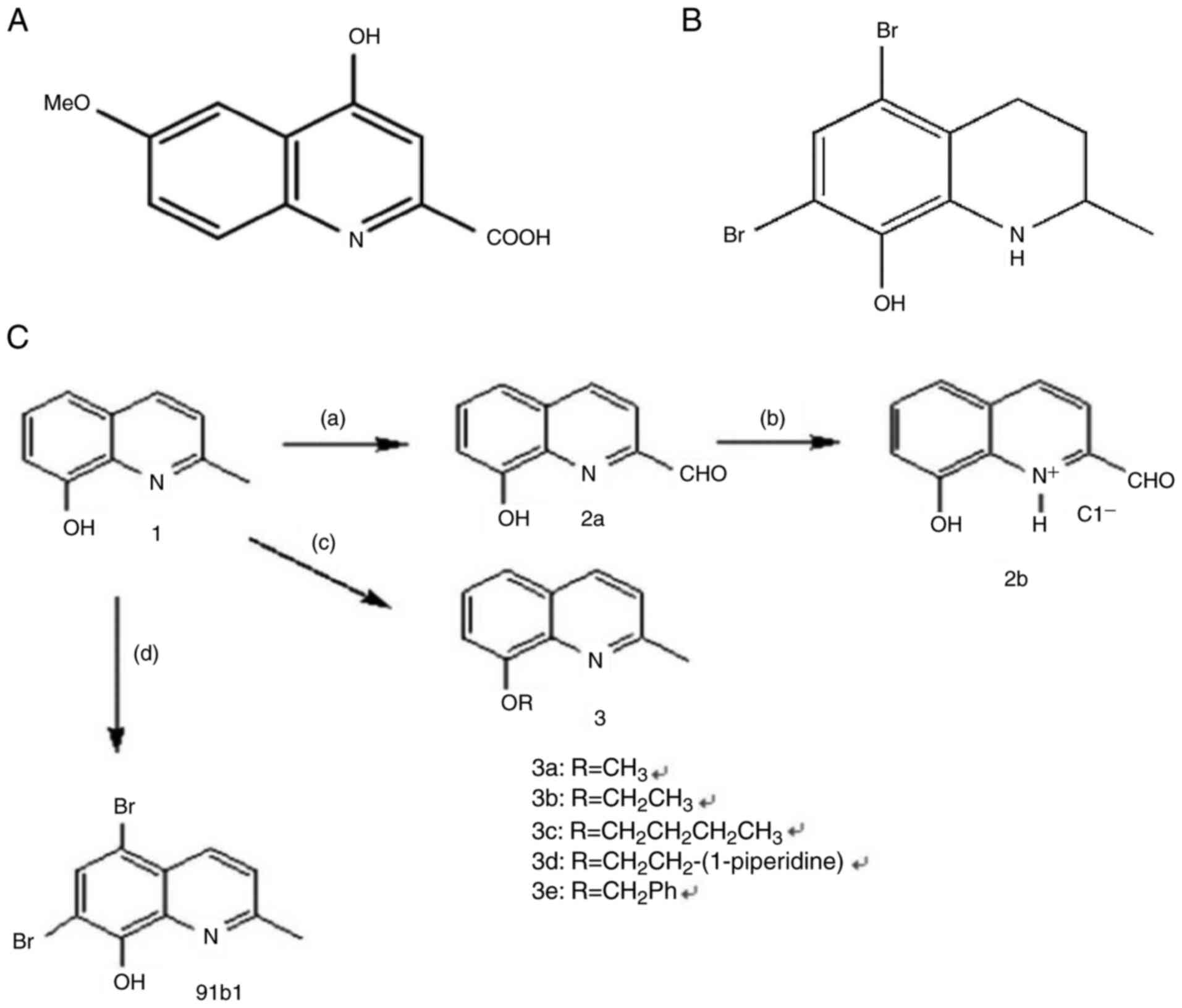
Introduction
Patients with esophageal squamous cell carcinoma
(ESCC) usually have poor prognosis and high mortality rate due to
the aggressiveness of the tumors (1). In 2020, it was responsible for
>604,100 new cases and ~544,076 mortalities worldwide (2). Currently, adjuvant chemotherapy
plays a central role in the treatment of esophageal cancer as one
third of patients are found with metastatic disease at the time of
diagnosis. Chemotherapy comprising cisplatin (CDDP) results in
response rates in esophageal carcinoma ≥40%. However, emergence of
chemoresistance to chemotherapeutic drugs and severe side effect
with toxicity lead to suboptimal survival rate (3,4),
underscoring the urgent need for novel and more effective
chemotherapeutic agents.
Chemokine ligand 5 (CCL5), part of the CC chemokine
family, is recognized by CCR1, CCR3, and CCR5 receptors (5). Predominantly expressed in T cells,
macrophages and cancer cells, CCL5 modulates the expression of the
surface receptors, the phenotypes of cancer cells, or the tumor
microenvironments reshaping to enhance cancer progression and
metastases (6-9). Although its involvement in liver
(10), prostate (11) and pancreatic cancer (12) is well documented, its role in
ESCC and its underlaying mechanisms remains to be elucidated.
According to the pan-cancer analysis and Immunohistochemical (IHC)
verification in ESCC cell lines and patients' samples in the
present study, CCL5 was usually highly expressed in cancer cells or
tumor tissue, suggesting its potential as a therapeutic target to
ESCC.
Natural products, particularly quinoline and its
derivatives, are well known for their diverse pharmacological
applications (13-18). For example,
4-hydroxy-6-methoxy-quinoline-2-carboxylic acid (Fig. 1A), which can be isolated from
Ephedra pachyclada ssp. Sinaica, has been traditionally used
to treating microbial, inflammation, allergy and cardiovascular
diseases (13). Another notable
quinoline derivative, quinine, from the bark of cinchona trees, is
renowned for its efficacy in malaria treatment. In addition, propyl
quinoline, which is one of the active ingredients in exudates of
bark of Galipea. longiflora (Rutaceae) trees, is
recognized for its effectiveness against leishmaniasis (14). Among different quinoline-based
compounds, 8-hydroxyquinoline derivatives, which can be extracted
from the root of a plant Commelina. Diffusa (15), stand out due to their broad range
of pharmacological efficacies (16-18). As prominent in vitro and
in vivo anticancer effects of 8-hydroxylquinoline
derivatives were also reported in our previous studies (19-23), a series of novel quinoline
derivatives were then synthesized by our group (23-26). A total of 27 compounds were
examined for anti-cancer activities against cancer cell lines of
hepatocellular carcinoma (Hep3B), lung carcinoma (A549), and
esophageal squamous cell carcinoma (HKESC-1, HKESC-4, and KYSE150).
Compound 91b1 (its original name in our published patent is
compound 2b) exhibited marked anti-cancer activity (25). Additionally, other compounds with
anti-cancer effect were also studied and some of them have been
reported. For example, quinoline compound 83b inhibits cancer
growth in esophageal squamous cell carcinoma by downregulating
COX-2 and PGE2 (22). The
cytotoxic potential 6 quinoline derivatives were examined in
vitro and in vivo (19). 2-formyl-8-hydroxy-qinolinium
chloride was prepared and its anti-cancer activity was evaluated
in vitro and in vivo (20).
Subsequent molecular docking analysis on the
synthesized 8-hydroxylquinoline derivatives was performed to
identity the potential protein targets involved in their anticancer
actions. Recently a novel quinoline derivative 91b1
(5,7-dibromo-1,2,3,4-tetrahydro-2-methylquinolin-8-ol) (Fig. 1B) was reported which suppressed
the tumor growth both in vitro and in vivo through
downregulating the expression of Lumican (27).
In the present study, the biological actions of the
most differentially expressed genes (DEGs) caused by 91b1 were
investigated based on the microarray analysis for gene expression
profiling. The protein expression levels of the most DEGs were also
examined on archival samples to underscore the therapeutic
relevance of 91b1 in the treatment of ESCC. The findings
highlighted that the 91b1-induced cytotoxicity was associated with
the downregulation of chemokine CCL5. Taken together with the
overexpression of CCL5 being a common event in the ESCC patient
samples, the overall results emphasized the potential of 91b1 as a
promising candidate for the treatment of ESCC.
Materials and methods
Bioinformatics analysis
Pan-cancer analysis of Ccl5 was conducted by
Tumor Immune Estimation Resource (TIMER) 2.0 (28). Ccl5 was entered into the
TIMER 2.0 web interface (http://timer.cistrome.org/), 'Gene_DE module' was
applied to study the differential expression between tumor and
adjacent normal tissues across all TCGA tumors. Distribution of
gene expression are displayed using box plots. The gene expression
profiling of Ccl5 influence was obtained from GEO datasets
GSE105042 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE105042).
Then two wild type macrophages and two Ccl5 knock out
macrophages was retrieved from a mouse dataset using the GPL21273
HiSeq X Ten platform (Illumina, Inc.). The original gene expression
profiles were analyzed to identify the upregulated or downregulated
DEGs in Ccl5 knock out samples, respectively. The criteria
for a DEG were |log2FC|>1 and adjusted P-value <0.05. Gene
Ontology (GO) enrichment analysis and Kyoto Encyclopedia or Genes
and Genomes (KEGG) pathway analysis were performed using the R
package cluster Profiler (v. 4.0.5; https://www.R-project.org) with identified DEGs
(29). GO enrichment analysis
and KEGG pathway analysis were performed with the thresholds of a
P-value <0.05.
