Introduction
Cartilage defects caused by trauma, congenital
malformations, or oncological resection are often devastating and
cannot be cured because of the intrinsically low regenerative
capacity of cartilage tissues (1). Autologous cartilage grafting as well
as modified operations, such as cartilage transplantation combined
with flap transfer have been developed to treat nasal, auricular,
and tracheal defects (2-4). However, cartilage tissue is
inevitably wasted during graft carving, and ensuring an accurate
shape of a graft may require extensive engraving techniques
(5). Tissue engineering offers
the possibility of replacing damaged chondral tissue as an
alternative to cartilage grafting. In this regard, stem cell-based
tissue engineering approaches have recently exhibited significant
potential for rapidly restoring injured cartilage tissues (6). For example, stem cells that are
transplanted successfully can differentiate into chondrocytes for
functional restoration (7).
However, stem cells frequently undergo apoptosis because of the
prevalence of oxidative stress and inflammation in the
microenvironment of injured sites (8). Hence, there is an urgent need to
find an effective method to abrogate apoptosis as survival in
hostile conditions is a prerequisite for cells to perform various
physiological functions.
Mitochondria are one of the most complex and
important organelles present in eukaryotes, which convert organic
matter into carbon dioxide and water through redox reactions
(9). Mitochondrial dysfunction
leads to the irregular transfer of electrons generated by redox
reactions to oxygen, water, or their intermediate states, thus
forming large quantities of reactive oxygen species (ROS),
eventually leading to cell death (10,11). Mitophagy, a special form of
autophagy, is a process of selective removal of excess or damaged
mitochondria that plays an important role in maintaining
mitochondrial homeostasis and regulating the number of mitochondria
in cells (12,13). Under various conditions of
cellular stress, including low oxygen conditions, oxidative stress,
and high glucose levels, mitophagy can be activated; the
upregulated mitophagy can promote cell survival by removing damaged
mitochondria (14). Recently,
there has been increasing evidence that induction of mitophagy
plays a crucial role in preventing chondrocyte apoptosis (15-17).
In healthy cartilage, chondrocytes exist in
relatively low oxygen conditions, in which hypoxia-inducible factor
1α (HIF-1α) plays a vital role in regulating chondrogenesis by
directing the differentiation of progenitor cells and maintaining
appropriate extracellular matrix production (18). Our previous study showed that
chondrocytes are well adapted to hypoxia and produce a more
functional extracellular matrix in low-oxygen environments in
vitro (19). This effect is
reversed in the presence of high oxygen concentrations, as HIF-1α
is degraded in high oxygen conditions, which in turn promotes
further apoptosis of chondrocytes (20). Therefore, the stabilization of
HIF-1α is key to the survival of chondrocytes. The prolyl
hydroxylase 2 (PHD2)-von Hippel Lindau (VHL) signaling cascade is
central to the regulation of HIF-1α (21). Under physiological conditions,
PHD2 hydroxylate residues are present on the oxygen-dependent
degradation domain of HIF-1α; VHL, as a part of the E3 ubiquitin
ligase complex, recognizes the motifs of the hydroxylated residues,
resulting in rapid degradation of HIF-1α. However, PHD2 is less
active during hypoxia, which leads to cytosolic accumulation and
nuclear translocation of HIF-1α, where, together with
transcriptional cofactors, it activates the expression of its
target genes in the HIF complex, such as BNIP3, an essential
molecule for mitophagy (22,23). It has been shown to stimulate
chondrogenesis of progenitor or stem cells by decreasing the oxygen
pressure locally within a biomaterial using various molecules or
simply by limiting oxygen diffusion (24,25).
However, regulating oxygen itself may not be ideal
because hypoxia is also known to cause oxidative stress, negatively
impact cell growth and viability, and boost potentially undesirable
effects on cell metabolism (26).
Recently, it was demonstrated that Ubiquitin C-terminal
hydrolase-L1 (UCHL1) protects against ischemic heart injury by
increasing the stability of HIF-1α (27). Moreover, UCHL1 was reported to
abrogate VHL-mediated ubiquitination of HIF-1α (28). However, the effect of UCHL1, with
its function of binding and stabilizing HIF-1α, on apoptosis in
chondrocytes is unclear.
In the present study, adipose-derived stem cells
(ADSCs) were utilized to differentiate into chondrocytes. Next, a
series of in vitro experiments were performed to assess
mitophagy, apoptosis, and mitochondrial function in the
chondrocytes. Moreover, the CRISPR activation (CRISPRa) system was
used to activate endogenous UCHL1 in chondrocytes. The results
revealed that activation of UCHL1 using CRISPRa inhibited apoptosis
and maintained mitochondrial function, resulting in the survival of
chondrocytes. This provides a theoretical basis for tissue
engineering strategies that can be used for the treatment of
cartilage defects.
Materials and methods
Cell culture and reagents
ADSCs were purchased from Procell Life Science
Technology Co., Ltd. (cat. no. CP-R147) and were cultured in α-MEM
medium (HyClone; Cytiva) supplemented with 10% (v/v) FBS (HyClone;
Cytiva), 100 U/ml penicillin, and 100 U/ml streptomycin at 37°C
with 5% CO2 in a humidified chamber. The cells were
passaged 3-5 times and used for subsequent experiments. To induce
hypoxia in cell cultures, the cells were subjected to a hypoxic
environment using a specialized incubator with an oxygen
concentration of 1% for a duration of 24 h. In contrast, the
control group was exposed to a normoxic environment maintaining a
regular oxygen concentration of 20%.
Insect cells (Sf-9) were purchased from the American
Type Culture Collection Cell Bank for baculovirus generation and
were cultured in TNM-FH medium (HyClone; Cytiva) supplemented with
10% FBS. Stock solutions of LDN (MilliporeSigma; cat. no. L4170)
and KC7F2 (MilliporeSigma; cat. no. SML1043) were prepared in DMSO
(10 mM and 30 μM, respectively) obtained from the Beyotime
Institute of Biotechnology. 3-Methyladenine (3-MA, MilliporeSigma;
cat. no. M9281) was prepared in DMEM solution (5 mM). In the
present study, cells were treated with the corresponding blocker
for 2 h, after which they were subsequently subjected to relevant
tests.
Characterization of ADSCs
To identify the phenotypes of ADSCs, flow cytometry
(FCM, CytoFLEX S; Beckman Coulter Life Sciences) was used to screen
for surface markers against CD29 (0.2 mg/ml; cat. no. 562153), CD34
(0.2 mg/ml; cat. no. 560233), CD44 (0.5 mg/ml; cat. no. 550974),
CD45 (0.2 mg/ml; cat. no. 561586), CD73 (0.5 mg/ml; cat. no.
551123), and CD90 (0.2 mg/ml; cat. no. 561409), which were all
purchased from BD Biosciences; Becton, Dickinson and Company.
Briefly, the cells were collected, washed and then suspended in
flow cytometry staining buffer (cat. no. 554656; BD Biosciences)
containing the aforementioned antibodies at 25°C for 30 min before
being subjected to flow cytometric analysis. Acquired data were
analyzed using FlowJo software (v.10.8.1; FlowJo, LLC). ADSCs were
induced to differentiate into chondrocytes, osteocytes, and
adipocytes using chondrogenic, osteogenic, and adipogenic media as
described in previous studies (29,30). After 21 days of induction by the
chondrogenic medium supplemented with 1×
insulin-transferrin-selenium (Corning, Inc.), ADSCs were fixed at
25°C with 4% (w/v) paraformaldehyde for 15 min and then stained for
30 min at 4°C with 0.5% alcian blue dye (MilliporeSigma) in 1 mol/l
HCl to detect the extracellular matrix of chondrocytes. For
osteogenic differentiation, the cells were cultured in an induction
medium supplemented with 10 nM dexamethasone (Beyotime Institute of
Biotechnology), 10 mM β-glycerophosphate (Beyotime Institute of
Biotechnology), and 50 μg/ml L-ascorbic acid (Beyotime
Institute of Biotechnology) for 7 days and stained using an
alkaline phosphatase (ALP) assay kit according to the
manufacturer's protocol (Beyotime Institute of Biotechnology; cat.
no. P0321S); while after 14 days the cells were stained using the
Alizarin Red S Staining kit according to the manufacturer's
protocol (Beyotime Institute of Biotechnology; cat. no. C0148S).
For adipogenic induction, cells were subjected to Oil Red O
staining (Beyotime Institute of Biotechnology; cat. no. C0157S)
after 21 days of induction in the adipogenic medium supplemented
with 10 μM dexamethasone (Beyotime Institute of
Biotechnology), 25 mM 3-isobutyl-1-methylxanthine (MilliporeSigma),
2 μM rosiglitazone (MilliporeSigma), and 1 μg/ml
insulin (Beyotime Institute of Biotechnology).
Construction and preparation of
baculovirus vectors
The construction of a viral vector (designated as
pBac-Sa-con) was divided into four steps. All primers used for the
construction of the virus are listed in Table SI. First, a DNA fragment composed
of tandem recombination sites (KpnI-loxP-NheI-X
hoI-NotI-PacI-EcoRI-BamHI-loxP-HindIII) was chemically synthesized
by Detai Bioscience, Inc. (the full sequence is listed in Fig. S1) and subcloned into pFastBac
Dual (Gibco; Thermo Fisher Scientific, Inc.) using KpnI/HindIII
(Beyotime Institute of Biotechnology) digestion to yield the
vector, pL. Second, the CMV enhancer-rEF-1α promoter fragment was
PCR-amplified from pVITRO1-neo-mcs (Invivogen; cat. no.
pvitro1-nmcs) and subcloned into pL between the XhoI and NotI sites
to generate the vector, pLE. Third, the cDNA of a woodchuck
hepatitis virus post-transcriptional regulatory element (WPRE),
which enhances mRNA stability, was amplified from
pENN.AAV.hSyn.Cre.WPRE.hGH (Addgene; cat. no. 105553) and subcloned
into pLE using EcoRI/BamHI (Beyotime Institute of Biotechnology) to
generate the vector, pLEW. Finally, a Sa-deadCas9-VPR
(Sa-dCas9-VPR) fragment was PCR-amplified from SadCas9VPR (Addgene,
cat. no. 188514) and inserted into pLEW between the PacI and EcoRI
sites to yield pBac-Sa-con.
