-catenin signaling pathway.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
JL conceived and designed the experiments. PX and YW
performed all the experiments with the assistance of LZ, and
analyzed the experimental data with the assistance of CM. PX and YW
confirm the authenticity of all the raw data. PX and YW wrote the
manuscript which was revised by JL. All authors read and approved
the final manuscript.
Ethics approval and consent to
participate
All procedures involving animals were approved by
the Animal Care and Use Committee of the First Affiliated Hospital
of Naval Medical University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
Funding
The present study was funded by the National Key clinical
specialist military construction project.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zheng RS, Sun KX, Zhang SW, Zeng HM and He
J: Report of cancer epidemiology in China, 2015. Zhonghua zhong liu
za zhi (Article in Chinese). 41:19–28. 2019.
|
|
3
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2020. CA Cancer J Clin. 70:7–30. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Prensner JR, Iyer MK, Sahu A, Asangani IA,
Cao Q, Patel L, Vergara IA, Davicioni E, Erho N, et al: The long
noncoding RNA SChLAP1 promotes aggressive prostate cancer and
antagonizes the SWI/SNF complex. Nat Genet. 45:1392–1398. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yang J, Hao T, Sun J, Wei P and Zhang H:
Long noncoding RNA GAS5 modulates α-Solanine-induced
radiosensitivity by negatively regulating miR-18a in human prostate
cancer cells. Biomed Pharmacother. 112:1086562019. View Article : Google Scholar
|
|
6
|
Wu M, Huang Y, Chen T, Wang W, Yang S, Ye
Z and Xi X: LncRNA MEG3 inhibits the progression of prostate cancer
by modulating miR-9-5p/QKI-5 axis. J Cell Mol Med. 23:29–38. 2019.
View Article : Google Scholar
|
|
7
|
Zhang Y, Zhang D, Lv J, Wang S and Zhang
Q: LncRNA SNHG15 acts as an oncogene in prostate cancer by
regulating miR-338-3p/FKBP1A axis. Gene. 705:44–50. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yuan Q, Chu H, Ge Y, Ma G, Du M, Wang M,
Zhang Z and Zhang W: LncRNA PCAT1 and its genetic variant rs1902432
are associated with prostate cancer risk. J Cancer. 9:1414–1420.
2018. View Article : Google Scholar :
|
|
9
|
Li X, Ren Y and Zuo T: Long noncoding RNA
LINC00978 promotes cell proliferation and invasion in non-small
cell lung cancer by inhibiting miR-6754-5p. Mol Med Rep.
18:4725–4732. 2018.PubMed/NCBI
|
|
10
|
Wang W, Xu Z, Wang J and Chen R: LINC00978
promotes bladder cancer cell proliferation, migration and invasion
by sponging miR-4288. Mol Med Rep. 20:1866–1872. 2019.PubMed/NCBI
|
|
11
|
Wang H, Wu M, Lu Y, He K, Cai X, Yu X, Lu
J and Teng L: LncRNA MIR4435-2HG targets desmoplakin and promotes
growth and metastasis of gastric cancer by activating Wnt/β-catenin
signaling. Aging (Albany NY). 11:6657–6673. 2019. View Article : Google Scholar
|
|
12
|
Shen X, Ding Y, Lu F, Yuan H and Luan W:
Long noncoding RNA MIR4435-2HG promotes hepatocellular carcinoma
proliferation and metastasis through the miR-22-3p/YWHAZ axis. Am J
Transl Res. 12:6381–6394. 2020.PubMed/NCBI
|
|
13
|
Nguyen K, Yan Y, Yuan B, Dasgupta A, Sun
V, Mu H, Do KA, Ueno NT, Andreeff M and Battula VL: ST8SIA1
regulates tumor growth and metastasis in TNBC by activating the
FAK-AKT-mTOR signaling pathway. Mol Cancer Ther. 17:2689–2701.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Shan Y, Liu Y, Zhao L, Liu B, Li Y and Jia
L: MicroRNA-33a and let-7e inhibit human colorectal cancer
progression by targeting ST8SIA1. Int J Biochem Cell Biol.