The Search Tool for the Retrieval of Interacting
Genes/Proteins (STRING) database (https://string-db.org/) was used to construct the PPI
network (30). DEGs were mapped
to a STRING list to perform a search for multiple proteins and
obtain a PPI network with interaction scores >0.4. Cytoscape (v.
3.9.0) was used to visualize the results from the PPI network and
perform module analysis (31).
Module analysis was performed using the molecular complex detection
(MCODE) plugin on the Cytoscape (v. 3.9.0) platform. The parameters
set to identify enriched functional modules were as follows: Degree
Cutoff=2, Node Score Cutoff=0.2, K-Core=2 and Maximum. Depth=100.
Modules with the MCODE score ≥4 were identified as significant
modules.
Preparation of Compound 91b1
(5,7-dibromo-1,2,3,4-tetrahydro-2-methylquinolin-8-ol)
Compound 91b1 (Fig.
1B) was prepared by addition of Br2 into
commercially available 8-hydroxy-2-methylquinolin-8-ol
(MilliporeSigma) in MeOH, followed by asymmetric hydrogenation
under optimal reaction condition as previously described (23). The structure of 91b1 has been
physically characterized using 1H- and 13C-NMR and liquid
chromatography mass spectrometry (LC-MS) as previously reported
(22). Dimethyl sulfoxide (DMSO)
was used to dissolve 91b1 for the in vitro biological assays
as described below.
Cell lines
Esophageal squamous cell carcinoma (ESCC) cell lines
of Japanese origin KYSE150, KYSE450, KYSE30 and KYSE510 (32) were purchased from
Leibniz-Institut DSMZ-Deutsche Sammlung von Mikroorganismen und
Zellkulturen GmbH. ESCC cell lines of Hong Kong origin SLMT-1
(33) and HKESC-4 (34) were kindly provided by Professor
Gopesh Srivastava in the Department of Pathology, The University of
Hong Kong, China. Non-neoplastic esophageal epithelial cell line
NE-3 (35) (immortalized by the
induction of genes E6 E7 of human papillomavirus type 18) was
kindly provided by Professor George S.W. Tsao in the Department of
Anatomy, The University of Hong Kong, China. Non-neoplastic
embryonic cell line 293 (36)
was purchased from the American Type Culture Collection (ATCC). The
culture medium for KYSE30, KYSE150, KYSE450, and KYSE510 was 45%
RPMI with 45% F-12 and 10% FBS; that for SLMT-1, HKESC4, and 293
was 90% MEMα with 10% FBS; and that for NE3 was KSFM with
complementary supplements. All the cells were cultured in media
supplemented with 100 units/ml penicillin G and 100 μg/ml
streptomycin and all cells were maintained in a humidified
atmosphere of 95% air and 5% CO2 at 37°C. The cultures
were passaged at preconfluent densities of ~80% using a solution of
0.25% trypsin (Invitrogen; Thermo Fisher Scientific, Inc.). Cells
were washed briefly with phosphate-buffered saline (PBS), treated
with 0.25% trypsin, and harvested by centrifugation (300 × g for 5
min) at room temperature for subculturing.
ESCC patient specimens
Paraffin-embedded specimens containing 26 cancer
tissues and 15 non-neoplastic tissues were collected after
esophagectomy, with the consent of patients, at the Department of
Surgery, Queen Mary Hospital, Hong Kong between January 1990 and
December 2001 and the ethics approval for working on the specimens
was obtained from the Hong Kong Polytechnic University (approval
no. HSEARS20171213007). All the specimens were collected from
patients who had received no prior treatment directed to the
primary ESCC. The age of the patients ranged from 45 to 77 years
old, with a median of 65, comprising 23 males and 3 females. Oral
informed consent was obtained in from all patients. The authors
confirm that all methods were carried out in accordance with
relevant guidelines and regulations. The clinical and histological
information of ESCC patient specimens were reported by a
pathologist (AKY Lam).
[3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium]
(MTS) cytotoxicity assay
The cytotoxic effect of 91b1 and CDDP on the ESCC
cell lines and non-neoplastic cell lines was examined by
CellTiter-96-AQueous One Solution Cell Proliferation Assay (Promega
Corporation) as previously described and expressed as the
MTS50 values (27).
Each assay was performed in triplicate.
cDNA microarray analysis
Total RNA was extracted from 2×108 cells
of KYSE150 treated with 91b1 at 9.5 μg/ml (based on the
MTS50 value of 91b1 on KYSE150 with the signal of MTS
cytotoxicity decreased by 50%) and DMSO (0.05%; MilliporeSigma) for
48 h using RNeasy Mini kit (Qiagen, Inc.). The cDNA microarray
analysis and the associated quality control using Human Genome U133
Plus 2.0 arrays (Affymetrix; Thermo Fisher Scientific, Inc.) were
performed at the Centre for Genomic Sciences of the University of
Hong Kong according to the manufacturer's protocol as previously
described (4). The signals of
the differentially expressed genes in 91b1-treated KYSE150 were
compared with the DMSO-treated KYSE150 control.
Reverse transcription-quantitative (RT-q)
PCR
The total RNA of non-neoplastic cells, ESCC parental
cells and 91b1-treated ESCC cells was extracted using RNeasy Mini
kit (Qiagen, Inc.) as previously described (4). cDNA was synthesized from total RNA
using the GoScript Reverse Transcription System (Promega
Corporation) according to manufacturer's instruction. The
expression level of Ccl5 in the tested cells was determined
by qPCR analysis using Go Taq qPCR Master Mix (Promega Corporation)
and Thermo Scientific PikoReal Real-Time PCR System (Thermo Fisher
Scientific, Inc.) according to manufacturer's protocol. The
synthesized cDNA (4 μl) was mixed with 10 μl of 2X
qPCR Mastermix (Promega Corporation), 2 μl of 2 μM
forward and reverse primers of either target gene or reference gene
(β-Actin was applied as reference gene), and 12 μl of RNase
free water to get a total volume of 20 μl in a PCR tube. All
20 μl of sample mixtures were transferred into the wells of
PikoReal 96-well strips (n=3). qPCR reactions were carried out by
PikoReal Real-Time PCR System (Thermo Fisher Scientific, Inc.). The
thermocycling conditions were: Polymerase activation at 95°C for 2
min, then followed by 40 cycles of denaturation at 95°C for 15 sec,
annealing and primer extension at 60°C for 1 min, then melt curve
data were identified by gradually increasing temperature from
60-95°C until the fluorescent signal dropped to zero. Cq (cycle of
quantification) of each sample was determined and recorded by the
program PikoReal Software 2.0 (Thermo Fisher Scientific, Inc.).
cDNA (~2 μg) produced by reverse transcription from the RNA
was amplified using a specific Ccl5 and β-Actin gene primer
pairs (IDT). Primers for Ccl5 were 5′-CGTGCCCACATCAAGGAG-3′
(forward) and 5′-GGACAAGAGCAAGCAGAAA-3′ (reverse). Primers for
β-Actin were 5′-ACCTTCTACAATGAGCTGCG-3′ (forward) and
5′-CCTGGATAGCAACGTACATGG-3′ (reverse). Relative Ccl5
expression was determined by comparing with vehicle DMSO (0.05%)
control, after being normalized with expression of β-Actin. For all
the qPCR reactions, the relative expression of target genes in
different samples were calculated and compared by using the
2−ΔΔCq method. The expression level of target genes was
normalized by the reference gene β-actin. Each independent
experiments is conducted three times.
The calculation of 2−ΔΔCq method was
(37):
ΔCq of target gene=Cq of target gene-Cq of reference
gene
ΔΔCq of target gene=ΔCq of the target gene in
treated group-ΔCq of the target gene in control group
Therefore, the fold change of gene expression
level=2−(ΔΔCq of target gene)
The expression level was regarded as overexpression
if the fold change of gene expression level ratio
was larger than 1.2; a ratio between 0.8 and 1.2 was considered as
no significant change, while a ratio smaller than 0.8 was
considered as under expression of the target gene (
38).
IHC staining
Paraffin-embedded cell-line blocks of KYSE150,
KYSE510, KYSE450, KYSE520 KYSE30, HKESC-3 HKESC-4, SLMT-1, NE-3,
DMSO-treated (0.05%, 48 h) and 91b1-treated (6.5, 9.5 and 21
μg/ml 91b1) for 48 h KYSE150 cells were prepared from
~5×106 cells of each respective cell line with
formalin-fixation (37% formalin and 15% methanol at room
temperature for 48 h). Flowing fixation, tissues were rinsed under
running water for 3 h. Then the tissues underwent a grade
dehydration process starting with 50% ethanol for 2 h, followed by
70% ethanol for 2 h, 80% ethanol overnight, 90 and 95% ethanol for
2 h each, and concluding twice in 100% ethanol for 2 h each time.