Single guide (sg)RNA cassettes of Sa-dCas9 were
synthesized using sequences of pGL3-U6-sgRNA-PGK-puromycin
(Addgene; cat. no. 51133), which contains a human U6 (hU6)
promoter, a spacer sequence, and a sgRNA scaffold. Spacer sequences
targeting UCHL1 with the highest targeting specificity scores
(5'-ACC GGT GAG ACC ACC ACC AGA TTA GCT CAC CGG CGA GTG GTC TCA GTT
TG-3') were designed using a guide RNA design tool (www.benchling.com). The resulting sgRNA sequences were
subcloned into pBac-Sa-con to yield pBac-Sa-UCHL1 using the NheI
reagent (Beyotime Institute of Biotechnology). The donor plasmids
pBac-Sa-con and pBac-Sa-UCHL1 were used to generate baculoviruses
Bac-Sa-con and Bac-Sa-UCHL1, respectively, using the
Bac-To-Bac® system (Invitrogen; Thermo Fisher
Scientific, Inc.). The recombinant BV vectors were amplified by
infecting Sf-9 insect cells and titrated using the end-point
dilution method (31).
Baculovirus transduction
For transduction, cells cultured overnight were
washed twice with PBS before being transduced with Bac-Sa-con and
Bac-Sa-UCHL1. Depending on the multiplicity of infection (MOI,
pfu/cell) and virus titer, a certain volume of the virus
supernatant was mixed with NaHCO3-free DMEM at a
volumetric ratio of 1:4 and added to the cells. The cells were
gently shaken on a rocking plate at 25°C for 6 h, after which the
solution was replaced with α-MEM medium containing 3 mM sodium
butyrate (MilliporeSigma), and the cells were further cultured. At
day 1 post-transduction (dpt), the medium was replaced with either
fresh α-MEM or chondroinductive medium. The chondroinductive medium
was replaced every 2-3 days until performing in vitro
analysis.
RNA extraction and reverse
transcription-quantitative (RT-q) PCR
Total RNA from ADSCs or chondrocytes in different
groups was measured using a Nanodrop 2000 spectrophotometer (Thermo
Fisher Scientific, Inc.) and reverse transcribed to cDNA using a
PrimeScript™ RT Master Mix according to the manufacturer's protocol
(Takara Bio, Inc.; cat. no. RR047A). qPCR was performed using the
PrimeScript RT-PCR kit (Takara Bio, Inc.; cat. no. RR820A), and the
primers used are listed in Table
SII. Relative mRNA expression of target genes was calculated
using the 2−ΔΔCq method (32). The thermocycling conditions were:
Initial denaturation for 30 sec at 95°C; followed by 40 cycles of
95°C for 5 sec, 60°C for 30 sec, and 95°C for 5 sec; melting at
65°C for 60 sec and 97°C for 1 sec; and cooling at 50°C for 30 sec.
The experiments were performed in triplicates and repeated three
times.
Immunofluorescence (IF) staining
To detect the expression of UCHL1 in hypoxia and
HIF-1α after activation of UCHL1, chondrocytes were stained with
DAPI (Beyotime Institute of Biotechnology; cat. no. C1005) at 25°C
for 5 min and simultaneously stained with antibodies against UCHL1
(1:200; Cell Signaling Technology; cat. no. 13179) or HIF-1α
(1:100; Cell Signaling Technology; cat. no. 48085) at a temperature
of 37°C for 1 h. To verify the levels of mitophagy in different
groups, chondrocytes were stained with DAPI at 25°C for 5 min and
simultaneously stained with antibodies against LC3B (1:200, Cell
Signaling Technology, cat. no. 3868) and Tom20 (1:200; Cell
Signaling Technology, cat. no. 42406) at 37°C for 1 h. Next, the
cells were stained with the corresponding secondary antibodies
(Alexa Fluor® 488- or 594-conjugated goat anti-rabbit or
anti-mouse IgG, all 1:1,000, Abcam; cat. nos. ab150077, ab150113,
ab150080, and ab150116) at 37°C for 1 h. Images were obtained using
a laser scanning confocal microscope (magnification, ×400; LSCM,
Zeiss GmbH, cat. no. LSM780). In addition, the fluorescence
intensity was measured using ImageJ version 2.1 (National
Institutes of Health).
TUNEL staining
Damaged DNA was detected using a TUNEL Cell
Apoptosis Detection Kit (Beyotime Institute of Biotechnology; cat.
no. C1088). Chondrocytes were fixed and stained with TUNEL test
solution for 30 min at 37°C according to the manufacturer's
instructions, and the nuclei were stained with DAPI at 25°C for 5
min. A total of three fields of view were randomly selected and
captured to count the number of TUNEL-positive cells.
Apoptosis
The Annexin V-APC Apoptosis Detection Kit (BioGems;
cat. no. 62700-80) was used to determine the apoptotic ratio,
according to the manufacturer's instructions. Briefly, chondrocytes
were washed twice with Cell Staining Buffer and resuspended in
Annexin V-Binding Buffer at a concentration of 1×107
cells/ml. A total of 100 μl cell suspension was transferred
to a 5 ml test tube, 5 μl APC Annexin V and 5 μl
7-AAD Viability Staining Solution were added in this order, and the
cells were gently vortexed and incubated for 15 min at 25°C in the
dark, and then 400 μl Annexin V Binding Buffer was added to
each tube. The apoptosis ratio of chondrocytes was analyzed using
FCM.
Mitochondrial membrane potential
(MMP)
A mitochondrial membrane potential assay kit with
JC-1 (Beyotime Institute of Biotechnology; cat. no. C2003S) was
used to assess the MMP. Chondrocytes were incubated with JC-1 (5
μM) for 30 min and DAPI for 5 min at 37°C, then washed three
times with PBS and observed using LSCM (magnification, ×400).
The MMP was detected using MitoTracker Red CMXRos
(Beyotime Institute of Biotechnology; cat. no. C1035). Chondrocytes
were incubated with 50 nM Mito-tracker probes for 30 min at 37°C
according to the manufacturer's instructions. After staining with
DAPI at 25°C for 5 min, the cells were washed thrice with PBS, and
images were captured using LSCM (magnification, ×400).
Measurement of mitochondrial and
intracellular ROS
Mito-SOX Red dye (Invitrogen; Thermo Fisher
Scientific, Inc.; cat. no. M36008) was used to assess mitochondrial
ROS levels. Chondrocytes from different groups were incubated with
Mito-Sox Red dye (5 μM) for 30 min at 37°C and stained with
DAPI. The cells were then washed twice with PBS and observed using
LSCM (magnification, ×400).
Intracellular ROS was detected using the ROS Assay
kit (Beyotime Institute of Biotechnology; cat. no. S0033M). After
adding the dichloro-dihydro-fluorescein diacetate (DCFH-DA) probe
at a final concentration of 10 μM, chondrocytes were
incubated in the dark for 30 min at 37°C and then stained with
DAPI. Images were captured using LSCM (magnification, ×400).
Cell proliferation
Chondrocyte growth was analyzed using a CCK-8 cell
viability kit (Dojindo Molecular Technologies, Inc. Molecular
Technologies, Inc.; cat. no. CK04-11). Briefly, cells were seeded
into 96-well plates, and DMEM containing different concentrations
of LDN was added for 1, 3, 5, or 7 days. CCK-8 reagent (10
μl) was added to each well and the plates were incubated at
37°C for 2 h. The absorbance of the supernatant was measured at 450
nm using a microplate reader (Thermo Fisher Scientific, Inc.).
Seahorse metabolic flux analysis
The oxygen consumption rate (OCR) was measured using
the Seahorse XF 96 Extracellular Flux Analyzer (Seahorse
Bioscience; Agilent Technologies, Inc.) with the Agilent Seahorse
XF Cell Mito Stress Test kit (Seahorse Bioscience; Agilent
Technologies, Inc.; cat. no. 103015-100). Briefly, chondrocytes
(2×104 cells/well) were seeded in a Seahorse XF 96-well
cell culture plate. The loaded sensor cartridge with the utility
plate was placed in the instrument for calibration, and oligomycin
(1.5 μM), fluoromethoxy carbonyl cyanide phenylhydrazone
(FCCP, 1.5 μM), rotenone, and antimycin A (1.25 μM)
were sequentially added to each well after 20-, 40-, and 60-min.
OCR data were assessed using the Seahorse XF-96 Wave software
version 2.6 (Seahorse Bioscience; Agilent Technologies, Inc.).
Statistical analysis
Data are presented as mean ± SD of at least three
independent experimental repeats. Following Shapiro-Wilk tests for
assessment of normality and Levene's test for equality of variance,
a paired Student's t-test was used to assess the differences
between the control and test groups. One- and two-way ANOVA
followed by a SNK post hoc test were used to analyze the
differences in CCK-8 assays, quantitative analysis of LC3B, TUNEL
staining, Cell apoptosis, and MMP between multiple groups after
treatment with LDN, CRISPRa, KC7F2, or 3-MA. P<0.05 was
considered to indicate a statistically significant difference.
Results
UCHL1 expression is increased under
hypoxic conditions
ADSCs exhibited typical spindle and elongated
fibroblast-like morphology (Fig.
1A). Mesenchymal stem cell markers (such as CD29, CD44, and
CD90) and the pluripotent marker CD73 were highly expressed,
whereas CD34 and CD45 (hematopoietic cell antigen) were
undetectable (Fig. 1B). Alcian
blue staining was used to demonstrate the chondrogenic
differentiation ability of ADSCs, which was also evidenced by
upregulation of chondrogenesis-related markers such as aggrecan,
SOX9, and COL2A1 (Fig. 1C and D).
The osteogenic differentiation ability of ADSCs was confirmed by
the strong ALP and Alizarin Red staining as well as by upregulation
of osteogenesis-related markers, such as RUNX2, ALP, and COL1
(Fig. 1E-G). Furthermore, Oil red
O staining revealed differentiation of adipocytes (Fig. 1H). These results verified the
potential of ADSCs to differentiate into multiple types of
cells.