90:48–58. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chen Y, Deng X, Chen W, Shi P, Lian M,
Wang H, Wang K, Qian D, Xiao D and Long H: Silencing of
microRNA-708 promotes cell growth and epithelial-to-mesenchymal
transition by activating the SPHK2/AKT/β-catenin pathway in glioma.
Cell Death Dis. 10:4482019. View Article : Google Scholar
|
|
16
|
Sun X, Xing G, Zhang C, Lu K, Wang Y and
He X: Knockdown of Trop2 inhibits proliferation and migration and
induces apoptosis of endometrial cancer cells via AKT/β-catenin
pathway. Cell Biochem Funct. 38:141–148. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wei B, Wang Y, Wang JW, Cai X, Xu L, Wu J,
Wang Y, Liu W, Gu Y, Guo W and Xu Q: Apatinib suppresses tumor
progression and enhances cisplatin sensitivity in esophageal cancer
via the Akt/β-catenin pathway. Cancer Cell Int. 20:1982020.
View Article : Google Scholar
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
19
|
Tosoian JJ, Gorin MA, Ross AE, Pienta KJ,
Tran PT and Schaeffer EM: Oligometastatic prostate cancer:
Definitions, clinical outcomes, and treatment considerations. Nat
Rev Urol. 14:15–25. 2017. View Article : Google Scholar
|
|
20
|
Pan B, Ye Y, Liu H, Zhen J, Zhou H, Li Y,
Qu L, Wu Y, Zeng C and Zhong W: URG11 regulates prostate cancer
cell proliferation, migration, and invasion. Biomed Res Int.
2018:40607282018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bu JY, Lv WZ, Liao YF, Xiao XY and Lv BJ:
Long non-coding RNA LINC00978 promotes cell proliferation and
tumorigenesis via regulating microRNA-497/NTRK3 axis in gastric
cancer. Int J Biol Macromol. 123:1106–1114. 2019. View Article : Google Scholar
|
|
22
|
Zhang Q, Cheng S, Cao L, Yang J, Wang Y
and Chen Y: LINC00978 promotes hepatocellular carcinoma
carcinogenesis partly via activating the MAPK/ERK pathway. Biosci
Rep. 40:BSR201927902020. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xu H, Zhang B, Yang Y, Yang Y, Li Z, Zhao
P, Wu W, Zhang H and Mao J: LncRNA MIR4435-2HG potentiates the
proliferation and invasion of glioblastoma cells via modulating
miR-1224-5p/TGFBR2 axis. J Cell Mol Med. 24:6362–6372. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Dong X, Yang Z, Yang H, Li D and Qiu X:
Long non-coding RNA MIR4435-2HG promotes colorectal cancer
proliferation and metastasis through miR-206/YAP1 axis. Front
Oncol. 10:1602020. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Shen H, Sun B, Yang Y, Cai X, Bi L, Deng L
and Zhang L: MIR4435-2HG regulates cancer cell behaviors in oral
squamous cell carcinoma cell growth by upregulating TGF-β1.
Odontology. 108:553–559. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Takashima S, Matsumoto T, Tsujimoto M and
Tsuji S: Effects of amino acid substitutions in the sialylmotifs on
molecular expression and enzymatic activities of
α2,8-sialyltransferases ST8Sia-I and ST8Sia-VI. Glycobiology.
23:603–612. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Rimoldi S, Papis E, Bernardini G, Prati M
and Gornati R: Molecular cloning and expression of
alpha2,8-sialyltransferase (ST8Sia I, GD3 Synthase) in xenopus. Mol
Cell Biochem. 301:143–153. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yamashiro S, Okada M, Haraguchi M and
Furukawa K, Lloyd KO, Shiku H and Furukawa K: Expression of alpha
2,8-sialyltransferase (GD3 synthase) gene in human cancer cell
lines: High level expression in melanomas and up-regulation in
activated T lymphocytes. Glycoconjugate J. 12:894–900. 1995.