Subsequently, the samples were placed in a mixture of
ethanol:xylene (1:1) for 30 min and cleared in xylene for 30 min
twice before being prepared for embedding in paraffin wax at 60°C
twice for 1 h each time. An automated embedding system was employed
to encapsulate the tissues in paraffin. Dewaxed paraffin sections
(8 μm) of cell-line blocks and samples from patients with
ESCC were immunostained using the streptavidin-biotin-peroxidase
complex method. As pretreatment, microwave-based antigen retrieval
was be performed in 10 mM citrate buffer (pH 6.0). CCL5 mouse
monoclonal antibody (1 mg/ml; Abnova) was applied at dilution of
1:100 for overnight incubation at 4°C. Images of stained samples
were captured under an inverted optical microscope (CKX41; Olympus
Corporation) at magnification, ×400 and four fields of images of
stained sections were examined for the percentage of positively
stained cells in cytoplasm by ImageJ (v 1.54; National Institutes
of Health). Immunostaining results of ESCC cell line were compared
with that of non-tumor cell line NE-3. Immunohistochemical staining
images of stained sections were examined and graded according to
the percentage of positively stained neoplastic cells as previously
described (4).
Enzyme-linked immuno-sorbent assay
(ELISA)
Protein expression levels of CCL5 in cells were
measured using RANTES (CCL5) Human SimpleStep ELISA kit (Abcam;
cat. no. ab174446) according to the manufacturer's instructions.
Cells receiving no treatment, DMSO-treatment (0.05%) or
91b1-treatment in the concentration of 6.5, 9.5 and 21 μg/ml
(with reference to the MTS50 value) were collected by
cell scrapper after 48 h. Total protein concentration of each
sample was determined using a Micro BCA Protein Assay kit (Thermo
Fisher Scientific, Inc.) according to manufacturer's manual for
normalization.
Transwell Matrigel invasion assay
The invasion ability of ESCC cells was evaluated
using chambers with Matrigel-coated membrane (8-μm pore
size; BD Biocoat; Corning, Inc.) in 24-well plate. The lower
chamber was filled with RPMI 1640 medium (Gibco; Thermo Fisher
Scientific, Inc.) containing 10% fetal bovine serum (FBS, Biosera)
with recombinant human CCL5 protein (rhCCL5; Abnova) at
concentration of 0, 50, 100 and 500 ng/ml. KYSE30 cells were
cultured in 200 μl serum free RPMI 1640 medium in the upper
chamber at a density of 2.5×105 cells/ml. The same
number of cells were cultured on an uncoated membrane (8-μm
pore size) chamber as control. After 24 h, the uninvaded cells on
the upper chamber were scraped off with a cotton swab. The
transmembrane cells which migrated to the opposite side of the
membrane were fixed in 100% methanol for 10 min and stained with
0.5% crystal violet solution (0.5 g crystal violet in 75 ml
methanol and 25 ml ddH2O) for 30 sec at room temperature
after washing twice with phosphate buffered saline (PBS). The
transmembrane cells were counted under microscope in five random
fields at magnification of ×100. The percentage of invasion was
calculated as follows:
Wound-healing assay
A wound-healing assay was performed to evaluate cell
migration and growth. KYSE150 cells (~1×106) were
cultured in a 6-well plate at 37°C with 5% CO2 overnight
to let the cells adhere and grow to 70-80% confluent monolayers. On
the second day, the monolayer was gently scratched with a new 1 ml
pipette tip across the center of the well to generate a wound area
without changing the medium. After scratching, the well was gently
washed twice with warm PBS buffer to remove detached cells, and the
well was replenished with serum-starved medium or different
concentrations of compound 91b. The cells were incubated at 37°C
with 5% CO2 again and observed by microscope (Olympus
CKX41; Olympus Corporation) at different time points (0, 12, and 24
h after scratching) for image capture. The number of cells invaded
across the wound area was counted by ImageJ (v 1.54, National
Institutes of Health).
Statistical analysis
The comparative ΔΔCq method was applied for relative
quantification in qPCR analysis. Statistical significance of the
differences among groups in MTS cytotoxicity assay, qPCR analysis,
ELISA, Transwell Matrigel invasion assay data were compared by
two-tailed t test or one-way analysis of variance (ANOVA) followed
by Dunnett's Correction using GraphPad Prism 7 (Dotmatics).
P<0.05 was considered to indicate a statistically significant
difference.
Results
Bioinformatics analysis of the function
of Ccl5 in cancers
According to the pan-cancer analysis in Fig. 2A, Ccl5 was differentially
expressed in several types of cancer, in which Ccl5 was
notably high expressed in breast invasive carcinoma, esophageal
carcinoma, glioblastoma multiforme, head and neck squamous cell
carcinoma, kidney renal clear cell carcinoma, kidney renal
papillary cell carcinoma and metastasis skin cutaneous melanoma,
suggesting the possible roles of Ccl5 in tumorigenesis and
tumor metastasis.
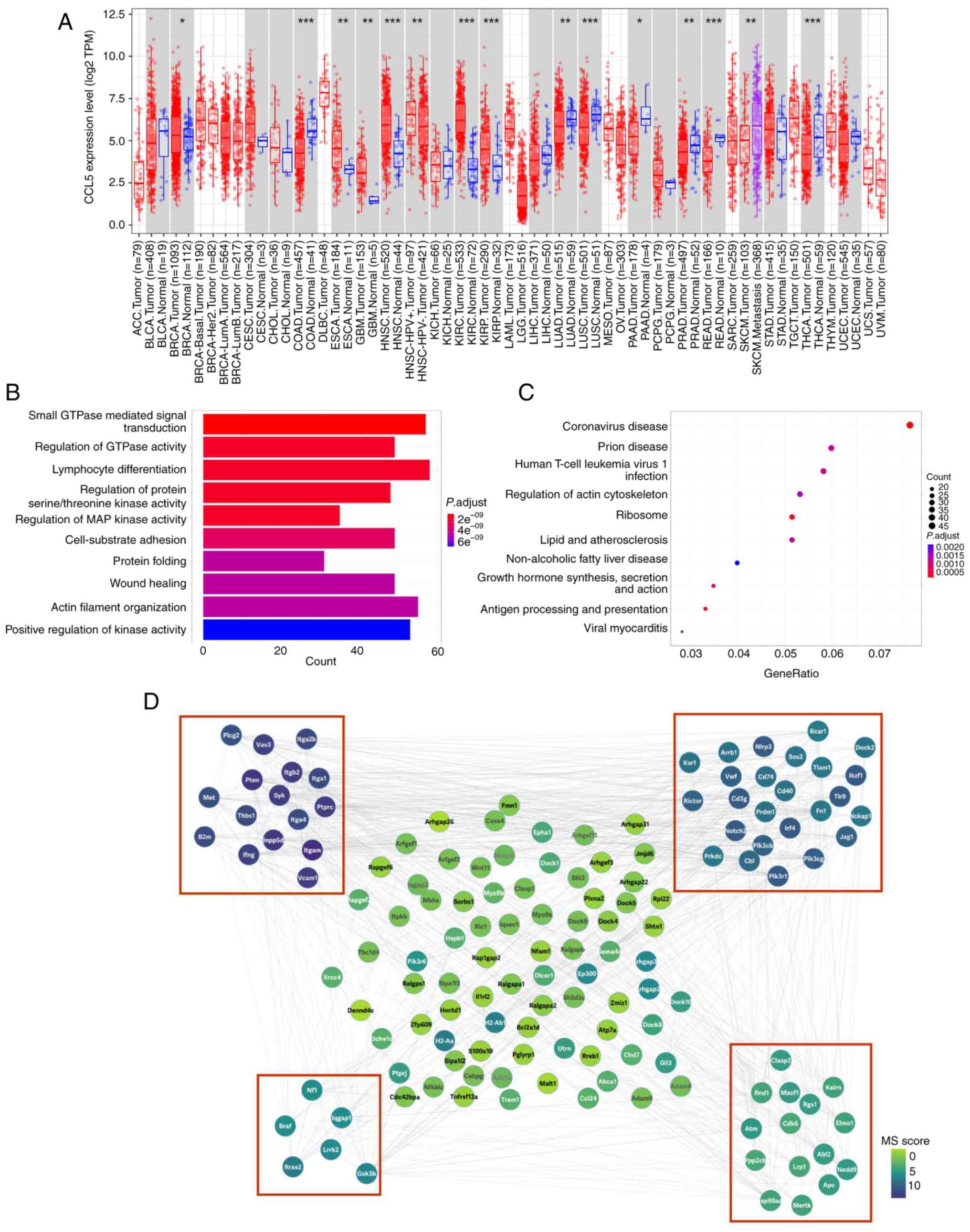 | Figure 2CCL5 plays a critical role in several
types of cancers, and its related functions mainly enriched in
metastasis and immunology. (A) Pan-cancer analysis of CCL5 by TIMER
2.0, upregulated or downregulated genes in the tumors were compared
with normal tissues for each cancer type, as displayed in gray
columns when normal data were available; red dots indicate tumor
tissue, blue dots indicate normal tissue, purple dots indicate
metastasis tumor tissue, *P<0.05; **P<0.01;
***P<0.001; (B) GO enrichment analysis; (C) KEGG
pathway analysis; (D) PPI analysis. PPI network for all the
overlapping DEGs was constructed and followed by module analysis
using the MCODE plugin on the Cytoscape (v. 3.9.0) platform.
Modules with a red border are significant modules. The size of
circles reflects the degree of connectivity. CCL5, chemokine (C-C
motif) ligand 5; PPI, protein-protein interaction; GO, Gene
Ontology; KEGG, Kyoto Encyclopedia or Genes and Genomes; DEGs,
differentially expressed genes. |
GO enrichment analysis results are in Fig. 2B. The most enriched GO molecular
functions were identified as 'small GTPase mediated signal
transduction', 'regulation of GTPase activity', 'lymphocyte
differentiation', 'regulation of protein serine/threonine kinase
activity', 'regulation of MAP kinase activity', 'cell-substrate
adhesion', 'protein folding', 'wound healing', 'actin filament
organization' and 'positive regulation of kinase activity'. KEGG
pathway analysis results are in Fig.