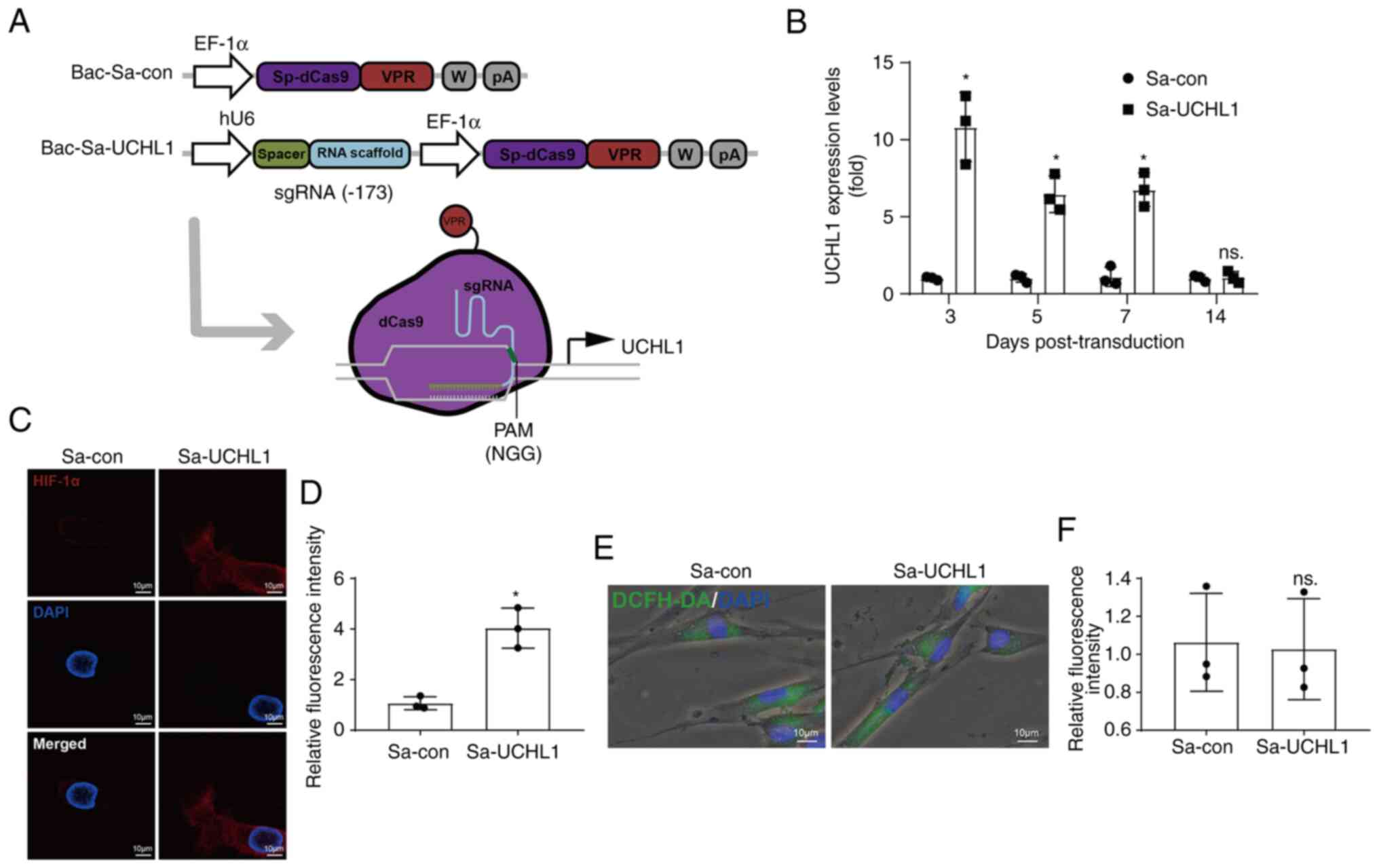
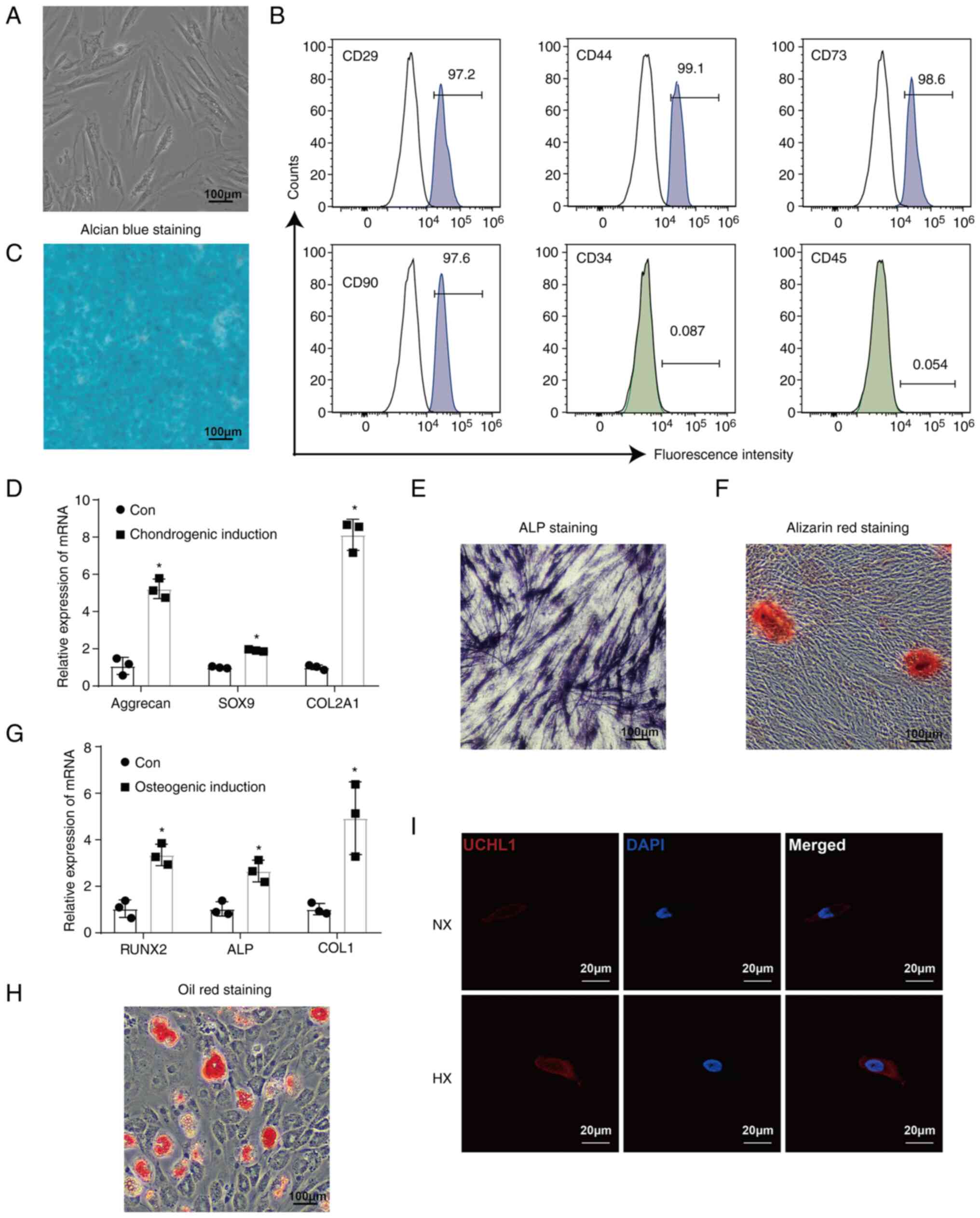 | Figure 1UCHL1 expression is increased under
hypoxic conditions. (A) Images of ADSCs observed under a microscope
(magnification, ×40). (B) Mesenchymal stem cell antigens (CD29,
CD44, CD73, and CD90) and hematopoietic cell antigens (CD34 and
CD45). expressed in ADSCs were detected by flow cytometry. (C)
Alcian blue staining of ADSCs after 21 days of chondrogenic
induction (magnification, ×40). (D) mRNA expression of chondrogenic
markers Aggrecan, SOX9, and COL2A1 after 14 days of chondrogenic
induction, as analyzed. (E) ALP staining of ADSCs after 7 days of
osteogenic induction (magnification, ×40). (F) Alizarin red
staining of ADSCs after 14 days of osteogenic induction
(magnification, ×40). (G) mRNA expression of chondrogenic markers
RUNX2, ALP, and COL1 after 14 days of osteogenic induction. (H) Oil
red O staining of ADSCs after 21 days of adipogenic induction
(magnification, ×40). (I) Immunofluorescence staining of UCHL1
(magnification, ×400). *P<0.05 vs. Con. NX, normoxia;
HX, hypoxia; Con, control group; UCHL1, Ubiquitin C-terminal
hydrolase-L1; ADSCs, adipose-derived stem cells; ALP, alkaline
phosphatase. |
To investigate the effects of UCHL1 on apoptosis in
chondrocytes, the chondrocytes differentiated from ADSCs were used
for subsequent experiments. The IF staining results indicated that
the levels of UCHL1 were enhanced under hypoxic conditions
(Fig. 1I).
Hypoxia regulates mitophagy and apoptosis
in chondrocytes
The results of IF staining indicated that the
protein levels of LC3B increased under hypoxic conditions (Fig. 2A and B). To detect apoptosis,
TUNEL assays and FCM were performed, and the results demonstrated
that hypoxia reduced apoptosis (Fig.
2C-F). As a fluorescent probe used to detect MMP, the
transformation of fluorescence of JC-1 from red to green
fluorescence is an early indicator of apoptosis. The results of
JC-1 staining also revealed that hypoxia inhibited apoptosis by
increasing MMP levels (Fig. 2G).
IF staining with MitoTracker Red revealed that MMP levels were also
increased under hypoxic conditions (Fig. 2H and I). In addition,
mitochondrial and intracellular ROS levels were significantly
increased under hypoxic conditions (Fig. 2J-M).