View Article : Google Scholar
|
|
29
|
Sarkar TR, Battula VL, Werden SJ, Vijay
GV, Ramirez-Peña EQ, Taube JH, Chang JT, Miura N, Porter W, Sphyris
N, et al: GD3 synthase regulates epithelial-mesenchymal transition
and metastasis in breast cancer. Oncogene. 34:2958–2967. 2015.
View Article : Google Scholar :
|
|
30
|
Cazet A, Lefebvre J, Adriaenssens E,
Julien S, Bobowski M, Grigoriadis A, Tutt A, Tulasne D, Le Bourhis
X and Delannoy P: GD3 synthase expression enhances
proliferation and tumor growth of MDA-MB-231 breast cancer cells
through c-Met activation. Mol Cancer Res. 8:1526–1535. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Birklé S, Gao L, Zeng G and Yu RK:
Down-regulation of GD3 ganglioside and its O-acetylated derivative
by stable transfection with antisense vector against GD3-synthase
gene expression in hamster melanoma cells: Effects on cellular
growth, melanogenesis, and dendricity. J Neurochem. 74:547–554.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kang NY, Kim CH, Kim KS, Ko JH, Lee JH,
Jeong YK and Lee YC: Expression of the human CMP-NeuAc:GM3
alpha2,8-sialyltransferase (GD3 synthase) gene through the
NF-kappaB activation in human melanoma SK-MEL-2 cells. Biochim
Biophys Acta. 1769:622–630. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Luo M and Guan JL: Focal adhesion kinase:
A prominent determinant in breast cancer initiation, progression
and metastasis. Cancer Lett. 289:127–139. 2010. View Article : Google Scholar :
|
|
34
|
Xia H, Nho RS, Kahm J, Kleidon J and Henke
CA: Focal adhesion kinase is upstream of phosphatidylinositol
3-kinase/Akt in regulating fibroblast survival in response to
contraction of type I collagen matrices via a beta 1 integrin
viability signaling pathway. J Biol Chem. 279:33024–33034. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Jindra PT, Jin YP, Rozengurt E and Reed
EF: HLA class I antibody-mediated endothelial cell proliferation
via the mTOR pathway. J Immunol. 180:2357–2366. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wan H, Li Z, Wang H, Cai F and Wang L:
ST8SIA1 inhibition sensitizes triple negative breast cancer to
chemotherapy via suppressing Wnt/β-catenin and FAK/Akt/mTOR. Clin
Transl Oncol. 23:902–910. 2020. View Article : Google Scholar
|
|
37
|
Qiao M, Iglehart JD and Pardee AB:
Metastatic potential of 21T human breast cancer cells depends on
Akt/protein kinase B activation. Cancer Res. 67:5293–5299. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhang X, Chen T, Zhang J, Mao Q, Li S,
Xiong W, Qiu Y, Xie Q and Ge J: Notch1 promotes glioma cell
migration and invasion by stimulating β-catenin and NF-κB signaling
via AKT activation. Cancer Sci. 103:181–190. 2012. View Article : Google Scholar
|
|
39
|
Liu J, Liu L, Chao S, Liu Y, Liu X, Zheng
J, Chen J, Gong W, Teng H, Li Z, et al: The role of
miR-330-3p/PKC-α signaling pathway in low-dose endothelial-monocyte
activating polypeptide-II increasing the permeability of
blood-tumor barrier. Front Cell Neurosci. 11:3582017. View Article : Google Scholar
|
|
40
|
Liu DC, Song LL, Liang Q, Hao L, Zhang ZG
and Han CH: Long noncoding RNA LEF1-AS1 silencing suppresses the
initiation and development of prostate cancer by acting as a
molecular sponge of miR-330-5p via LEF1 repression. J Cell Physiol.
234:12727–12744. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Li Q, Wang W, Zhang M, Sun W, Shi W and Li
F: Circular RNA circ-0016068 promotes the growth, migration, and
invasion of prostate cancer cells by regulating the
miR-330-3p/BMI-1 axis as a competing endogenous RNA. Front Cell Dev
Biol. 8:8272020. View Article : Google Scholar : PubMed/NCBI
|