2C, indicating that the DEGs were enriched in nine pathways,
'coronavirus disease', 'prion disease', 'human T-cell leukemia
virus 1 infection', 'regulation of actin cytoskeleton', 'ribosome',
'lipid and atheroselerosis', 'non-alcoholic fatty liver disease',
'growth hormone synthesis, secretion and action', 'antigen
processing and presentation' and 'viral myocarditis'.
A total of 1,579 interactions were obtained with
interaction scores >0.4 using the STRING database. The PPI
network was then constructed and presented with the Cytoscape (v.
3.9.0) platform (Fig. 2D). The
top 10 hub genes included Itgb2, Pik3cg, Pik3r1, Cbl, Pten, Syk,
Pik3cb, Ptprc, Met and Inpp5d. A total of four modules
were obtained through MCODE analysis. The function of key proteins,
such as PIK3, PTEN, and ATM were associated with metastases, IFNG,
CD74, CD40, and CD3g were associated with immune response.
CCL5 is usually high expressed in ESCC
cell lines and patient samples
Immunostaining was employed to detect the
cytoplasmic protein expression level of CCL5 in eight ESCC cell
lines and non-tumor esophageal cell line NE-3. Upregulation of CCL5
was detected in 7/8 (87.5%) ESCC cell lines (KYSE510, KYSE450,
KYSE520, KYSE150, HKESC-3, HKESC-4 and SLMT-1) when compared with
NE-3 (Fig. 3A and B). In
addition, high expression level of CCL5 was also observed in 20/26
(76.9%) of ESCC tissues and only 6/15 (40%) of non-neoplastic
esophageal tissues. ESCC expressing CCL5 protein in high level was
more frequently observed than non-neoplastic esophageal mucosa
(P=0.018). Immunohistochemical staining showed high expression and
low expression of CCL5 in ESCC (Fig.
3C).
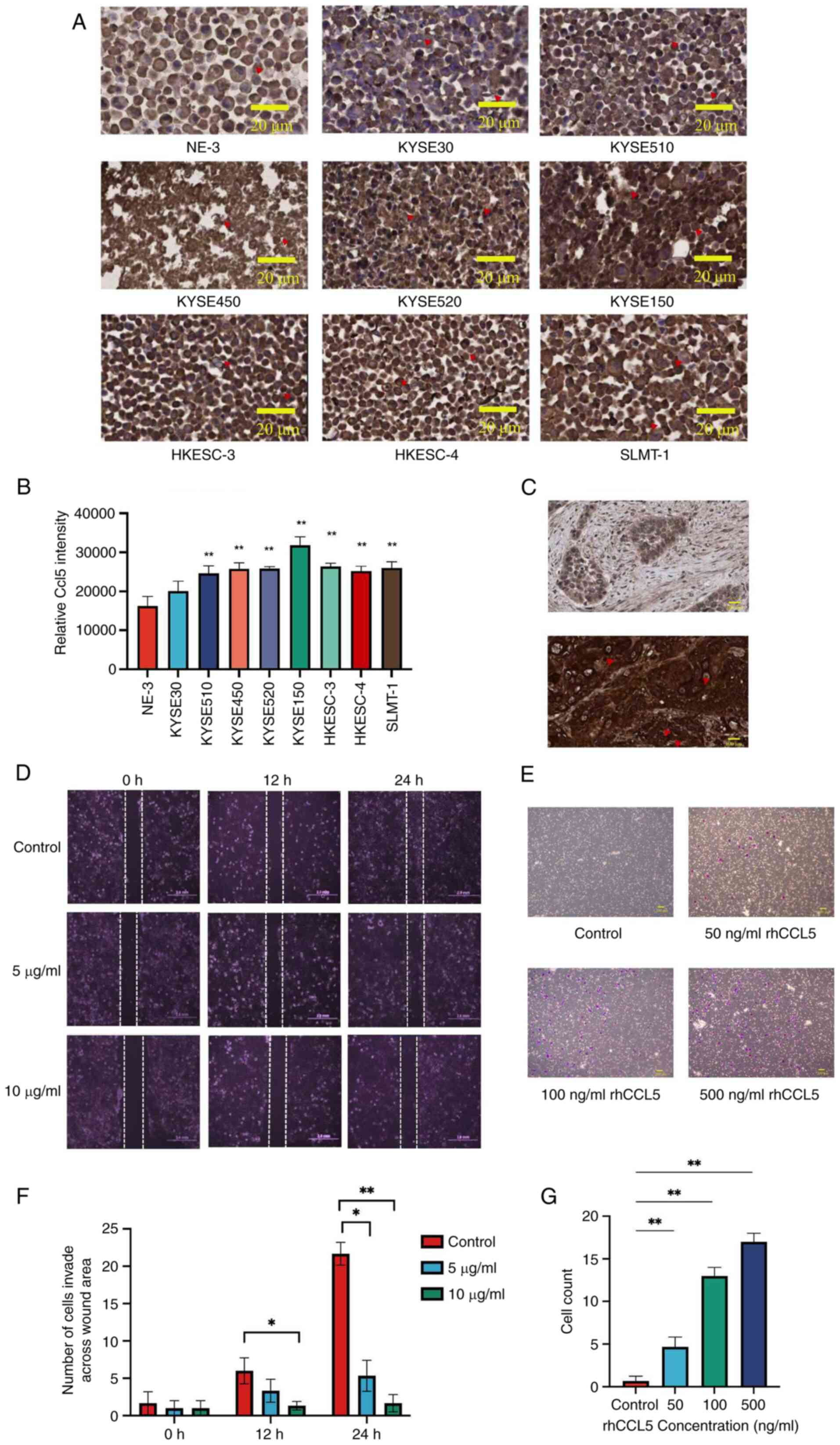 | Figure 3CCL5 is usually highly expressed in
ESCC cell lines and patient samples, enhanced cancer cell invasion
which can be suppressed by compound 91b1. (A) Immunohistochemical
staining of CCL5 in eight ESCC cell lines, non-tumor cell line NE-3
with (B) quantitative analysis, Original magnification, ×400; scale
bar, 200 μm. (C) Representative images of
immunohistochemical staining for CCL5 in ESCC specimens graded as
low expression (upper photo) and high expression (lower photo),
Original magnification, ×400; scale bar, 200 μm. Red
triangles represent IHC staining of CCL5 protein in cancer cells or
specimens. (D) Images of wound healing progress of KYSE150 cells
under 5 μg/ml or 10 μg/ml compound 91b1 at 0, 12 and
24 h respectively with average cell count invaded across the wound
area, Original magnification, ×100; scale bar, 2 mm (F); (E) Cell
Invasion assay using Transwell Matrigel chamber with 0 ng/ml rhCCL5
(control), 50 ng/ml rhCCL5, 100 ng/ml rhCCL5, and 500 ng/ml rhCCL5.
Transmembrane cells were stained by crystal violet, Original
magnification, ×200; scale bar, 200 μm; (G) Average invaded
cell count of KYSE150 co-cultured with different concentrations of
rhCCL5 (0, 50, 100, 500 ng/ml). The invaded cells were counted
under a microscope in four random fields. *P<0.05;
**P<0.01. CCL5, chemokine (C-C motif) ligand 5; ESCC,
esophageal squamous cell carcinoma. |
The effect of CCL5 on the invasion ability of KYSE30
was examined by Transwell Matrigel invasion assay. As shown in
Fig. 3E, no transmembrane cells
were detected in the invasion assay after 24 h without the addition
of recombinant human CCL5 protein (rhCCL5). Increasing numbers of
transmembrane cells were detected with increasing rhCCL5
concentration. The percentage of invasion increased with the
concentration of rhCCL5 (Fig.
3G), suggesting that CCL5 enhanced the invasion ability of ESCC
cells. Moreover, to evaluate the cell migration and growth
properties affected by compound 91b1, wound healing analysis was
performed on KYSE510 cell line. A wound gap was created by
scratching, and healing progress was captured at different time
points. Cancer cells were treated with low (5 μg/ml) or high
(10 μg/ml) dose of compound 91b1 (Fig. 3D and F). After 12-h, or 24-h
incubation, fewer cells of compound 91b1 treatment groups migrated
into the scratched area than the control groups, indicating the
reduction of cell migration of the cancer cells following 91b1
treatment.