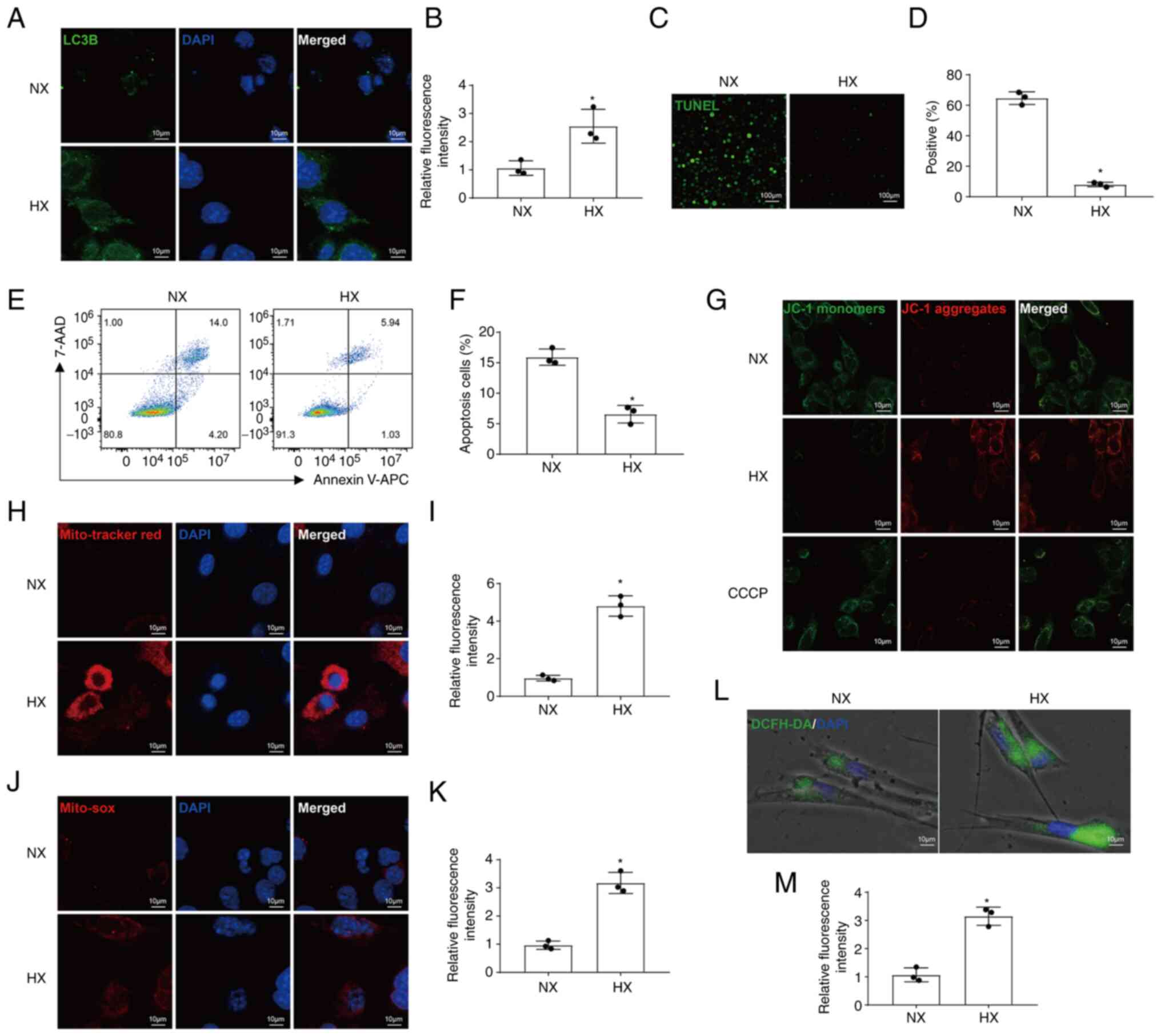 | Figure 2Hypoxia regulates mitophagy and
apoptosis in chondrocytes. (A and B) IF images and quantification
of LC3B expression (magnification, ×400). (C and D) Representative
IF images and quantification of TUNEL staining (magnification,
×40). (E and F) Apoptosis was analyzed using the Annexin-APC/7-AAD
kit and measured using FCM. (G) MMP was measured by IF using the
JC-1 dye (magnification, ×400). CCCP was used as a positive
control. (H and I) Representative IF images and quantitative
analysis of MitoTracker Red staining (magnification, ×400). (J and
K) Representative IF images and quantification of MitoSOX
(magnification, ×400). (L and M) Representative IF images and
quantitative analysis of DCFH-DA (magnification, ×400).
*P<0.05 vs. NX. NX, normoxia; HX, hypoxia; FCM, flow
cytometry; CCCP, carbonyl cyanide 3-chlorophenylhydrazone; IF,
immunofluorescence. |
UCHL1 mediates the mitophagy regulated by
hypoxia in chondrocytes
To further confirm whether UCHL1 plays a role in
hypoxia-induced mitophagy, chondrocytes were treated with LDN, a
specific UCHL1 inhibitor. LDN was not cytotoxic to chondrocytes at
0, 1, 2, 4, and 8 μM concentrations after treatment for 1,
3, 5, and 7 days as observed using CCK-8 assays. However, the
proliferation of the cells was significantly inhibited when treated
with 16 μM LDN for 5 days (Fig. 3A). Thus, 8 μM LDN was
utilized for 1 day in subsequent experiments. The increase in LC3B
expression under hypoxic conditions was reversed by LDN treatment
(Fig. 3B and C). The proportion
of apoptotic cells, which had decreased under hypoxic conditions,
increased when UCHL1 was inhibited (Fig. 3D-G). The increase in MMP observed
under hypoxic conditions was also reversed by LDN (Fig. 3H-J). These results suggest that
UCHL1 mediates mitophagy and apoptosis regulated by hypoxia in
chondrocytes.
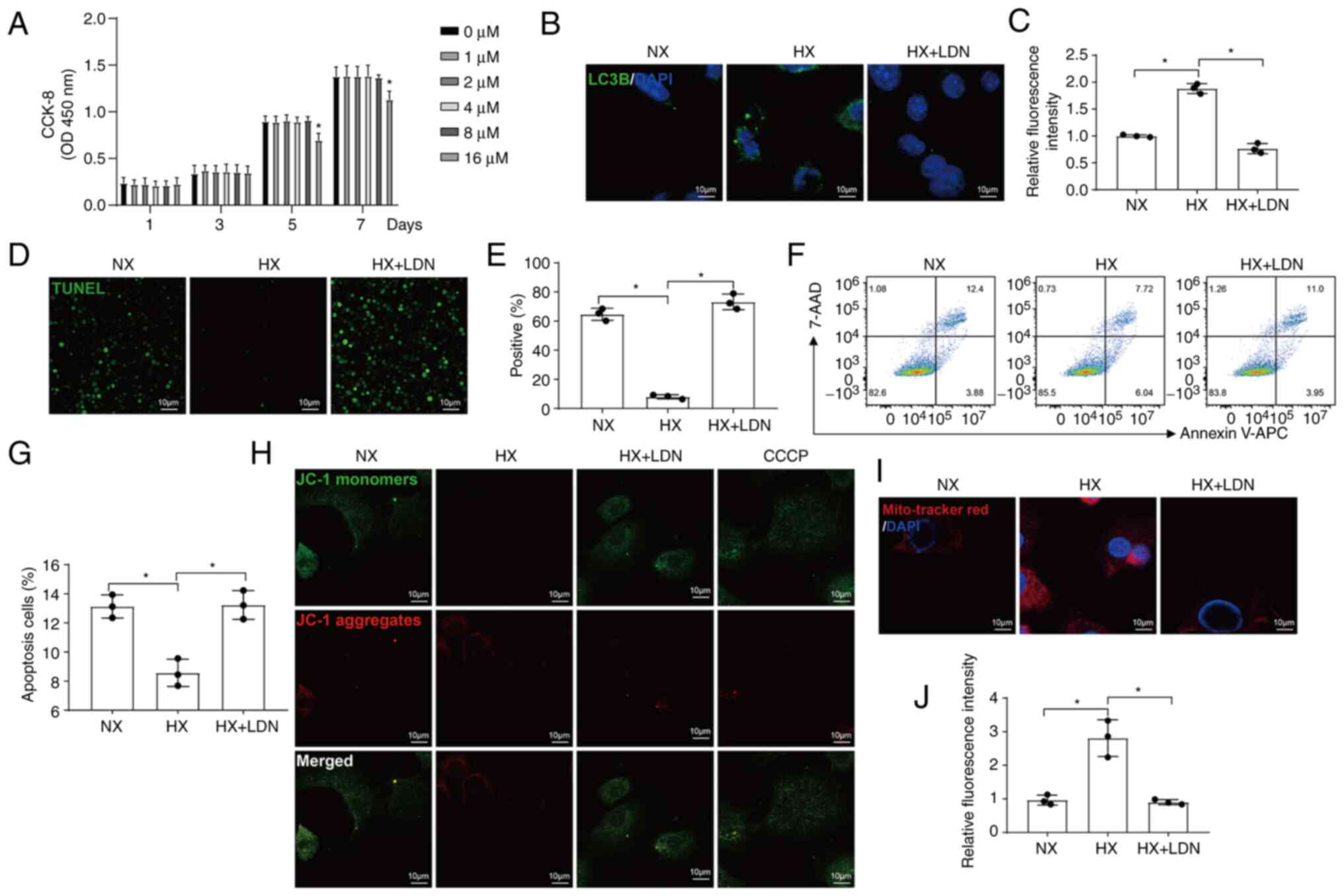 | Figure 3UCHL1 mediates the mitophagy
regulated by hypoxia in chondrocytes. (A) CCK-8 assays of
LDN-treated cells at the indicated concentrations. (B and C)
Representative IF images and quantitative analysis of LC3B
(magnification, ×400). (D and E) Representative IF images and
quantitative analysis of the TUNEL staining (magnification, ×40).
(F and G) Cell apoptosis analyzed using the Annexin-APC/7-AAD kit
was measured by FCM. (H) The MMP was measured by IF using a JC-1
dye. CCCP was used as the positive control (magnification, ×400).
(I and J) Representative IF images and quantitative analysis of
MitoTracker Red staining (magnification, ×400).
*P<0.05 vs. 0 μM. NX, normoxia; HX, hypoxia;
UCHL1, Ubiquitin C-terminal hydrolase-L1; FCM, flow cytometry;
CCCP, carbonyl cyanide 3-chlorophenylhydrazone; IF,
immunofluorescence; LDN, LDN-57444. |
The CRISPRa system effectively activates
UCHL1
Next, the CRISPRa module was used to activate UCHL1
expression. BV was designed to express the CRISPRa module, which
expressed sgRNA under the human U6 promoter and dCas9-VPR under the
rat EF-1α promoter (Fig. 4A).
dCas9 is derived from Staphylococcus aureus (Sa) and has a
protospacer-adjacent motif (PAM, NNGRRT). Bac-Sa-UCHL1 expressed
the Sa-dCas9-VPR and its associated sgRNA. As a control, Bac-Sa-con
that expressed Sa-dCas9-VPR but not sgRNA was constructed (Fig. 4A). Chondrocytes were transduced
with Bac-Sa-con or Bac-Sa-UCHL1 (designated as Sa-con and Sa-UCHL1
groups, respectively). UCHL1 expression was analyzed by RT-qPCR at
3, 5, 7, and 14 dpt. Compared with the Sa-con group, Sa-UCHL1
triggered significant UCHL1 upregulation for 7 days (Fig. 4B), suggesting that the CRISPRa
system activated UCHL1 expression for at least 7 days. The
expression levels of HIF-1α were increased in the Sa-UCHL1 group
(Fig. 4C and D), while the
intracellular ROS levels remained unchanged (Fig. 4E and F), indicating that
endogenous activation of UCHL1 could induce the expression of
HIF-1α without affecting intracellular ROS levels.