Compound 91b1 downregulated the
expression of Ccl5 in ESCC cells
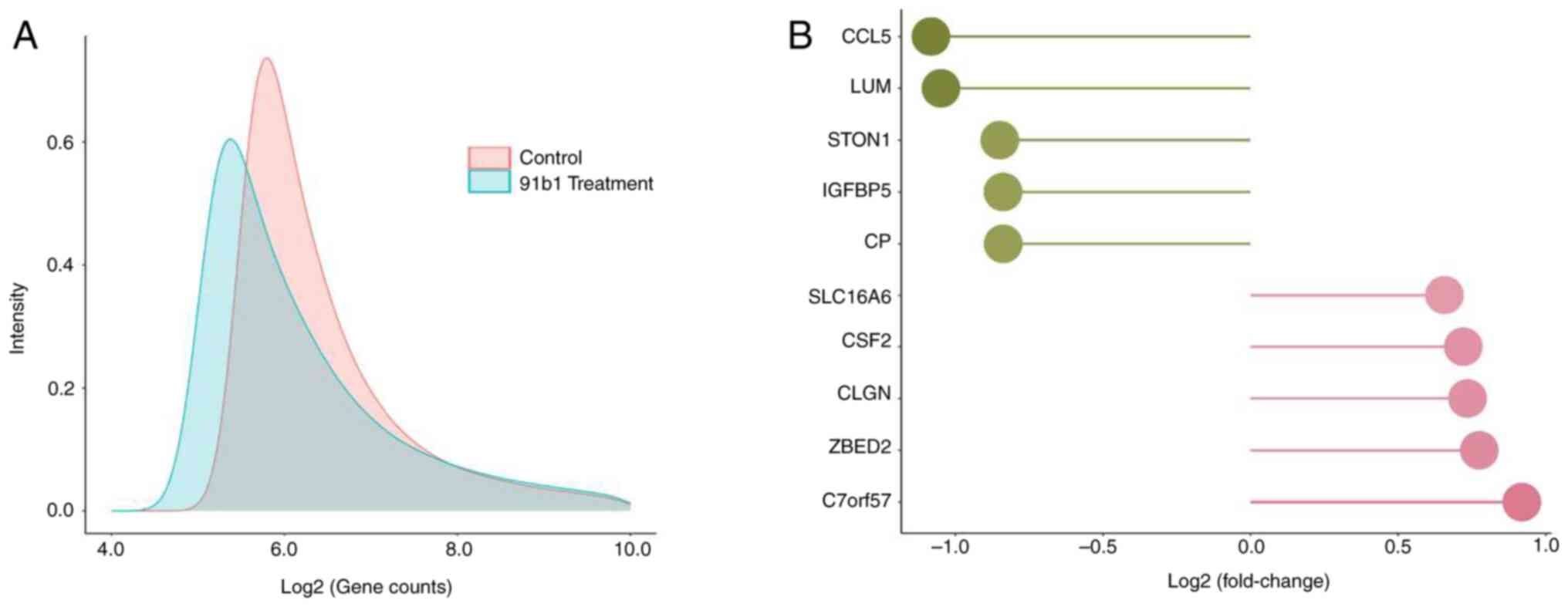
cDNA microarray analysis was performed to study the
changes of gene expression caused by compound 91b1 in cancer cells.
Density plots (Fig. 4A) showed
the different expression profile of KYSE150 cells treated with
compound 91b1 compared with a blank control. The fold changes of
normalized signal intensity of each gene obtained from the
microarray analysis were evaluated. The five most downregulated
genes were Ccl5, Lumican, Ston1, Igfb5 and Cp while
the five most upregulated genes were C7orf57, Zbed2, Clgn,
Csf2 and Slc16a6 (Table
I and Fig. 4B). Ccl5
was found to be downregulated in 91b1-treated KYSE150 cells with
the highest fold change (-2.12 times), thus 91b1 was a promising
anti-cancer compound to inhibit metastasis by targeting
Ccl5.
 | Table IList of the five most down- and
upregulated genes in KYSE150 cells treated with 91b1 (9.5
μg/ml) for 48 h compared with the vehicle control. |
Table I
List of the five most down- and
upregulated genes in KYSE150 cells treated with 91b1 (9.5
μg/ml) for 48 h compared with the vehicle control.
A, Genes
downregulated in 91b1-treated KYSE150 cells
|
|---|
| Probe set ID | Gene name | Fold-change |
|---|
| 1405_i_at | Ccl5,
chemokine (C-C motif) ligand 5 | -2.12 |
| 201744_s_at | LUM,
Lumican | -2.07 |
| 213413_at | STON1,
Stonin 1 | -1.80 |
| 211959_at | Igfbp5,
insulin-like growth factor binding protein 5 | -1.79 |
| 1558034_s_at | Cp,
ceruloplasmin | -1.79 |
|
| B, Genes
upregulated in 91b1-treated KYSE150cells |
|
| 1557636_a_at | C7orf57,
chromosome 7 open reading frame 57 | 1.89 |
| 219836_at | Zbed2, zinc
finger, BED-type containing 2 | 1.71 |
| 205830_at | Clgn,
calmegin | 1.66 |
| 210229_s_at | Csf2, colony
stimulating factor 2 | 1.65 |
| 230748_at | Slc16a6,
solute carrier family 16, member 6 | 1.58 |
The effect of 91b1 on mRNA expression and protein
expression of CCL5 in ESCC cells were examined by qPCR analysis and
ELISA. All the tested ESCC cell lines showed the reduction in CCL5
mRNA (Fig. 5A-D) and protein
expression (Fig. 5E-H) after the
treatment with 91b1, which is in agreement with the microarray
results using KYSE150. In general, the suppressing effect of 91b1
on CCL5 expression was dose dependent. The results thus confirmed
that CCL5 is one of the affected downstream candidates for the
cytotoxicity induced by 91b1. The suppression effect of 91b1 on
CCL5 protein expression was also demonstrated by IHC staining with
CCL5 antibody on KYSE150 cells. The number of cells with
cytoplasmic staining signals revealed the relative protein
expression level of CCL5. Dark-brownish stained cells in the
vehicle-control revealed the high CCL5 protein expression (Fig. 5I) with quantitative analysis
(Fig. 5J). Fewer CCL5 positive
cells were evident after being treated with increasing
concentration of 91b1 for 48 h, suggesting that the protein
expression of CCL5 was reduced with 91b1 treatment in a
dose-dependent manner.
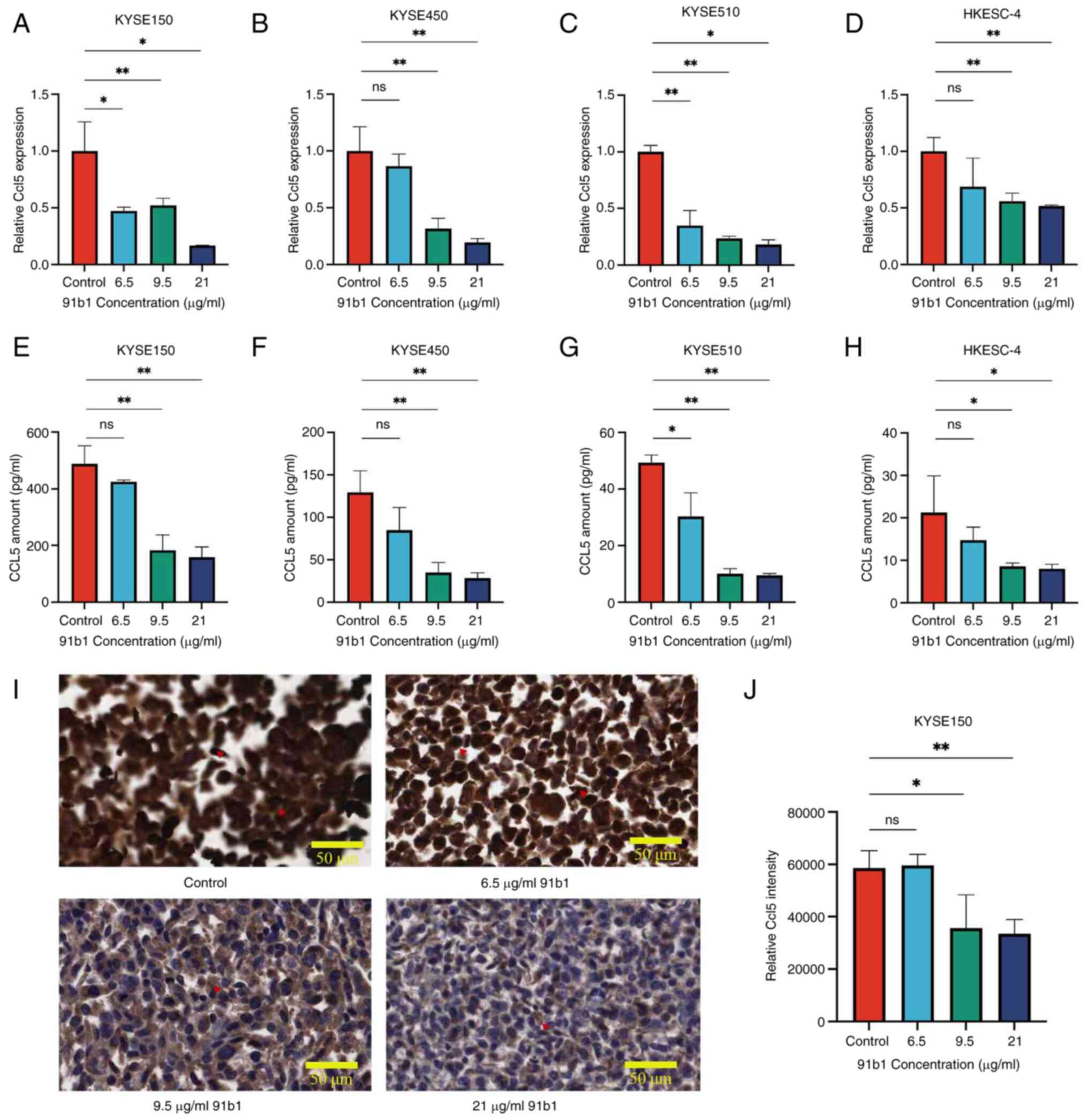 | Figure 5Compound 91b1 downregulates the
expression of CCL5 in ESCC cell lines. CCL5 mRNA expression
levels in ESCC cell lines (A) KYSE150, (B) KYSE450, (C) KYSE510 and
(D) HKESC-4 after 48-h treatment of 91b1 in different
concentrations. Each test was performed in triplicate and relative
CCL5 expression levels were determined by comparing with
cells treated with vehicle DMSO (0.05%) following normalized with
the expression of β-actin. CCL5 protein expression of (E) KYSE150,
(F) KYSE450, (G) KYSE510 and (H) HKESC-4 after 48-h treatment of
91b1 in different concentrations vs. vehicle. Each assay was
performed in triplicate. (I) Immunohistochemical staining of CCL5
for KYSE150 treated with vehicle control (DMSO), 6.5, 9.5 and 21
μg/ml 91b1 for 48 h; original magnification, ×400; (J) The
staining signals were quantitatively analyzed by ImageJ (National
Institutes of Health). *P<0.05;
**P<0.01; ns, not significant. CCL5, chemokine (C-C
motif) ligand 5; ESCC, esophageal squamous cell carcinoma. |
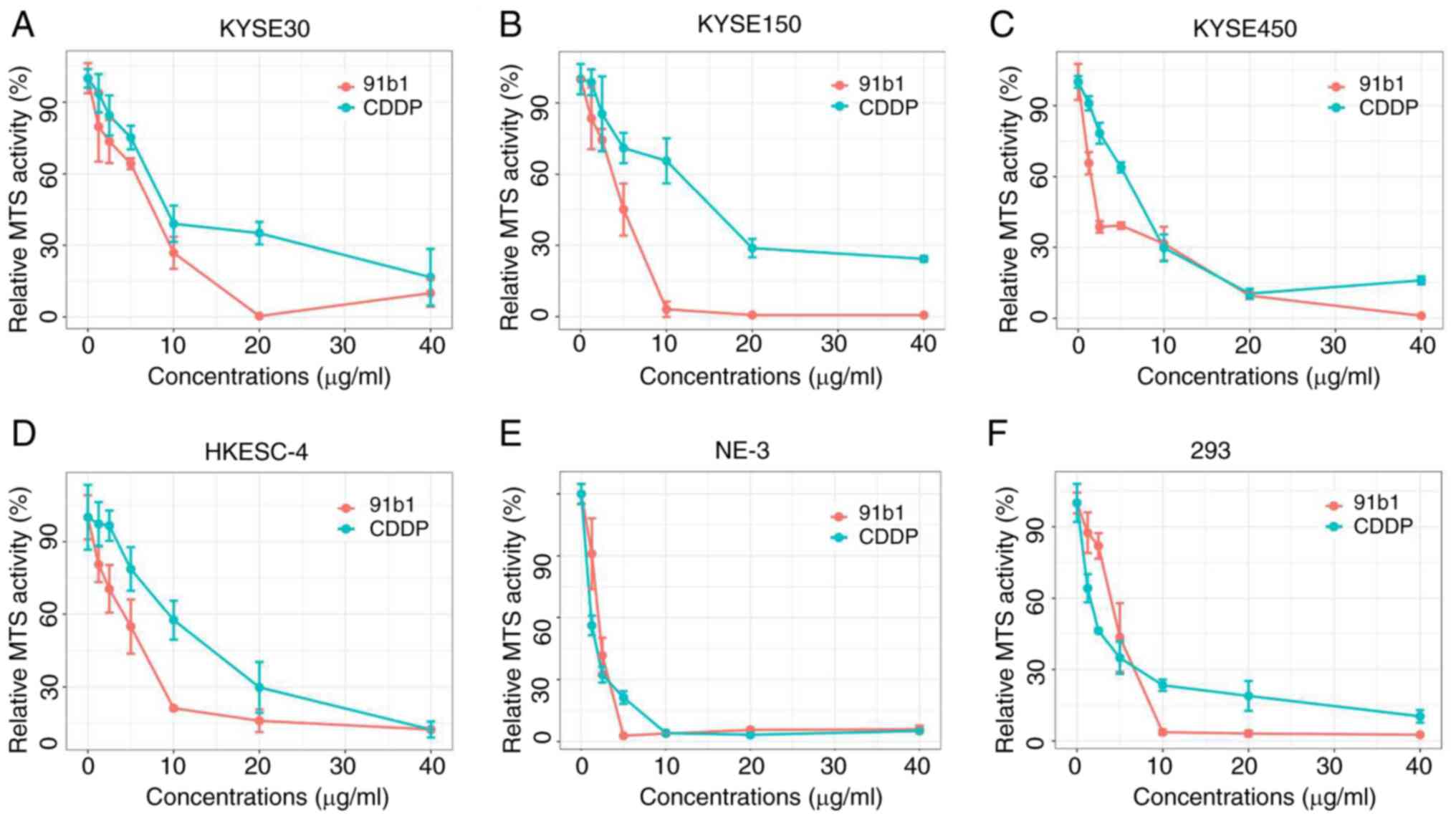
In vitro cytotoxicity assay of compound
91b1
The anticancer effect of 91b1 on the four ESCC cell
lines (KYSE30, KYSE150, KYSE450 and HKESC-4) and two non-tumor cell
lines (NE-3 and 293) was evaluated using MTS cytotoxicity assay
using CDDP as the positive control (Fig. 6). The MTS50 values
(concentration of tested compounds that had 50% inhibition on MTS
activity; Table II). 91b1
showed stronger cytotoxic effect in ESCC cell lines and lesser
cytotoxic effect in non-neoplastic cells (NE-3 and 293) than
CDDP.
 | Table IIMTS50 (μg/ml) of
91b1 and CDDP for four ESCC cell lines and two non-tumor cell
lines. Results were expressed with mean ± SD from triplicate.
experiments. |
Table II
MTS50 (μg/ml) of
91b1 and CDDP for four ESCC cell lines and two non-tumor cell
lines. Results were expressed with mean ± SD from triplicate.
experiments.
| Cell lines | MTS50
|
|---|
| 91b1
(μg/ml) | CDDP
(μg/ml) |
|---|
| KYSE30 | 6.50±0.41 | 8.00±1.08 |
| KYSE150 | 4.55±0.77 | 13.16±2.54 |
| KYSE450 | 1.80±0.23 | 6.69±0.34 |
| HKESC-4 | 4.75±1.83 | 11.88±1.52 |
| NE-3 | 1.94±0.29 | 1.18±0.21 |
| 293 | 4.55±0.87 | 2.19±0.25 |
Discussion
Quinoline derivatives have frequently demonstrated
anticancer properties in previous studies (19,20,39). In the current study, a novel
quinoline derivative 91b1 was prepared from the naturally occurring
core structure of the 8-hydroxyquinoline. The anticancer effect of
91b1 on esophageal cancer and its effect on the gene expression
profile of esophageal cancer cells were evaluated to assess its
potential to be explored as a novel anticancer agent.
The present study confirmed by MTS cytotoxicity
assay that cytotoxic effects of 91b1 on the five ESCC cells
(KYSE30, KYSE150, KYSE450, KYSE510 and HKESC-4) were comparable to
that of the first-line chemotherapeutic drug CDDP. CDDP is a
well-known chemotherapeutic drug to treat non-small cell lung
cancer, ESCC and gastrointestinal cancer (40-42), which was applied as the positive
control to assess the anti-cancer potential of compound 91b1. In
addition, 91b1 demonstrated less cytotoxicity to non-neoplastic
cell lines (NE-3 and 293) than CDDP by 1.6 and 2.1 times
respectively. Hence, the current results provided the first
evidence about the anticancer potential of 91b1 with lesser
cytotoxicity induced on non-tumor cells.
Among the five ESCC cell lines tested, KYSE150
showed sensitivity to CDDP treatment (MTS50; 13.16
μg/ml) but responded positively to the novel quinoline
derivative 91b1 (MTS50; 4.55 μg/ml). Subsequent
cDNA microarray analysis of KYSE150 identified Ccl5 as the
most downregulated in 91b1-treated KYSE150 cells. Ccl5, also
known as Human Regulated on Activation in Normal secreted T-cell
Expression and Secreted, is one of the members in CC-chemokine
family. It is also a well-known chemokine to stimulate tumor
progression (5). It mediates its
biological effect by activating G protein-coupled receptors CCR1,
CCR3 and CCR5 with CCR5 as the dominant receptor (43). The most important role for the
interaction between CCL5 and its receptor CCR5 in tumor development
is the regulation of metastasis process. The mechanism of
metastasis mediated by elevated level of CCL5 remains to be
elucidated. However, studies have demonstrated the influence of the
CCL5/CCR5 activity on invasion. Secretion of CCL5 by stromal cells
in bone marrow was found to enhance the invasion ability of
hepatocellular carcinoma cells (44-46).
In the present study, qPCR analysis, ELISA and
immunostaining collectively demonstrated the dose-dependent
suppression of CCL5 induced by of 91b1, suggesting the anticancer
effect is strongly related to the expression of CCL5. Notably, for
the expression status of CCL5 in ESCC, 87.5% (7/8) of ESCC cell
lines were found to overexpress compared with non-neoplastic
esophageal epithelial cells, as revealed by immunostaining. The
elevated expression of CCL5 was also observed in ESCC specimens by
immunohistochemistry with 76.9% of ESCC tissues showed high
expression level for CCL5. It has been reported that CCL5 may be
involved in the early stage of carcinogenesis of ESCC, playing the
role in transformation of pre-invasive lesions to cancer as
reported in oral squamous cell carcinoma (45). Future investigations could be
conducted to elaborate the roles of CCL5 overexpression in
pre-malignant lesions of ESCC and other cancers.