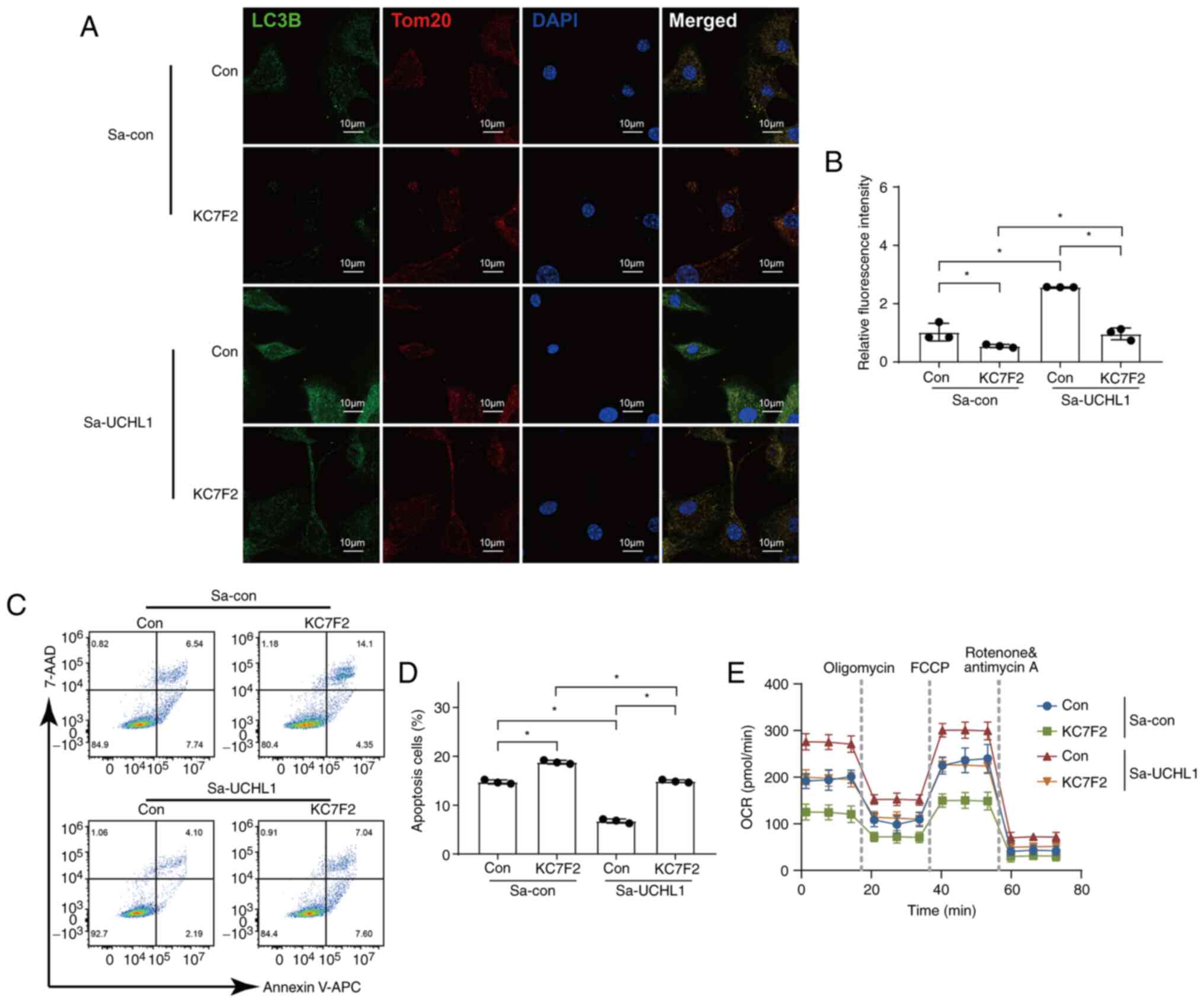
UCHL1 accelerates mitophagy, maintains
mitochondrial function, and inhibits apoptosis
Next, the effects of endogenous activation of UCHL1
on mitophagy, mitochondrial function, and apoptosis were studied.
The protein levels of LC3B were increased in the Sp-UCHL1 group
(Fig. 5A and B), indicating the
occurrence of intensive mitophagy. The proportion of apoptotic
cells was reduced in the Sa-UCHL1 group, as measured by the TUNEL
assay and FCM (Fig. 5C-F). IF
images of cells stained with MitoTracker Red and JC-1 dyes revealed
that the MMP increased in the Sa-UCHL1 group (Fig. 5G-J). To investigate the mechanism
by which endogenous activation of UCHL1 orchestrated mitochondrial
function, the OCR of chondrocytes was measured according to the
manufacturer's instructions. The results revealed that the OCR in
chondrocytes was enhanced by the endogenous activation of UCHL1
(Fig. 5K). Specifically, UCHL1
increased basal respiration, ATP production, maximal respiration,
and spare-respiratory capacity to maintain mitochondrial functions
(Fig. 5L).
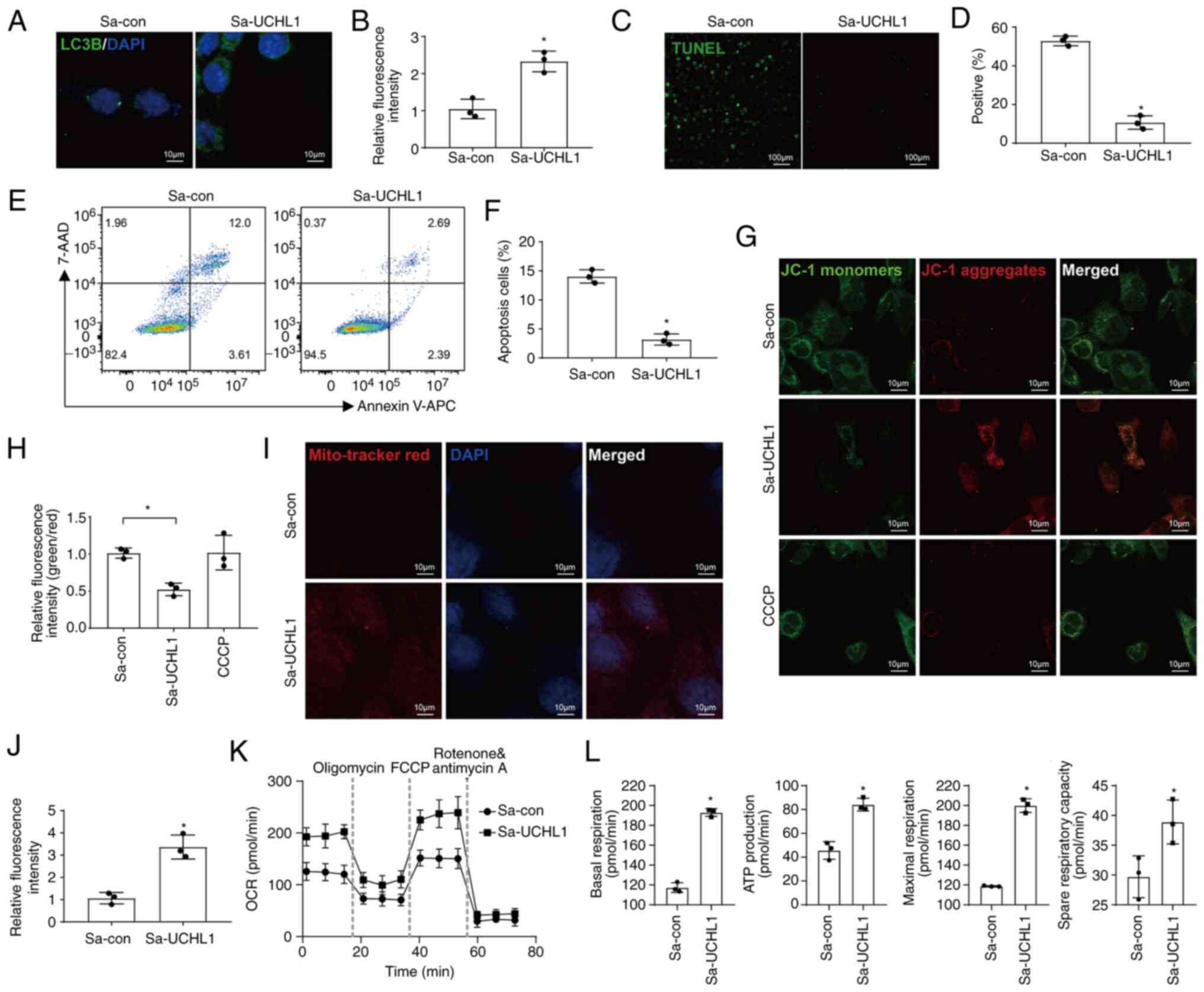 | Figure 5UCHL1 accelerates mitophagy,
maintains mitochondrial function, and inhibits apoptosis. (A and B)
IF images and quantification of LC3B expression (magnification,
×400). (C and D) Representative IF images and quantification of
TUNEL staining (magnification, ×40). (E and F) Apoptosis was
analyzed using the Annexin-APC/7-AAD kit and assessed using FCM. (G
and H) MMP was measured by IF using JC-1 dye (magnification, ×400).
CCCP was used as a positive control. (I and J) Representative IF
images and quantitative analysis of MitoTracker Red staining
(magnification, ×400). (K) OCR of chondrocytes following activation
of UCHL1 using the CRISPRa system. (L) Effects of UCHL1 activated
by CRISPRa on basal respiration, ATP production, maximal
respiration, and spare respiratory capacity estimated using the OCR
assay are shown. *P<0.05 vs. Sa-con. UCHL1, Ubiquitin
C-terminal hydrolase-L1; Sa, Staphylococcus aureus; CCCP,
carbonyl cyanide 3-chlorophenylhydrazone; IF, immunofluorescence;
OCR, oxygen consumption rate; FCCP, fluoromethoxy carbonyl cyanide
phenylhydrazone. |
HIF-1α mediates mitochondrial functions
modulated by UCHL1
To determine whether the effect of UCHL1 on
mitophagy is dependent on HIF-1α, a specific inhibitor of HIF-1α,
KC7F2, was used to ascertain the mechanism of action of UCHL1 in
mitophagy. As shown in Fig. 6A,
the expression of LC3B was increased by UCHL1 in chondrocytes.