Previous reports demonstrate the role of CCL5 in
tumor progression including increased invasive abilities (44,46). The ability of CCL5 to enhance the
invasion of ESCC cells was further examined by Transwell Matrigel
invasion assay, reinforcing its role in tumor progression. Notably,
CCL5 (50-500 ng/ml) induced the invasion ability of KYSE30 in the
Transwell Matrigel invasion assay, implying that CCL5 can enhance
the invasion ability of ESCC cells. To strengthen the association
of CCL5 treatment and cell invasion, more experiments, including
knock down Ccl5 by siRNA in a series of ESCC cells, will be
performed in the future studies. From the present study, CCL5
expression in the tumor cells can be suppressed by 91b1 and the
inhibition of tumor progression based on the suppression of CCL5
expression has been postulated (5). Thus, the overall findings
illustrated, for the first time to the best of the authors'
knowledge, the potential of 91b1 in suppressing the invasion and
progression of ESCC cells through CCL5 suppression.
Some previous studies also suggest that different
organs or tissues may secrete different chemokines (including CCL5)
along with specific types of lectins and integrins. These molecules
facilitate the adhesion of cancer cells to organs, potentially
leading to metastasis (47,48). This suggests that the
overexpression of CCL5 in other tissues may also increase the risk
of tumor metastasis. According to the results of the present study,
91b1 can suppress the expression of CCL5 which may also involve the
suppression of metastasis of ESCC cells to other tissue sites.
Further studies should explore the anti-metastatic effects of 91b1
on a large across different cancer types.
The present study demonstrated that a novel
8-hydroxyquinoline derivative
(5,7-dibromo-1,2,3,4-tetrahydro-2-methylquinolin-8-ol; 91b1) showed
a cytotoxic effect on ESCC cells with relatively lower cytotoxicity
to non-tumor cells compared with CDDP. 91b1 induced the suppression
of mRNA and protein expression of the most downregulated target
Ccl5 in ESCC cells in the dose-dependent manner. CCL5 also
enhanced invasive ability of ESCC cells in vitro and was
found frequently upregulated in ESCC cell lines and tumor tissues.
Considering the critical function of CCL5 in cancer development and
metastasis, the strategy of suppressing the expression of CCL5
opens a new path for studies on the possible treatment of ESCC
using 91b1 and possibly such an approach can be extended to other
types of cancer in future. By contrast, the application of
quinoline compound is limited by its solubility, retention time, or
multi-drug resistance. With the development of novel biological
materials, integrating this compound with innovative biomaterials
presents a viable pathway to enhance its therapeutic potential and
applicability in anti-cancer therapy. Nanomaterials can be employed
as carriers to enhance solubility and bioavailability, or designed
with specific function for tumor cells to reduce systemic side
effects (49). Hydrogels can act
as scaffolds due to its biocompatibility and controlled degradation
rates for sustained drug delivery (50). Loading 91b1 within hydrogel might
offer controlled release for maintaining therapeutic concentrations
of drug in tumor or tumor microenvironment over extended period.
These strategies aim to address the challenges associated with the
clinical application of novel small molecule anticancer drugs such
as 91b1 with the unique properties of advanced biomaterials.
The novel quinoline compound 91b1 demonstrated
promising anticancer effect to ESCC cells compared with CDDP
through the downregulation of CCL5 expression with suppression of
tumor invasion. CCL5 was found frequently upregulated in ESCC cell
lines and tumor tissues, indicating the high potential use of
compound 91b1 for the treatment of ESCC in future. Furthermore, in
the present study, while in vitro data provide valuable
insights, they do not always predict in vivo behavior due to
the complexity of living organisms. Hence, subsequent in
vivo studies are crucial for confirming the present findings
and understanding the biological relevance of the gene functions
studied in ESCC or other types of cancers. Future research should
focus on expanding the scope of animal experiments to include
mechanism such as related signaling pathway, long-term studies and
the evaluation of potential side effects, which will enhance the
translational potential of our findings into clinical
applications.
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author. All the sequencing data
have been deposited in the NCBI GEO depository and are accessible
under accession number GEO: GSE273055; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE273055.
Authors' contributions
JT, DC, and YZ participated in study design and
drafted the article. DC and ZY collected samples, performed the
experiments and carried out the initial analysis. PC, YL, KL, and
AL performed the further analysis. SL, WH and AC participated in
study design. JT and DC confirm the authenticity of all raw data.
All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Paraffin-embedded specimens containing 26 cancer
tissues and 15 non-neoplastic tissues were collected after
esophagectomy, with the consent of patients, at the Department of
Surgery, Queen Mary Hospital, Hong Kong between January 1990 and
December 2001 and the ethics approval for working on the specimens
was obtained from the Hong Kong Polytechnic University (approval
no. HSEARS20171213007).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgements
Not applicable.
Funding
The present study was supported by the research fund of
Guangzhou Huashang College (grant no. 2024HSDS09) and the Research
in Chirosciences and Chemical Biology (grant no. 1-BBX8) offered by
the Hong Kong Polytechnic University It was also supported by the
Innovative Technology Commission (HKSAR Government), which
established the State Key Laboratory of Chemical Biology and Drug
Discovery at Hong Kong Polytechnic University (grant no. ZE20). The
present study was also supported by an MOU signed by Hong Kong
Baptist University and Griffith University in Australia.
References
|
1
|
Then EO, Lopez M, Saleem S, Gayam V,
Sunkara T, Culliford A and Gaduputi V: Esophageal cancer: An
updated surveillance epidemiology and end results database
analysis. World J Oncol. 11:55–64. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Tong D and Law S: Hong Kong experience.
Ando N: Esophageal Squamous Cell Carcinoma. Springer; Tokyo: pp.
261–278. 2015, View Article : Google Scholar
|
|
4
|
Chan D, Zhou Y, Chui CH, Lam KH, Law S,
Chan AS, Li X, Lam AK and Tang JCO: Expression of insulin-like
growth factor binding protein-5 (IGFBP5) reverses
cisplatin-resistance in esophageal carcinoma. Cells. 7:1432018.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Aldinucci D, Borghese C and Casagrande N:
The CCL5/CCR5 axis in cancer progression. Cancers (Basel).
12:17652020. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Schall TJ, Bacon K, Toy KJ and Goeddel DV:
Selective attraction of monocytes and T lymphocytes of the memory
phenotype by cytokine RANTES. Nature. 347:669–671. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Brett E, Duscher D, Pagani A, Daigeler A,
Kolbenschlag J and Hahn M: Naming the barriers between Anti-CCR5
therapy, breast cancer and its microenvironment. Int J Mol Sci.
23:141592022. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ding H, Zhao L, Dai S, Li L, Wang F and
Shan B: CCL5 secreted by tumor associated macrophages may be a new
target in treatment of gastric cancer. Biomed Pharmacother.
77:142–149. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang XF, Zhang XL, Wang YJ, Fang Y, Li
ML, Liu XY, Luo HY and Tian Y: The regulatory network of the
chemokine CCL5 in colorectal cancer. Ann Med. 55:22051682023.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xu H, Zhao J, Li J, Zhu Z, Cui Z, Liu R,
Lu R, Yao Z and Xu Q: Cancer associated fibroblast-derived CCL5
promotes hepatocellular carcinoma metastasis through activating
HIF1α/ZEB1 axis. Cell Death Dis. 13:4782022. View Article : Google Scholar
|
|
11
|
Huang R, Wang S, Wang N, Zheng Y, Zhou J,
Yang B, Wang X, Zhang J, Guo L, Wang S, et al: CCL5 derived from
tumor-associated macrophages promotes prostate cancer stem cells
and metastasis via activating β-catenin/STAT3 signaling. Cell Death
Dis. 11:2342020. View Article : Google Scholar
|
|
12
|
Chen K, Wang Y, Hou Y, Wang Q, Long D, Liu
X, Tian X and Yang Y: Single cell RNA-seq reveals the CCL5/SDC1
receptor-ligand interaction between T cells and tumor cells in
pancreatic cancer. Cancer Lett. 545:2158342022. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Michael JP: Quinoline, quinazoline and
acridone alkaloids. Nat Prod Rep. 15:595–606. 1998. View Article : Google Scholar
|
|
14
|
Balaraman K, Vieira NC, Moussa F, Vacus J,
Cojean S, Pomel S, Bories C, Figadère B, Kesavan V and Loiseau PM:
In vitro and in vivo antileishmanial properties of a
2-n-propylquinoline hydroxypropyl β-cyclodextrin formulation and
pharmacoki-netics via intravenous route. Biomed Pharmacother.