However, the augmented LC3B expression was inhibited by treatment
with KC7F2 (Fig. 6A and B). The
reduced ratio of apoptosis in chondrocytes via activation of UCHL1
was augmented by treatment with KC7F2 (Fig. 6C and D). OCR data indicated that
chondrocytes displayed a higher OCR of cellular metabolism due to
the activation of UCHL1. However, this increase in OCR was
inhibited by treatment with FC7F2 (Fig. 6E). These results showed that
HIF-1α mediated mitochondrial functions modulated by UCHL1.
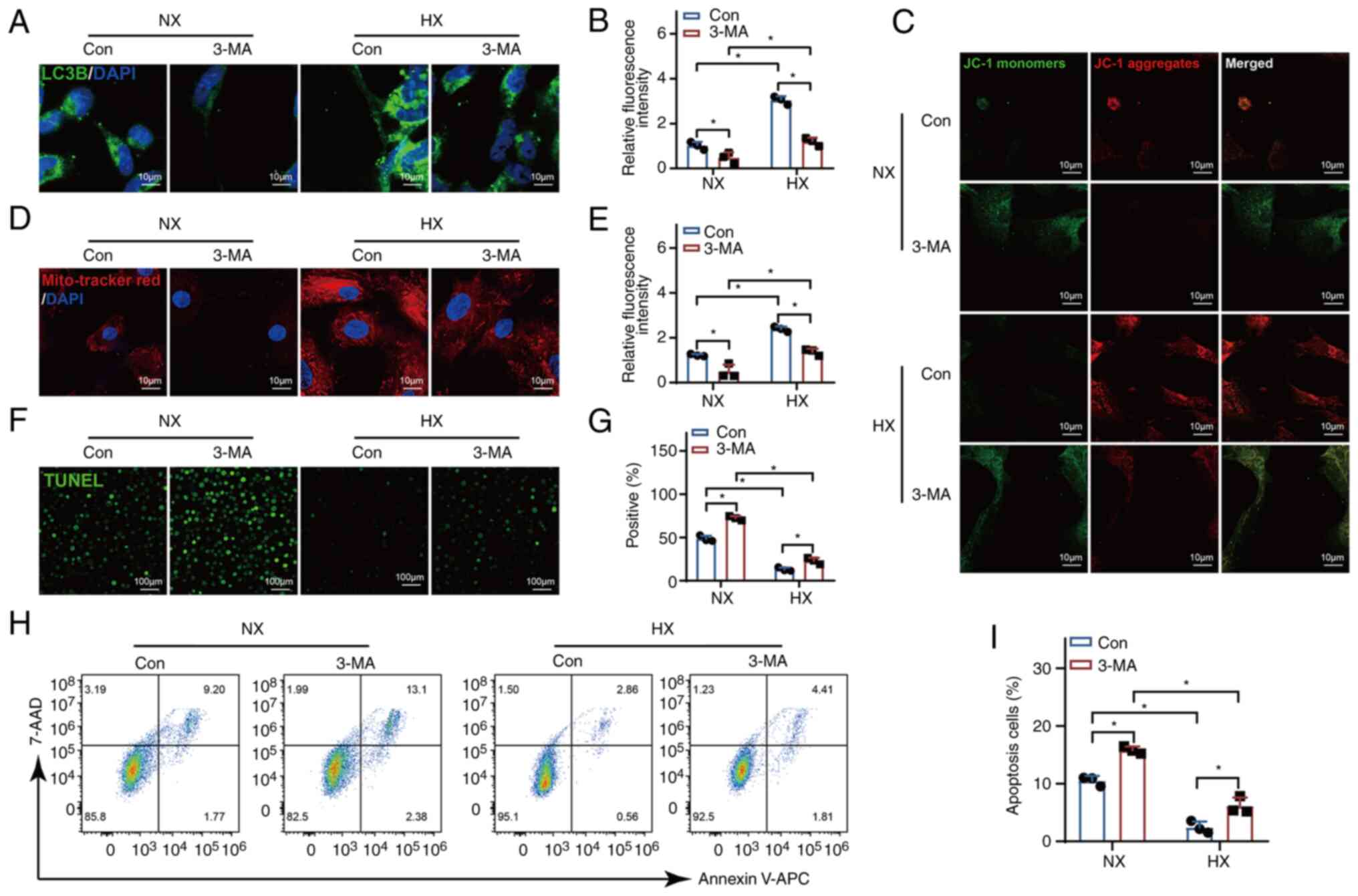
Mitophagy mediates the inhibition of
apoptosis by UCHL1
To examine the correlation between autophagy and
apoptosis, cells in both the normoxia and hypoxia groups were
subjected to treatment with 3-MA. The inhibition of mitophagy by
3-MA resulted in the cessation of LC3B accumulation induced by
hypoxia (Fig. 7A and B).
Additionally, the inhibition of mitophagy eliminated the
hypoxia-induced increase in MMP (Fig.
7C-E) and suppressed apoptosis (Fig. 7F-I). These observations signify
that the inhibition of apoptosis is contingent upon enhanced
mitophagy.
Discussion
ADSCs have been widely used to treat diabetic foot
disease (33), knee
osteoarthritis (34), and
cirrhosis (35) given their
abundance, convenience of access, and low immunogenicity (36). A growing body of research has
verified that ADSCs have significant potential in restoring the
structure and functions of damaged tissues and may thus serve as
novel treatment approaches for various refractory diseases such as
cartilage defects (31). In the
current study, it was found that UCHL1 expression was notably
increased under hypoxic conditions in ADSC-derived chondrocytes.
Given that UCHL1 abrogates VHL-mediated ubiquitination of HIF-1α
(28) and that HIF-1α can
alleviate apoptosis and senescence in chondrocytes through
mitophagy (16), it was
hypothesized that UCHL1 could alleviate apoptosis in chondrocytes
via HIF-1α. Consistent with this hypothesis, the results of the
current study suggested that UCHL1 attenuated apoptosis in
chondrocytes derived from ADSCs via upregulation of HIF-1α-mediated
mitophagy.
During mitophagy, cytosolic LC3B is recruited to the
mitochondria, forming LC3B-positive puncta. The presence of LC3B
puncta indicates the initiation of mitophagy (37). Moreover, efficient mitophagy helps
maintain optimal MMP levels by facilitating the removal of damaged
mitochondria, preventing their accumulation and associated
metabolic defects (38).
Therefore, LC3B and MMP were chosen as markers for mitophagy and
detected in this study.
The response of chondrocytes to hypoxia-mediated by
HIF-1α plays a vital role in regulating chondrogenesis by
maintaining appropriate extracellular matrix production (39) and directing progenitor cell
differentiation (21). In the
present study, the in vitro experiments illustrated that
hypoxia attenuated apoptosis and induced mitophagy, suggesting that
hypoxia plays a role not only in regulating chondrogenesis, but
also in cell survival. As hypoxia aids in the maintenance of
chondral tissue, controlling the oxygen pressure may be an
effective strategy for engineering chondral tissues. However,
hypoxia has been reported to cause oxidative stress and promote
potentially undesirable effects on cell metabolism which may be
detrimental to the formation of healthy chondral tissues (26). Consistent with these results, the
results of the present study demonstrated that mitochondrial and
intracellular ROS levels in chondrocytes were significantly
increased under hypoxic conditions. Therefore, stabilization of
HIF-1α under normoxic conditions has the potential to provide
beneficial pro-chondrogenic effects of hypoxia in a potentially
less deleterious and controlled manner.
Cofactors, such as p300 and the CREB-binding protein
(CBP), are required to be recruited to the HIF transcriptional
complex when HIF-1α binds to the Hypoxia response element in target
gene promoters. The key residue present on HIF-1α that is involved
in its binding with p300/CBP is asparagine-803 (Asn-803), which is
also a target of 2-oxoglutarate (2-OG). As a factor inhibiting
HIF-1α, 2-OG hydroxylates Asn-803 on HIF-1α, thereby preventing the
binding of p300/CBP to HIF-1α, and in turn, disrupting the function
of the HIF transcriptional complex (40-42). Certain compounds are reported to
stabilize HIF-1α and enhance its binding through transcriptional
cofactors under normoxic conditions. Among these compounds,
dimethyloxalylglycine (DMOG) acts by competing with 2-OG by
engaging the binding pocket of the prolyl hydroxylase active site
(43). Recent research revealed
that DMOG-loaded grafts promote vascular regeneration by
stabilizing HIF-1α (44).
Moreover, DMOG-doped zeolitic imidazolate frameworks-8
significantly promoted vascularized bone regeneration, primarily
through the activation of HIF-1α (45). Nevertheless, although promising,
DMOG lacks a high degree of specificity and may also target
similarly structured enzymes that are essential for the formation
of the collagen triple helix (46). In the present study, to stabilize
HIF-1α under normoxic conditions in a controlled manner, endogenous
UCHL1 was activated using the CRISPRa system. The results
demonstrated that the intracellular ROS levels in chondrocytes were
not increased following activation of UCHL1. Furthermore, UCHL1
accelerated mitophagy, maintained mitochondrial functions, and
inhibited apoptosis by stabilizing HIF-1α.
In 1981, UCHL1 was initially described as a soluble
nervous system-specific protein (47). Additional studies extended these
observations by showing that UCHL1 was not only present in neurons
of the central nervous system, but also in the heart (27), kidney (48), intervertebral disc (49), and periodontium (50). IF staining demonstrated that
subcellular localization of UCHL1 is closely associated with the
endoplasmic reticulum and mitochondria in neuroblastoma cells
(51). Moreover, UCHL1 was shown
to influence the morphology and respiratory functions of
mitochondria in skeletal muscles, suggesting the existence of a
link between UCHL1 and mitochondria of vital organelles (52). Recent reports suggest that UCHL1
can stabilize HIF-1α via abrogating ubiquitination of HIF-1α
(27,28). Based on these experiments, the
role of UCHL1 on mitochondrial functions was assessed and it was
found that UCHL1 induced mitophagy by abrogating the ubiquitination
of HIF-1α in chondrocytes derived from ADSCs. In addition to
regulating mitophagy, UCHL1 plays a role in mitochondrial dynamics
and bioenergetics (53).
Downregulation of UCHL1 reduces the levels of the mitochondrial
fusion protein Mitofusin-2, resulting in mitochondrial enlargement
and disruption of the tubular network in various cell lines
(51). In addition, the
respiratory function of the mitochondria was enhanced by the
activation of UCHL1 in the present study. Thus, the effects of
UCHL1 on mitochondria are multifaceted and require further
investigation.
The CRISPRa system is an RNA-guided gene editing
system repurposed from CRISPR-Cas9 and comprises gRNA and
catalytically dead Cas9 (dCas9) (54). The gRNA was composed of a scaffold
sequence responsible for dCas9 binding and a spacer sequence to
recognize the target DNA. dCas9 is derived from mutated Cas9, the
orthologs of which are derived from different bacteria such as
Streptococcus pyogenes (SpCas9), Staphylococcus
aureus (SaCas9), and Neisseria meningitides (NmCas9),
among which SpCas9 is the most widely used (35). As the dCas9-VPR from
Staphylococcus aureus (SadCas9-VPR) is more efficient than
that from Streptococcus pyogenes in ADSCs (31), SadCas9-VPR was used in the present
study. The SpdCas9 protein can be fused with a transcription
activator (such as VP64) for CRISPRa of the target gene. In the
present study, for more robust gene activation, SpdCas9 was fused
with a tripartite activator, VPR, consisting of VP64, p65, and Rta
to form SpdCas9-VPR to activate UCHL1, and the results demonstrated
that UCHL1 expression was activated for at least 7 days. The size
of SpdCas9-VPR is ~5.8 kb, which exceeds the packaging capacity of
commonly used adeno-associated virus (55). Baculoviruses can deliver large
amounts of genetic cargo (at least 38 kb) into ADSCs with an
efficiency of >95%. As a non-pathogenic insect virus,
baculoviruses neither replicate nor integrate their genome into the
chromosomes of transduced cells, thereby minimizing their potential
genotoxicity (31). Therefore, a
baculovirus was employed to deliver the CRISPRa system to ADSCs in
the current study.
Improving the safety profile of BV leads to
short-term transgene expression, which is insufficient to maintain
the long-term survival of ADSCs. The Cre/loxP-based hybrid BV
system consists of a vector expressing Cre recombinase and another
vector carrying a transgene cassette flanked by loxP sites,
enabling the formation of DNA minicircles that can persist
independent of chromosomes and prolong transgene expression
(29). This is an area of ongoing
research in our laboratory.
The present study investigated the impact of UCHL1
on chondrocytes, and shed light on its ability to prevent cell
apoptosis via upregulation of HIF-1α-mediated mitophagy. By
suppressing apoptosis, UCHL1 provides innate protection to
chondrocytes and potentially contributes to sustaining cartilage
integrity. This finding highlights novel avenues for cartilage
tissue engineering by identifying UCHL1 as a molecular target for
therapeutic interventions aimed at promoting cartilage repair.
Additionally, the observed effects of UCHL1 on HIF-1α mediated
mitophagy provide valuable insights into potential mechanisms for
maintaining cellular homeostasis within cartilage tissues. These
results not only contribute to our understanding of the underlying
processes involved in cartilage health but also suggest a novel
method for future cartilage repair. By harnessing the effect of
UCHL1 and its ability to inhibit apoptosis and preserve
mitochondrial function, researchers may develop innovative
therapies to rejuvenate or regenerate damaged cartilage.
Supplementary Data
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
QY contributed to the conception, design, data
acquisition, analysis and interpretation, and drafting of the
manuscript, and critically revised the manuscript. SS and YG
contributed to the data acquisition and analysis, and drafted and
critically revised the manuscript. SW and ML contributed to the
data acquisition and critically revised the manuscript. ML
contributed to the conception and design of the study. All authors
have read and approved the final manuscript. ML and QY confirm the
authenticity of all the raw data.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Abbreviations:
|
3-MA
|
3-Methyladenine
|
|
ROS
|
reactive oxygen species
|
|
HIF-1α
|
hypoxia-inducible factor 1α
|
|
PHD2
|
prolyl hydroxylase 2
|
|
VHL
|
von hippel-lindau
|
|
UCHL1
|
Ubiquitin C-terminal hydrolase-L1
|
|
CRISPRa
|
CRISPR activation
|
|
ADSCs
|
adipose-derived stem cells
|
|
FCM
|
flow cytometry
|
|
ALP
|
alkaline phosphatase
|
|
dpt
|
day post-transduction
|
|
MMP
|
Mitochondrial membrane potential
|
|
DCFH-DA
|
dichloro-dihydro-fluorescein
diacetate
|
|
OCR
|
oxygen consumption rate
|
|
FCCP
|
fluoromethoxy carbonyl cyanide
phenylhydrazone
|
|
Sa
|
Staphylococcus aureus
|
|
CBP
|
CREB-binding protein
|
|
DMOG
|
dimethyloxalylglycine
|
Acknowledgments
Not applicable.
Funding
This study was funded by the Science Research Cultivation
Program of Stomatological Hospital, Southern Medical University
(grant nos. PY2021028 and RC202202) and Guangzhou Basic and Applied
Basic Research Foundation (grant no. 2023A04J0427).
References
|
1
|
Jiang Y and Tuan RS: Origin and function
of cartilage stem/progenitor cells in osteoarthritis. Nat Rev
Rheumatol. 11:206–212. 2015. View Article : Google Scholar
|
|
2
|
Vila PM, Jeanpierre LM, Rizzi CJ, Yaeger
LH and Chi JJ: Comparison of autologous vs Homologous costal
cartilage grafts in dorsal augmentation rhinoplasty: A systematic
review and Meta-analysis. JAMA Otolaryngol Head Neck Surg.
146:347–354. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhang L, Wang JW, Ding J, Zhang X, Wang
XM, Zhang ZZ and Yu RZ: A new technique for Asian nasal tip
shaping: 'Twin tower' folding ear cartilage transplantation. Case
Reports Plast Surg Hand Surg. 9:207–213. 2022. View Article : Google Scholar :
|
|
4
|
Eftekhar N, Borjian A, Rafieian S, Borjian
MA and Sahebi MA: Successful tracheal necrosis management using a
pedicle pectoralis flap: A case report. Turk Gogus Kalp Damar
Cerrahisi Derg. 28:547–551. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Calvert JW, Patel AC and Daniel RK:
Reconstructive rhinoplasty: Operative revision of patients with
previous autologous costal cartilage grafts. Plast Reconstr Surg.
133:1087–1096. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang S, Yang L, Cai B, Liu F, Hou Y, Zheng
H, Cheng F, Zhang H, Wang L, Wang X, et al: Injectable hybrid
inorganic nanoscaffold as rapid stem cell assembly template for
cartilage repair. Natl Sci Rev. 9:nwac0372022. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Johnson K, Zhu S, Tremblay MS, Payette JN,
Wang J, Bouchez LC, Meeusen S, Althage A, Cho CY, Wu X and Schultz
PG: A stem cell-based approach to cartilage repair. Science.
336:717–721. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Watanabe J, Yamada M, Niibe K, Zhang M,
Kondo T, Ishibashi M and Egusa H: Preconditioning of bone
marrow-derived mesenchymal stem cells with N-acetyl-L-cysteine
enhances bone regeneration via reinforced resistance to oxidative
stress. Biomaterials. 185:25–38. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hughes CE, Coody TK, Jeong MY, Berg JA,
Winge DR and Hughes AL: Cysteine Toxicity Drives Age-Related
Mitochondrial Decline by Altering Iron Homeostasis. Cell.
180:296–310.e18. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
He K, Nie L, Ali T, Liu Z, Li W, Gao R,
Zhang Z, Liu J, Dai Z, Xie Y, et al: Adiponectin deficiency
accelerates brain aging via mitochondria-associated
neuroinflammation. Immun Ageing. 20:152023. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Akter M, Ma H, Hasan M, Karim A, Zhu X,
Zhang L and Li Y: Exogenous L-lactate administration in rat
hippocampus increases expression of key regulators of mitochondrial
biogenesis and antioxidant defense. Front Mol Neurosci Mar.
16:11171462023. View Article : Google Scholar
|
|
12
|
Qu F, Wang P, Zhang K, Shi Y, Li Y, Li C,
Lu J, Liu Q and Wang X: Manipulation of Mitophagy by 'All-in-One'
nanosensitizer augments sonodynamic glioma therapy. Autophagy.
16:1413–1435. 2020. View Article : Google Scholar
|
|
13
|
Tang C, Han H, Yan M, Zhu S, Liu J, Liu Z,
He L, Tan J, Liu Y, Liu H, et al: PINK1-PRKN/PARK2 pathway of
mitophagy is activated to protect against renal
ischemia-reperfusion injury. Autophagy. 14:880–897. 2018.
View Article : Google Scholar :
|
|
14
|
Kubli DA and Gustafsson ÅB: Mitochondria
and mitophagy: The yin and yang of cell death control. Circ Res.
111:1208–1221. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu L, Zhang W, Liu T, Tan Y, Chen C, Zhao
J, Geng H and Ma C: The physiological metabolite α-ketoglutarate
ameliorates osteoarthritis by regulating mitophagy and oxidative
stress. Redox Biol. 62:1026632023. View Article : Google Scholar
|
|
16
|
Hu S, Zhang C, Ni L, Huang C, Chen D, Shi
K, Jin H, Zhang K, Li Y, Xie L, et al: Stabilization of HIF-1α
alleviates osteoarthritis via enhancing mitophagy. Cell Death Dis.
11:4812020. View Article : Google Scholar
|
|
17
|
Wang FS, Kuo CW, Ko JY, Chen YS, Wang SY,
Ke HJ, Kuo PC, Lee CH, Wu JC, Lu WB, et al: Irisin mitigates
oxidative stress, chondrocyte dysfunction and osteoarthritis
development through regulating mitochondrial integrity and
autophagy. Antioxidants (Basel). 9:8102020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Taheem DK, Jell G and Gentleman E: Hypoxia
inducible factor-1α in osteochondral tissue engineering. Tissue Eng
Part B Rev. 26:105–115. 2020. View Article : Google Scholar :
|
|
19
|
Li M, Ning J, Wang J, Yan Q, Zhao K and
Jia X: SETD7 regulates chondrocyte differentiation and glycolysis
via the Hippo signaling pathway and HIF-1α. Int J Mol Med.