76:127–133. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Vivanco JM, Bais HP, Stermitz FR, Thelen
GC and Callaway RM: Biogeographical variation in community response
to root allelochemistry: Novel weapons and exotic invasion. Ecol
Lett. 7:285–292. 2004. View Article : Google Scholar
|
|
16
|
Huang XQ, Wu RC, Liang JM, Zhou Z, Qin QP
and Liang H: Anticancer activity of
8-hydroxyquinoline-triphenylphosphine rhodium(III) complexes
targeting mitophagy pathways. Eur J Med Chem. 272:1164782024.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Prajapati AK, Bhattacharya A and Choudhary
S: Inhibiting the activity of malarial drug target Plasmepsin V by
quinolines in aqueous medium. J Mol Liq. 397:1241582024. View Article : Google Scholar
|
|
18
|
Joaquim AR, Gionbelli MP, Gosmann G,
Fuentefria AM, Lopes MS and Fernandes de Andrade S: Novel
antimicrobial 8-hydroxyquinoline-based agents: Current development,
structure-activity relationships, and perspectives. J Med Chem.
64:16349–16379. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chan SH, Chui CH, Chan SW, Kok SH, Chan D,
Tsoi MY, Leung PH, Lam AK, Chan AS, Lam KH and Tang JC: Synthesis
of 8-hydroxyquinoline derivatives as novel antitumor agents. ACS
Med Chem Lett. 4:170–174. 2012. View Article : Google Scholar
|
|
20
|
Lam KH, Lee KK, Gambari R, Kok SH, Kok TW,
Chan AS, Bian ZX, Wong WY, Wong RS, Lau FY, et al: Anti-tumour and
pharmacokinetics study of 2-Formyl-8-hydroxy-quinolinium chloride
as Galipea longiflora alkaloid analogue. Phytomedicine. 21:877–882.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lam KH, Lee KK, Kok SH, Wong RS, Lau FY,
Cheng GY, Wong WY, Tong SW, Chan KW, Chan RY, et al: Antiangiogenic
activity of 2-formyl-8-hydroxy-quinolinium chloride. Biomed
Pharmacother. 80:145–150. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Pun IH, Chan D, Chan SH, Chung PY, Zhou
YY, Law S, Lam AK, Chui CH, Chan AS, Lam KH and Tang JC:
Anti-cancer Effects of a Novel Quinoline Derivative 83b1 on human
esophageal squamous cell carcinoma through down-regulation of COX-2
mRNA and PGE2. Cancer Res Treat. 49:219–229. 2017.
View Article : Google Scholar
|
|
23
|
Lam KH, Lee KK, Gambari R, Wong RS, Cheng
GY, Tong SW, Chan KW, Lau FY, Lai PB, Wong WY, et al: Preparation
of Galipea officinalis Hancock type tetrahydroquinoline alkaloid
analogues as anti-tumour agents. Phytomedicine. 20:166–171. 2013.
View Article : Google Scholar
|
|
24
|
Chan ASC, Tang JCO, Lam KH, Chui CH, Kok
SHL, Chan SH, Cheung F, Chor RG and Cheng H: Method of making and
administering quinoline derivatives as anti-cancer agents. The Hong
Kong Polytechnic University; 2016
|
|
25
|
Tang JCO, Chan ASC, Lam KH and Chan SH:
Quinoline derivatives as anti-cancer agents. Hong Kong Polytechnic
University; 2016
|
|
26
|
Chung PY, Lam PL, Zhou YY, Gasparello J,
Finotti A, Chilin A, Marzaro G, Gambari R, Bian ZX, Kwok WM, et al:
Targeting DNA binding for NF-κB as an anticancer approach in
hepatocellular carcinoma. Cells. 7:1772018. View Article : Google Scholar
|
|
27
|
Zhou Y, Zhou Z, Chan D, Chung PY, Wang Y,
Chan ASC, Law S, Lam KH and Tang JCO: The Anticancer effect of a
novel quinoline derivative 91b1 through downregulation of Lumican.
Int J Mol Sci. 23:131812022. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li T, Fu J, Zeng Z, Cohen D, Li J, Chen Q,
Li B and Liu XS: TIMER2.0 for analysis of tumor-infiltrating immune
cells. Nucleic Acids Res. 48(W1): W509–W514. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z,
Feng T, Zhou L, Tang W, Zhan L, et al: clusterProfiler 4.0: A
universal enrichment tool for interpreting omics data. Innovation
(Camb). 2:1001412021.
|
|
30
|
Szklarczyk D, Gable AL, Nastou KC, Lyon D,
Kirsch R, Pyysalo S, Doncheva NT, Legeay M, Fang T, Bork P, et al:
The STRING database in 2021: Customizable protein-protein networks,
and functional characterization of user-uploaded gene/measurement
sets. Nucleic Acids Res. 49(D1): D605–D612. 2021. View Article : Google Scholar
|
|
31
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Shimada Y, Imamura M, Wagata T, Yamaguchi
N and Tobe T: Characterization of 21 newly established esophageal
cancer cell lines. Cancer. 69:277–284. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Tang JC, Wan TS, Wong N, Pang E, Lam KY,
Law SY, Chow LM, Ma ES, Chan LC, Wong J and Srivastava G:
Establishment and characterization of a new xenograft-derived human
esophageal squamous cell carcinoma cell line SLMT-1 of Chinese
origin. Cancer Genet Cytogenet. 124:36–41. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Cheung LC, Tang JC, Lee PY, Hu L, Guan XY,
Tang WK, Srivastava G, Wong J, Luk JM and Law S: Establishment and
characterization of a new xenograft-derived human esophageal
squamous cell carcinoma cell line HKESC-4 of Chinese origin. Cancer
Genet Cytogenet. 178:17–25. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhang H, Jin Y, Chen X, Jin C, Law S, Tsao
SW and Kwong YL: Cytogenetic aberrations in immortalization of
esophageal epithelial cells. Cancer Genet Cytogenet. 165:25–35.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Graham FL, Smiley J, Russell WC and Nairn
R: Characteristics of a human cell line transformed by DNA from
human adenovirus type 5. J Gen Virol. 36:59–74. 1997. View Article : Google Scholar
|
|
37
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative C(T) method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhou C, Liu S, Zhou X, Xue L, Quan L, Lu
N, Zhang G, Bai J, Wang Y, Liu Z, et al: Overexpression of human
pituitary tumor transforming gene (hPTTG), is regulated by
beta-catenin/TCF pathway in human esophageal squamous cell
carcinoma. Int J Cancer. 113:891–898. 2005. View Article : Google Scholar
|
|
39
|
Kumar S, Bawa S and Gupta H: Biological
activities of quinoline derivatives. Mini Rev Med Chem.
9:1648–1654. 2009. View Article : Google Scholar
|
|
40
|
Li S, Shen XY, Ouyang T, Qu Y, Luo T and
Wang HQ: Synergistic anticancer effect of combined crocetin and
cisplatin on KYSE-150 cells via p53/p21 pathway. Cancer Cell Int.
17:982017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Cesna V, Sukovas A, Jasukaitiene A,
Naginiene R, Barauskas G, Dambrauskas Z, Paskauskas S and Gulbinas
A: Narrow line between benefit and harm: Additivity of hyperthermia
to cisplatin cytotoxicity in different gastrointestinal cancer
cells. World J Gastroenterol. 24:1072–1083. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Kryczka J, Kryczka J, Czarnecka-Chrebelska
KH and Brzeziańska-Lasota E: Molecular mechanisms of
chemoresistance induced by cisplatin in NSCLC cancer therapy. Int J
Mol Sci. 22:88852021. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Martin L, Blanpain C, Garnier P, Wittamer
V, Parmentier M and Vita C: Structural and functional analysis of
the RANTES-glycosaminoglycans interactions. Biochemistry.
40:6303–6318. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Singh SK, Mishra MK, Rivers BM, Gordetsky
JB, Bae S and Singh R: Biological and clinical significance of the
CCR5/CCL5 axis in hepatocellular carcinoma. Cancers (Basel).
12:8832020. View Article : Google Scholar
|
|
45
|
Chen D, Yang K, Mei J, Zhang G, Lv X and
Xiang L: Screening the pathogenic genes and pathways related to
DMBA (7,12-dimethylbenz[a]anthracene)-induced transformation of
hamster oral mucosa from precancerous lesions to squamous cell
carcinoma. Oncol Lett. 2:637–642. 2011. View Article : Google Scholar
|
|
46
|
González-Arriagada WA, Coletta RD,
Lozano-Burgos C, García C, Maripillán J, Alcayaga-Miranda F,
Godínez-Pacheco B, Oyarce-Pezoa S, Martínez-Flores R and García IE:
CR5/CCL5 axis is linked to a poor outcome, and inhibition reduces
metastasis in oral squamous cell carcinoma. J Cancer Res Clin
Oncol. 149:17335–17346. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Karmakar S and Mukherjee R: Integrin
receptors and ECM proteins involved in preferential adhesion of
colon carcinoma cells to lung cells. Cancer Lett. 196:217–227.
2003. View Article : Google Scholar
|
|
48
|
Langley RR and Fidler IJ: The seed and
soil hypothesis revisited-the role of tumor-stroma interactions in
metastasis to different organs. Int J Cancer. 128:2527–2535. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Wang C, Zhou H, Kurboniyon MS, Tang Y, Cai
Z, Ning S, Zhang L and Liang X: Chemodynamic PtMn nanocubes for
effective photothermal ROS storm a key anti-tumor therapy in-vivo.
Int J Nanomedicine. 19:5045–5056. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Bordbar-Khiabani A and Gasik M: Smart
hydrogels for advanced drug delivery systems. Int J Mol Sci.
23:36652022. View Article : Google Scholar : PubMed/NCBI
|