48:2102021. View Article : Google Scholar
|
|
20
|
Xiaoshi J, Maoquan L, Jiwei W, Jinqiu N
and Ke Z: SETD7 mediates the vascular invasion in articular
cartilage and chondrocytes apoptosis in osteoarthriis. FASEB J.
35:e212832021. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang H, Wang L, Cui J, Wang S, Han Y,
Shao H, Wang C, Hu Y, Li X, Zhou Q, et al: Maintaining hypoxia
environment of subchondral bone alleviates osteoarthritis
progression. Sci Adv. 9:eabo78682023. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lampert MA, Orogo AM, Najor RH, Hammerling
BC, Leon LJ, Wang BJ, Kim T, Sussman MA and Gustafsson ÅB:
BNIP3L/NIX and FUNDC1-mediated mitophagy is required for
mitochondrial network remodeling during cardiac progenitor cell
differentiation. Autophagy. 15:1182–1198. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Deng Z, Ou H, Ren F, Guan Y, Huan Y, Cai H
and Sun B: LncRNA SNHG14 promotes OGD/R-induced neuron injury by
inducing excessive mitophagy via miR-182-5p/BINP3 axis in HT22
mouse hippocampal neuronal cells. Biol Res. 53:382020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ashammakhi N, Darabi MA, Kehr NS, Erdem A,
Hu SK, Dokmeci MR, Nasr AS and Khademhosseini A: Advances in
controlled oxygen generating biomaterials for tissue engineering
and regenerative therapy. Biomacromolecules. 21:56–72. 2020.
View Article : Google Scholar
|
|
25
|
Montesdeoca CYC, Stocco TD, Marciano FR,
Webster TJ and Lobo AO: 3D bioprinting of smart oxygen-releasing
cartilage scaffolds. J Funct Biomater. 13:2522022. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Majmundar AJ, Wong WJ and Simon MC:
Hypoxia-inducible factors and the response to hypoxic stress. Mol
Cell. 40:294–309. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Geng B, Wang X, Park KH, Lee KE, Kim J,
Chen P, Zhou X, Tan T, Yang C, Zou X, et al: UCHL1 protects against
ischemic heart injury via activating HIF-1α signal pathway. Redox
Biol. 52:1022952022. View Article : Google Scholar
|
|
28
|
Goto Y, Zeng L, Yeom CJ, Zhu Y, Morinibu
A, Shinomiya K, Kobayashi M, Hirota K, Itasaka S, Yoshimura M, et
al: UCHL1 provides diagnostic and antimetastatic strategies due to
its deubiquitinating effect on HIF-1α. Nat Commun. 6:61532015.
View Article : Google Scholar
|
|
29
|
Truong VA, Lin YH, Nguyen NTK, Hsu MN,
Pham NN, Chang YH, Chang CW, Shen CC, Lee HS, Lai PL, et al:
Bi-directional gene activation and repression promote ASC
differentiation and enhance bone healing in osteoporotic rats. Mol
Ther. 30:92–104. 2022. View Article : Google Scholar :
|
|
30
|
Hsu MN, Yu FJ, Chang YH, Huang KL, Pham
NN, Truong VA, Lin MW, Kieu Nguyen NT, Hwang SM and Hu YC: CRISPR
interference-mediated noggin knockdown promotes BMP2-induced
osteogenesis and calvarial bone healing. Biomaterials.
252:1200942020. View Article : Google Scholar
|
|
31
|
Nguyen NTK, Chang YH, Truong VA, Hsu MN,
Pham NN, Chang CW, Wu YH, Chang YH, Li H and Hu YC: CRISPR
activation of long non-coding RNA DANCR promotes bone regeneration.
Biomaterials. 275:1209652021. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
33
|
Moon KC, Suh HS, Kim KB, Han SK, Young KW,
Lee JW and Kim MH: Potential of allogeneic adipose-derived stem
cell-hydrogel complex for treating diabetic foot ulcers. Diabetes.
68:837–846. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wiggers TG, Winters M, Van den Boom NA,
Haisma HJ and Moen MH: Autologous stem cell therapy in knee
osteoarthritis: A systematic review of randomised controlled
trials. Br J Sports Med. 55:1161–1169. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Seki A, Sakai Y, Komura T, Nasti A,
Yoshida K, Higashimoto M, Honda M, Usui S, Takamura M, Takamura T,
et al: Adipose tissue-derived stem cells as a regenerative therapy
for a mouse steatohepatitis-induced cirrhosis model. Hepatology.
58:1133–1142. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Qin Y, Ge G, Yang P, Wang L, Qiao Y, Pan
G, Yang H, Bai J, Cui W and Geng D: An update on adipose-derived
stem cells for regenerative medicine: Where challenge meets
opportunity. Adv Sci (Weinh). 10:e22073342013. View Article : Google Scholar
|
|
37
|
He G, Nie JJ, Liu X, Ding Z, Luo P, Liu Y,
Zhang BW, Wang R, Liu X, Hai Y and Chen DF: Zinc oxide
nanoparticles inhibit osteosarcoma metastasis by downregulating
β-catenin via HIF-1α/BNIP3/LC3B-mediated mitophagy pathway. Bioact
Mater. 19:690–702. 2022. View Article : Google Scholar :
|
|
38
|
Onishi M, Yamano K, Sato M, Matsuda N and
Okamoto K: Molecular mechanisms and physiological functions of
mitophagy. EMBO J. 40:e1047052021. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Stegen S, Laperre K, Eelen G, Rinaldi G,
Fraisl P, Torrekens S, Van Looveren R, Loopmans S, Bultynck G,
Vinckier S, et al: HIF-1α metabolically controls collagen synthesis
and modification in chondrocytes. Nature. 565:511–515. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Sonoda K, Bogahawatta S, Katayama A, Ujike
S, Kuroki S, Kitagawa N, Hirotsuru K, Suzuki N, Miyata T, Kawaguchi
SI and Tsujita T: Prolyl Hydroxylase domain protein inhibitor not
harboring a 2-Oxoglutarate scaffold protects against hypoxic
stress. ACS Pharmacol Transl Sci. 5:362–372. 2022. View Article : Google Scholar
|
|
41
|
Usui-Ouchi A, Aguilar E, Murinello S,
Prins M, Gantner ML, Wright PE, Berlow RB and Friedlander M: An
allosteric peptide inhibitor of HIF-1α regulates hypoxia-induced
retinal neovascularization. Proc Natl Acad Sci USA.
117:28297–28306. 2020. View Article : Google Scholar
|
|
42
|
Elvidge GP, Glenny L, Appelhoff RJ,
Ratcliffe PJ, Ragoussis J and Gleadle JM: Concordant regulation of
gene expression by hypoxia and 2-oxoglutarate-dependent dioxygenase
inhibition: The role of HIF-1alpha, HIF-2alpha, and other pathways.
J Biol Chem. 281:15215–15226. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Nguyen LK, Cavadas MA, Scholz CC,
Fitzpatrick SF, Bruning U, Cummins EP, Tambuwala MM, Manresa MC,
Kholodenko BN, Taylor CT and Cheong A: A dynamic model of the
hypoxia-inducible factor 1α (HIF-1α) network. J Cell Sci.
126:1454–1463. 2013.PubMed/NCBI
|
|
44
|
Rafique M, Wei T, Sun Q, Midgley AC, Huang
Z, Wang T, Shafiq M, Zhi D, Si J, Yan H, et al: The effect of
hypoxia-mimicking responses on improving the regeneration of
artificial vascular grafts. Biomaterials. 271:1207462021.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zhang X, Chen JY, Pei X, Li YH, Feng H, He
ZH, Xie WJ, Pei XB, Zhu Z, Wan QB and Wang J: One-Pot facile
encapsulation of dimethyloxallyl glycine by nanoscale zeolitic
imidazolate frameworks-8 for enhancing vascularized bone
regeneration. Adv Healthc Mater. 12:e22023172023. View Article : Google Scholar
|
|
46
|
Myllyharju J: Prolyl 4-hydroxylases, the
key enzymes of collagen biosynthesis. Matrix Biol. 22:15–24. 2003.
View Article : Google Scholar
|
|
47
|
Jackson P and Thompson RJ: The
demonstration of new human brain-specific proteins by
high-resolution two-dimensional polyacrylamide gel electrophoresis.
J Neurol Sci. 49:429–438. 1981. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Hu Y, Qi C, Shi J, Tan W, Adiljan
Abdurusul, Zhao Z, Xu Y, Wu H and Zhang Z: Podocyte-specific
deletion of ubiquitin carboxyl-terminal hydrolase L1 causes
podocyte injury by inducing endoplasmic reticulum stress. Cell Mol
Life Sci. 80:1062023. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Zhu Z, He Z, Tang T, Wang F, Chen H, Li B,
Chen G, Wang J, Tian W, Chen D, et al: Integrative bioinformatics
analysis revealed mitochondrial dysfunction-related genes
underlying intervertebral disc degeneration. Oxid Med Cell Longev.
2022:13724832022. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Lin L, Li S, Hu S, Yu W, Jiang B, Mao C,
Li G, Yang R, Miao X, Jin M, et al: UCHL1 impairs periodontal
ligament stem cell osteogenesis in periodontitis. J Dent Res.
102:61–71. 2023. View Article : Google Scholar
|
|
51
|
Cerqueira FM, von Stockum S, Giacomello M,
Goliand I, Kakimoto P, Marchesan E, De Stefani D, Kowaltowski AJ,
Ziviani E and Shirihai OS: A new target for an old DUB: UCH-L1
regulates mitofusin-2 levels, altering mitochondrial morphology,
function and calcium uptake. Redox Biol. 37:1016762020. View Article : Google Scholar :
|
|
52
|
Gao H, Antony R, Srinivasan R, Wu P, Wang
X and Li Y: UCHL1 regulates oxidative activity in skeletal muscle.
PLoS One. 15:e02417162020. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Bouron A, Aubry L, Loreth D, Fauvarque MO
and Meyer-Schwesinger C: Role of the deubiquitinating enzyme UCH-L1
in mitochondrial function. Front Cell Neurosci. 17:11499542023.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Komor AC, Badran AH and Liu DR:
CRISPR-Based technologies for the manipulation of eukaryotic
genomes. Cell. 168:20–36. 2017. View Article : Google Scholar :
|
|
55
|
Li C and Samulski RJ: Engineering
adeno-associated virus vectors for gene therapy. Nat Rev Genet.
21:255–272. 2020. View Article : Google Scholar : PubMed/NCBI
|